Recombinant Silk Proteins with Additional Polyalanine Have Excellent Mechanical Properties
Abstract
:1. Introduction
2. Results
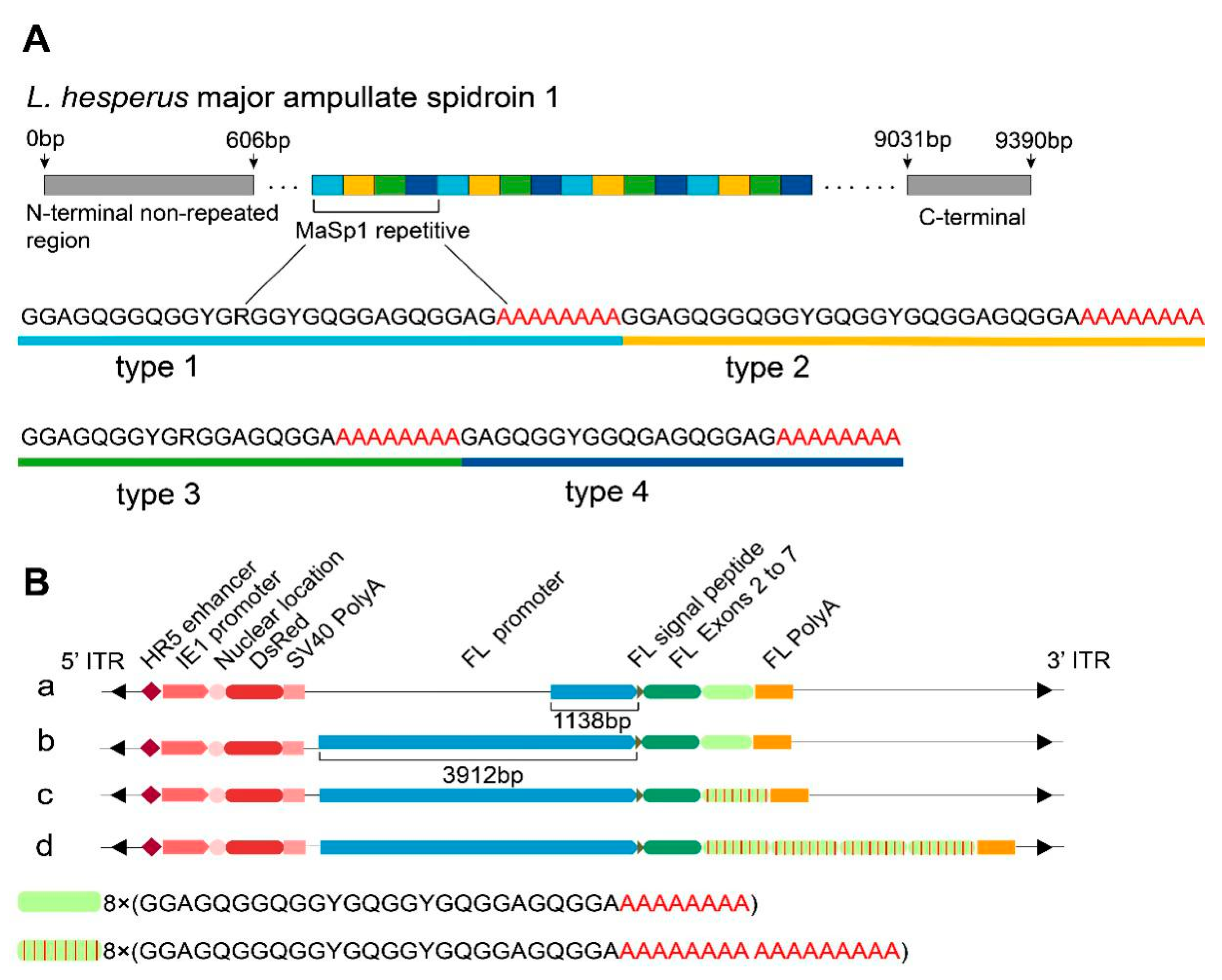
2.1. Construction of the Transgenic Vector and Screening for Positive Silkworms
2.2. Analysis of Foreign Gene Insertion Sites in the Transgenic Silkworms
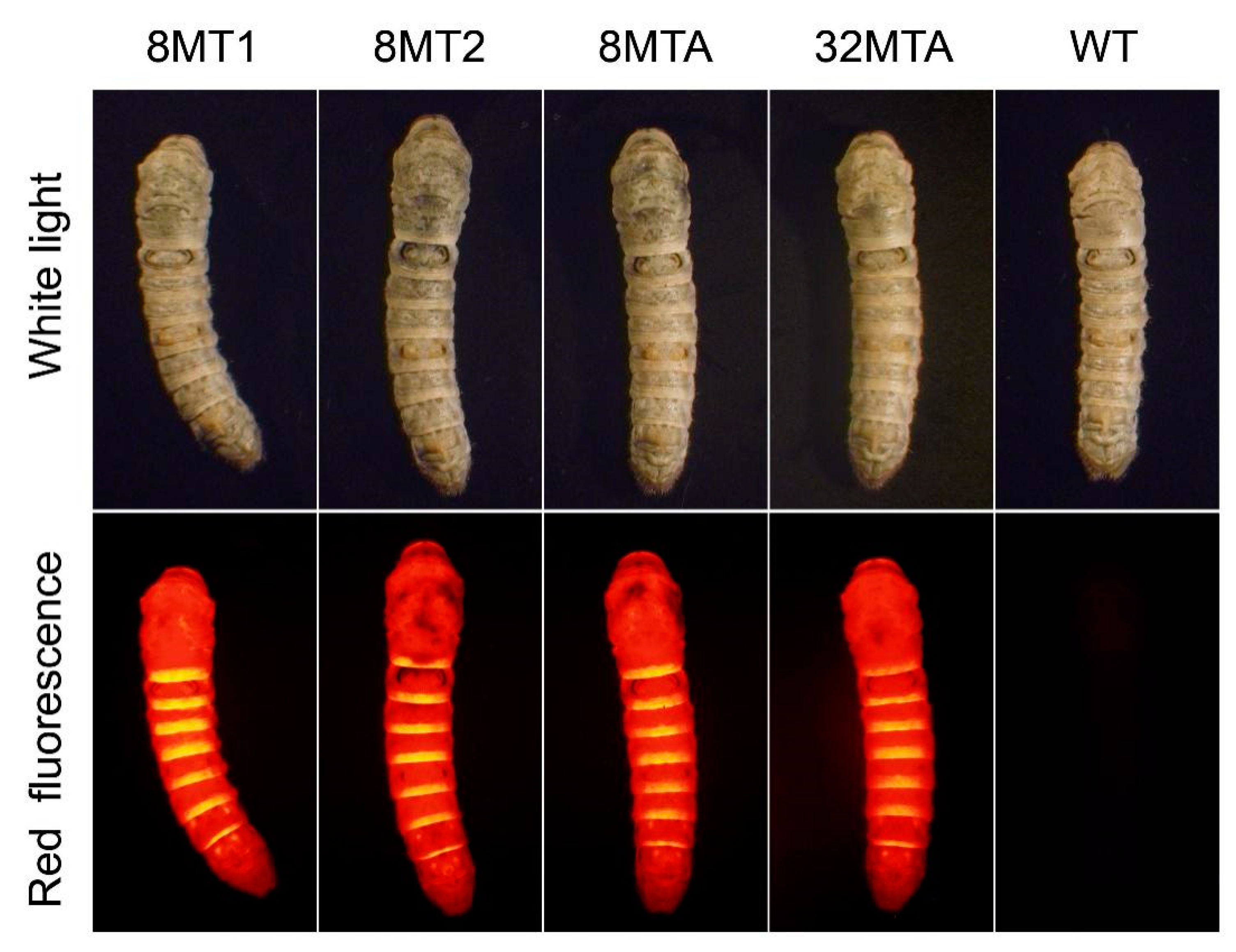
2.3. Identification of Foreign Gene Expression in the Transgenic Silkworms
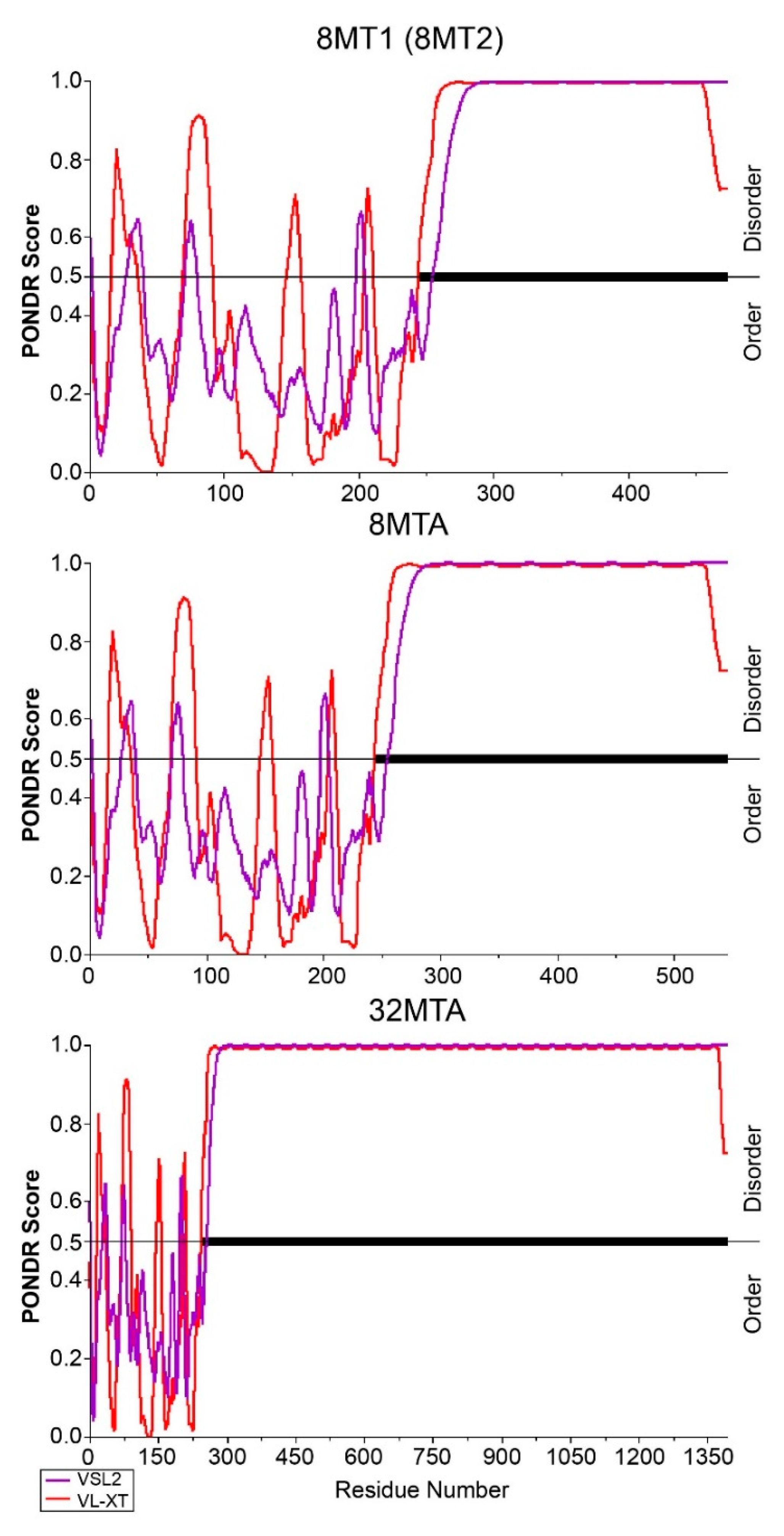
2.4. Foreign Protein Structure Predictions
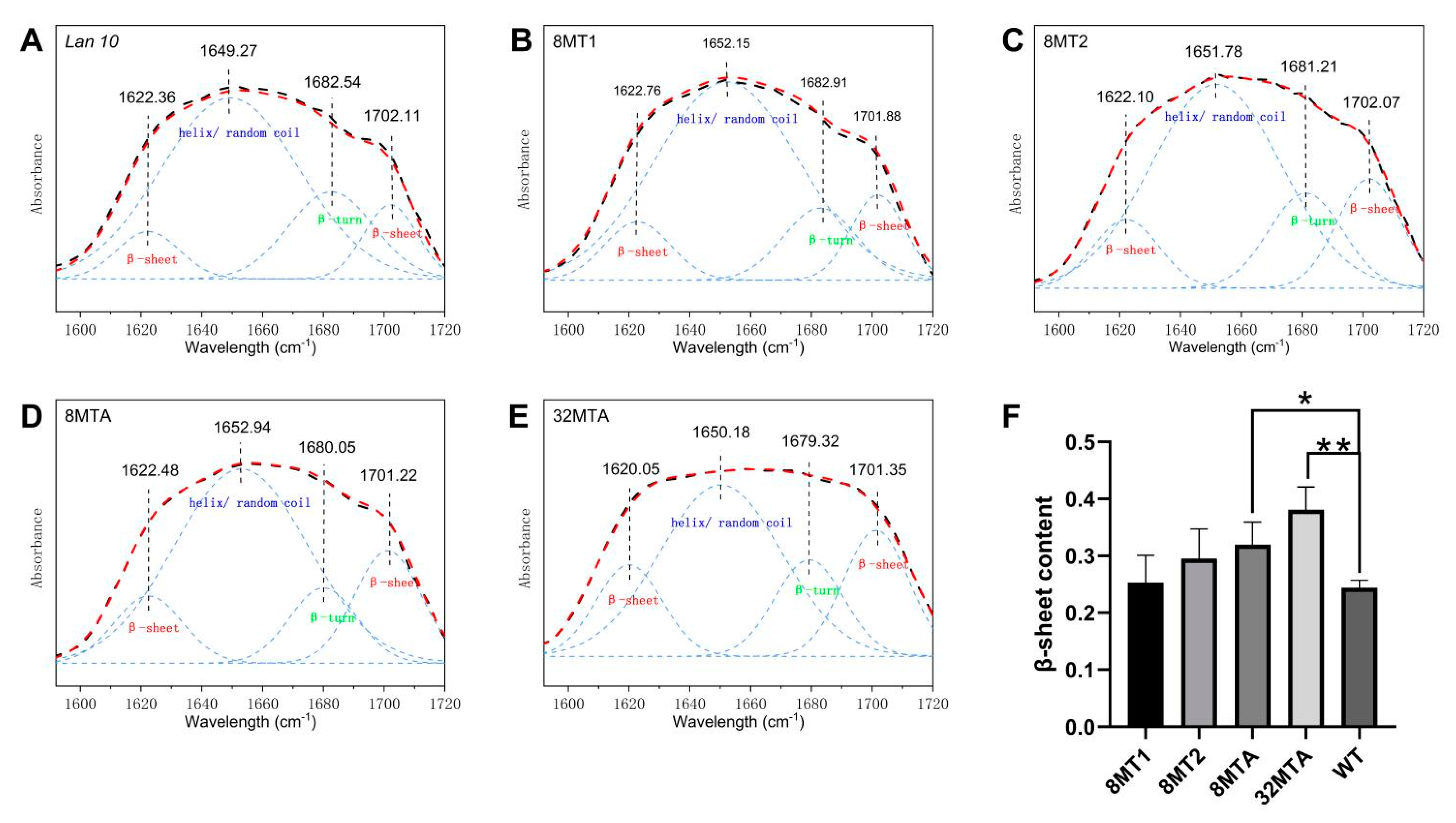
2.5. Secondary Structure Analyses of the Transgenic Fibers
2.6. Mechanical Properties of the Transgenic Fibers
3. Discussion
4. Materials and Methods
4.1. Vector Construction
4.2. Silkworm Transformation and Identification
4.3. Western Blotting
4.4. Prediction of the Protein Disordered Regions and FTIR Spectroscopy Analysis
4.5. Mechanical Properties Testing of the Transgenic Silk
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Confalonieri, F.; Medina, N.; Zivanovic, Y.; Esnault, C.; Yang, T.; Jacquet, M.; Janin, J.; Duguet, M.; Perasso, R.; et al. Fine organization of Bombyx mori fibroin heavy chain gene. Nucleic Acids Res. 2000, 28, 2413–2419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Xia, L.; Day, B.A.; Harris, T.I.; Oliveira, P.; Knittel, C.; Licon, A.L.; Gong, C.; Dion, G.; Lewis, R.V.; et al. CRISPR/Cas9 Initiated Transgenic Silkworms as a Natural Spinner of Spider Silk. Biomacromolecules 2019, 20, 2252–2264. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Kikuchi, Y.; Takagi, T.; Kikuchi, A.; Oyama, F.; Shimura, K.; Mizuno, S. Primary Structure of the Silk Fibroin Light Chain Determined by cDNA Sequencing and Peptide Analysis. J. Mol. Biol. 1989, 210, 127–139. [Google Scholar] [CrossRef]
