Thanatin: An Emerging Host Defense Antimicrobial Peptide with Multiple Modes of Action
Abstract
:1. Introduction
2. Thanatin
3. In Vivo Antibacterial Activity of Thanatin
Engineering Thanatin for Superior Activity
4. Effect of Thanatin on Bacterial Cells, LPS, and Liposome Integrity
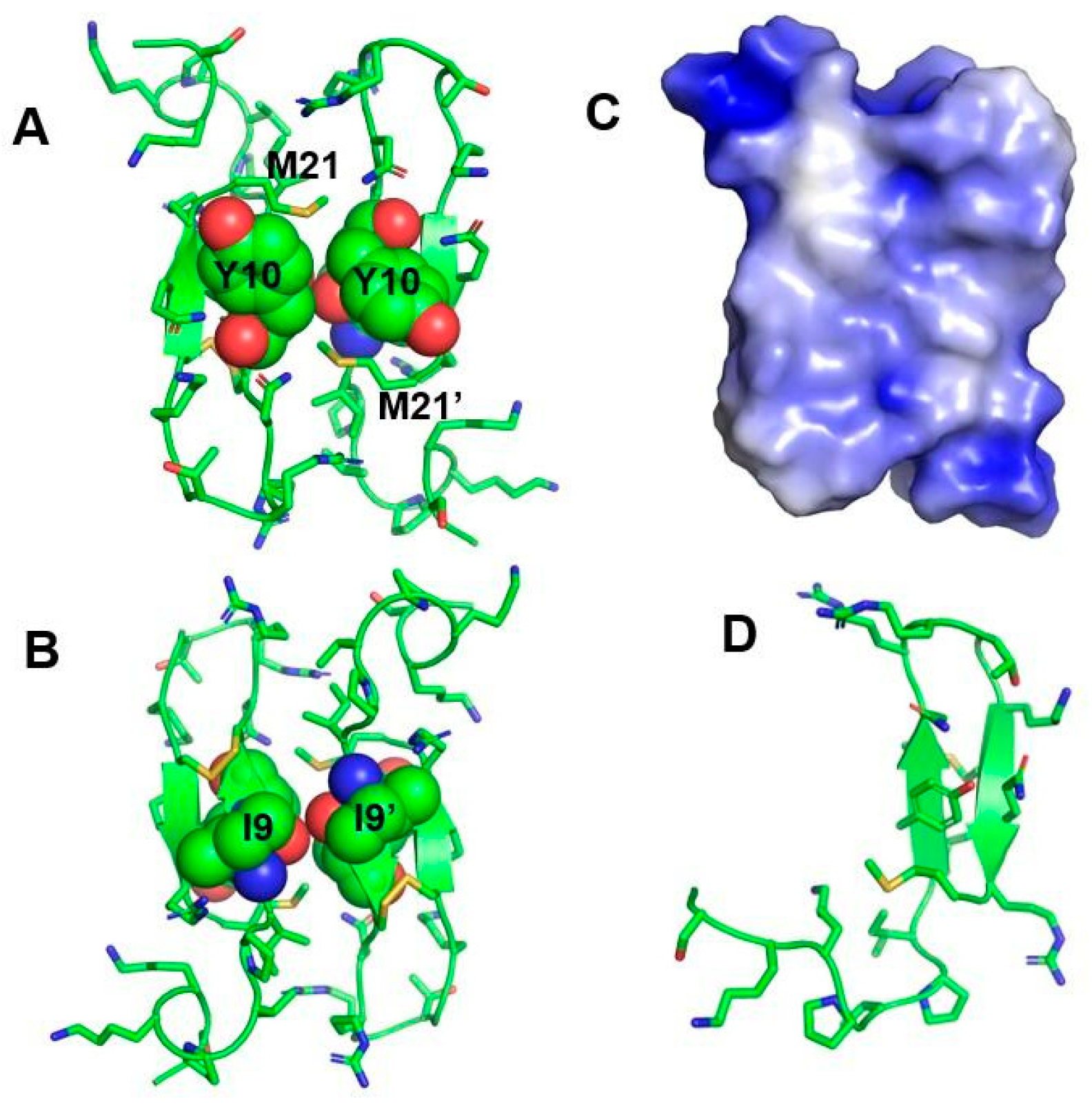
5. Atomic-Resolution Structure of Thanatin in LPS Outer Membrane
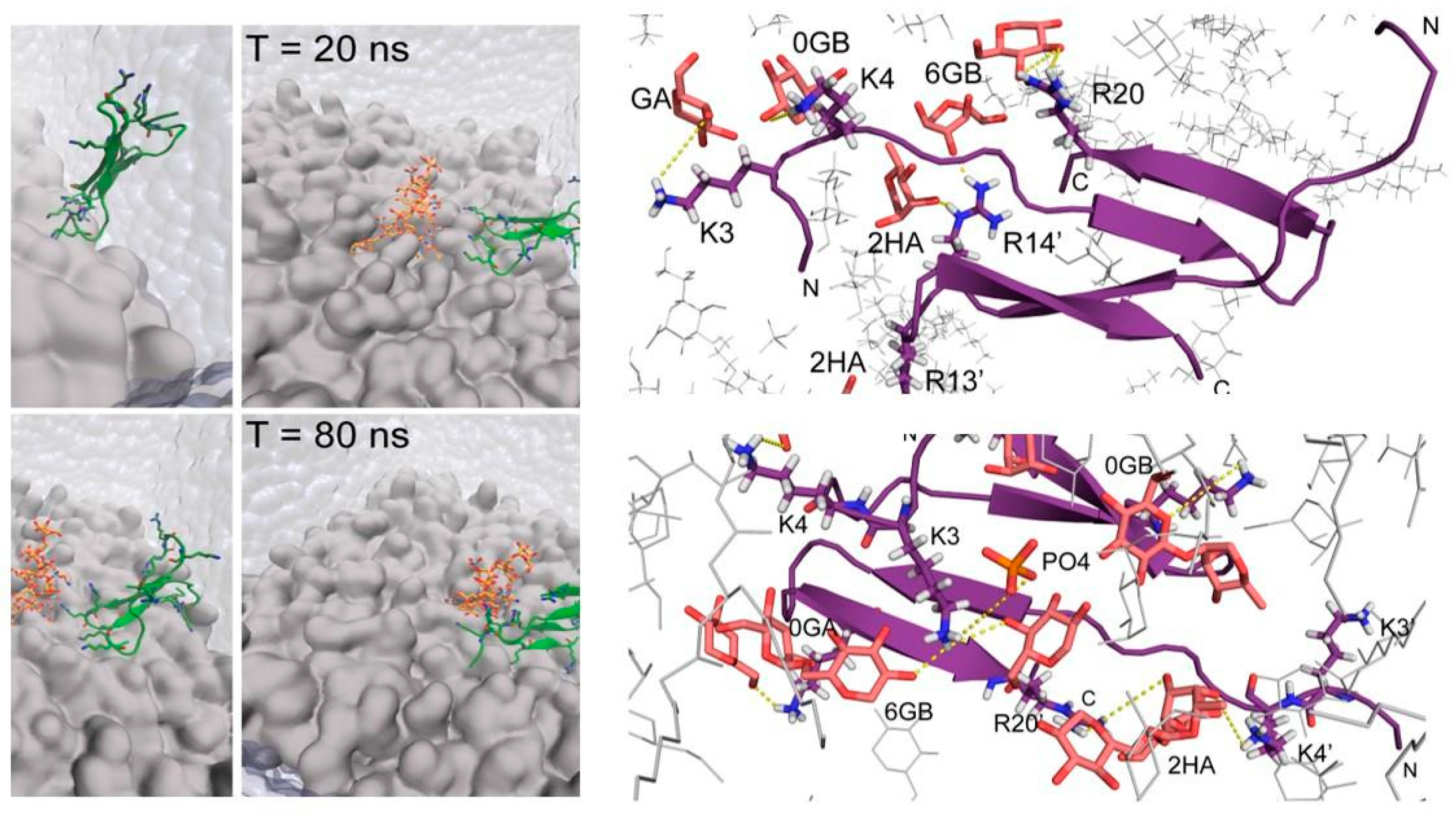