- Yang, K.; Guan, J.; Shao, Z.; Ritchie, R.O. Mechanical properties and toughening mechanisms of natural silkworm silks and their composites. J. Mech. Behav. Biomed. Mater. 2020, 110, 103942. [Google Scholar] [CrossRef]
- Xu, J.; Dong, Q.; Yu, Y.; Niu, B.; Ji, D.; Li, M.; Huang, Y.; Chen, X.; Tan, A. Mass spider silk production through targeted gene replacement in Bombyx mori. Proc. Natl. Acad. Sci. USA 2018, 115, 8757–8762. [Google Scholar] [CrossRef] [Green Version]
- You, Z.; Ye, X.; Ye, L.; Qian, Q.; Wu, M.; Song, J.; Che, J.; Zhong, B. Extraordinary Mechanical Properties of Composite Silk Through Hereditable Transgenic Silkworm Expressing Recombinant Major Ampullate Spidroin. Sci. Rep. 2018, 8, 15956. [Google Scholar] [CrossRef] [Green Version]
- Ling, S.; Qin, Z.; Li, C.; Huang, W.; Kaplan, D.L.; Buehler, M.J. Polymorphic regenerated silk fibers assembled through bioinspired spinning. Nat. Commun. 2017, 8, 1387. [Google Scholar] [CrossRef] [Green Version]
- Tokareva, O.; Jacobsen, M.; Buehler, M.; Wong, J.; Kaplan, D.L. Structure-function-property-design interplay in biopolymers: Spider silk. Acta Biomater. 2014, 10, 1612–1626. [Google Scholar] [CrossRef] [Green Version]
- Lefèvre, T.; Auger, M. Spider silk as a blueprint for greener materials: A review. Int. Mater. Rev. 2016, 61, 127–153. [Google Scholar] [CrossRef] [Green Version]
- Blamires, S.J.; Blackledge, T.A.; Tso, I.M. Physicochemical Property Variation in Spider Silk: Ecology, Evolution, and Synthetic Production. Annu. Rev. Entomol. 2017, 62, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Yarger, J.L.; Cherry, B.R.; Vaart, A.V.D. Uncovering the structure–function relationship in spider silk. Nat. Rev. Mater. 2018, 3, 1–11. [Google Scholar] [CrossRef]
- Kono, N.; Nakamura, H.; Ohtoshi, R.; Tomita, M.; Numata, K.; Arakawa, K. The bagworm genome reveals a unique fibroin gene that provides high tensile strength. Commun. Biol. 2019, 2, 148. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, T.; Tsubota, T.; Tashiro, K.; Jouraku, A.; Kameda, T. A study of the extraordinarily strong and tough silk produced by bagworms. Nat. Commun. 2019, 10, 1469. [Google Scholar] [CrossRef] [PubMed]
- Teule, F.; Miao, Y.G.; Sohn, B.H.; Kim, Y.S.; Hull, J.J.; Fraser, M.J., Jr.; Lewis, R.V.; Jarvis, D.L. Silkworms transformed with chimeric silkworm/spider silk genes spin composite silk fibers with improved mechanical properties. Proc. Natl. Acad. Sci. USA 2012, 109, 923–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuwana, Y.; Sezutsu, H.; Nakajima, K.; Tamada, Y.; Kojima, K. High-toughness silk produced by a transgenic silkworm expressing spider (Araneus ventricosus) dragline silk protein. PLoS ONE 2014, 9, e105325. [Google Scholar]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Yang, X.; Liu, C.; Dong, Z.; Wang, F.; Wang, X.; Hu, W.; Zhang, X.; Zhao, P.; Xia, Q. Structural and Mechanical Properties of Silk from Different Instars of Bombyx mori. Biomacromolecules 2019, 20, 1203–1216. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, J.; Jordan, J.S.; Wang, X.; Henning, R.W.; Yarger, J.L. Structural Comparison of Various Silkworm Silks: An Insight into the Structure-Property Relationship. Biomacromolecules 2018, 19, 906–917. [Google Scholar] [CrossRef]
- Guo, Z.; Xie, W.; Gao, Q.; Wang, D.; Gao, F.; Li, S.; Zhao, L. In situ biomineralization by silkworm feeding with ion precursors for the improved mechanical properties of silk fiber. Int. J. Biol. Macromol. 2018, 109, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Liu, Q.; Chen, Q.; Xia, Q.; Zhao, P. In vivo effects of metal ions on conformation and mechanical performance of silkworm silks. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Tanaka, K.; Tanaka, H.; Ohtomo, K.; Kanda, T.; Imamura, M.; Quan, G.X.; Kojima, K.; Yamashita, T.; Nakajima, T.; et al. Assembly of the silk fibroin elementary unit in endoplasmic reticulum and a role of L-chain for protection of alpha1,2-mannose residues in N-linked oligosaccharide chains of fibrohexamerin/P25. Eur. J. Biochem. 2004, 271, 356–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Z.; Long, D.; Zhang, Y.; Umuhoza, D.; Dai, J.; Xu, Z.; Zhang, G.; Meng, W.; Xiang, Z.; Zhao, A. New insight into the mechanism of in vivo fibroin self-assembly and secretion in the silkworm, Bombyx mori. Int. J. Biol. Macromol. 2020, 169, 473–479. [Google Scholar] [CrossRef]
- Bernacki, J.P.; Murphy, R.M. Length-dependent aggregation of uninterrupted polyalanine peptides. Biochemistry 2011, 50, 9200–9211. [Google Scholar] [CrossRef]
- Jenkins, J.E.; Holland, G.P.; Yarger, J.L. High resolution magic angle spinning NMR investigation of silk protein structure within major ampullate glands of orb weaving spiders. Soft Matter 2012, 8, 1947–1954. [Google Scholar] [CrossRef]
- Giesa, T.; Perry, C.C.; Buehler, M.J. Secondary Structure Transition and Critical Stress for a Model of Spider Silk Assembly. Biomacromolecules 2016, 17, 427–436. [Google Scholar] [CrossRef] [Green Version]
- He, Y.X.; Zhang, N.N.; Li, W.F.; Jia, N.; Chen, B.Y.; Zhou, K.; Zhang, J.; Chen, Y.; Zhou, C.Z. N-Terminal domain of Bombyx mori fibroin mediates the assembly of silk in response to pH decrease. J. Mol. Biol. 2012, 418, 197–207. [Google Scholar] [CrossRef]
- Blamires, S.J.; Kasumovic, M.M.; Tso, I.M.; Martens, P.J.; Hook, J.M.; Rawal, A. Evidence of Decoupling Protein Structure from Spidroin Expression in Spider Dragline Silks. Int. J. Mol. Sci. 2016, 17, 1294. [Google Scholar] [CrossRef] [Green Version]
- Domigan, L.J.; Andersson, M.; Alberti, K.A.; Chesler, M.; Xu, Q.; Johansson, J.; Rising, A.; Kaplan, D.L. Carbonic anhydrase generates a pH gradient in Bombyx mori silk glands. Insect Biochem. Mol. Biol. 2015, 65, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Blamires, S.J.; Liao, C.P.; Chang, C.K.; Chuang, Y.C.; Wu, C.L.; Blackledge, T.A.; Sheu, H.S.; Tso, I.M. Mechanical performance of spider silk is robust to nutrient-mediated changes in protein composition. Biomacromolecules 2015, 16, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Johansson, J.; Rising, A. Silk Spinning in Silkworms and Spiders. Int. J. Mol. Sci. 2016, 17, 1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.; Chen, G.; Otikovs, M.; Landreh, M.; Nordling, K.; Kronqvist, N.; Westermark, P.; Jornvall, H.; Knight, S.; Ridderstrale, Y.; et al. Carbonic anhydrase generates CO2 and H+ that drive spider silk formation via opposite effects on the terminal domains. PLoS Biol. 2014, 12, e1001921. [Google Scholar] [CrossRef] [PubMed]
- Filippidi, E.; Cristiani, T.R.; Eisenbach, C.D.; Waite, J.H.; Israelachvili, J.N.; Ahn, B.K.; Valentine, M.T. Toughening elastomers using mussel-inspired iron-catechol complexes. Science 2017, 358, 502–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moseti, K.O.; Yoshioka, T.; Kameda, T.; Nakazawa, Y. Structure Water-Solubility Relationship in alpha-Helix-Rich Films Cast from Aqueous and 1,1,1,3,3,3-Hexafluoro-2-Propanol Solutions of S. c. ricini Silk Fibroin. Molecules 2019, 24, 3945. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, K.; Yamamoto, H.; Yoshioka, T.; Ninh, T.H.; Tasaki, M.; Shimada, S.; Nakatani, T.; Iwamoto, H.; Ohta, N.; Masunaga, H. Hierarchical Structural Change in the Stress-Induced Phase Transition of Poly(tetramethylene terephthalate) As Studied by the Simultaneous Measurement of FTIR Spectra and 2D Synchrotron Undulator WAXD/SAXS Data. Macromolecules 2014, 47, 2052–2061. [Google Scholar] [CrossRef]
- Yoshioka, T.; Tashiro, K.; Ohta, N. Molecular Orientation Enhancement of Silk by the Hot-Stretching-Induced Transition from alpha-Helix-HFIP Complex to beta-Sheet. Biomacromolecules 2016, 17, 1437–1448. [Google Scholar] [CrossRef]
- Giesa, T.; Arslan, M.; Pugno, N.M.; Buehler, M.J. Nanoconfinement of spider silk fibrils begets superior strength, extensibility, and toughness. Nano Lett. 2011, 11, 5038–5046. [Google Scholar] [CrossRef] [Green Version]
- Andersson, M.; Holm, L.; Ridderstrale, Y.; Johansson, J.; Rising, A. Morphology and composition of the spider major ampullate gland and dragline silk. Biomacromolecules 2013, 14, 2945–2952. [Google Scholar] [CrossRef]
- Guan, J.; Porter, D.; Vollrath, F. Silks cope with stress by tuning their mechanical properties under load. Polymer 2012, 53, 2717–2726. [Google Scholar] [CrossRef]
- Chen, X.; Shao, Z.; Vollrath, F. The spinning processes for spider silk. Soft Matter 2006, 2, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Holland, C.; O’Neil, K.; Vollrath, F.; Dicko, C. Distinct structural and optical regimes in natural silk spinning. Biopolymers 2012, 97, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Teulé, F.; Cooper, A.R.; Furin, W.A.; Bittencourt, D.; Rech, E.L.; Brooks, A.; Lewis, R.V. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat. Protoc. 2009, 4, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, T.; Thibert, C.; Royer, C.; Kanda, T.; Abraham, E.; Kamba, M.; Kômoto, N.; Thomas, J.L.; Mauchamp, B.; Chavancy, G.; et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat. Biotechnol. 2000, 18, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Zhong, B.; Li, J.; Chen, J.; Ye, J.; Yu, S. Comparison of transformation efficiency of piggyBac transposon among three different silkworm Bombyx mori Strains. Acta Biochim. Biophys. Sin. 2007, 39, 117–122. [Google Scholar] [CrossRef]
- Zhao, H.-P.; Feng, X.-Q.; Shi, H.-J. Variability in mechanical properties of Bombyx mori silk. Mater. Sci. Eng. C 2007, 27, 675–683. [Google Scholar] [CrossRef]
- Pérez-Rigueiro, J.; Viney, C.; Llorca, J.; Elices, M. Mechanical properties of single-brin silkworm silk. J. Appl. Polym. Sci. 2000, 75, 1270–1277. [Google Scholar] [CrossRef]

| Silkworm Strains | Max Stress (MPa) | Young’s Modulus (MPa) |
|---|---|---|
| WT | 179.62 ± 25.78 | 3283.97 ± 1045.32 |
| 8MT1 | 183.56 ± 19.59 | 3379.44 ± 783.01 |
| 8MT2 | 190.95 ± 15.04 | 3538.23 ± 503.71 |
| 8MTA | 214.02 ± 25.03 | 4321.43 ± 1380.56 |
| 32MTA | 238.72 ± 13.98 | 4631.16 ± 611.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Ye, X.; Wu, M.; Ruan, J.; Wang, X.; Tang, X.; Zhong, B. Recombinant Silk Proteins with Additional Polyalanine Have Excellent Mechanical Properties. Int. J. Mol. Sci. 2021, 22, 1513. https://doi.org/10.3390/ijms22041513
Zhao S, Ye X, Wu M, Ruan J, Wang X, Tang X, Zhong B. Recombinant Silk Proteins with Additional Polyalanine Have Excellent Mechanical Properties. International Journal of Molecular Sciences. 2021; 22(4):1513. https://doi.org/10.3390/ijms22041513
Chicago/Turabian StyleZhao, Shuo, Xiaogang Ye, Meiyu Wu, Jinghua Ruan, Xiaoxiao Wang, Xiaoli Tang, and Boxiong Zhong. 2021. "Recombinant Silk Proteins with Additional Polyalanine Have Excellent Mechanical Properties" International Journal of Molecular Sciences 22, no. 4: 1513. https://doi.org/10.3390/ijms22041513
APA StyleZhao, S., Ye, X., Wu, M., Ruan, J., Wang, X., Tang, X., & Zhong, B. (2021). Recombinant Silk Proteins with Additional Polyalanine Have Excellent Mechanical Properties. International Journal of Molecular Sciences, 22(4), 1513. https://doi.org/10.3390/ijms22041513








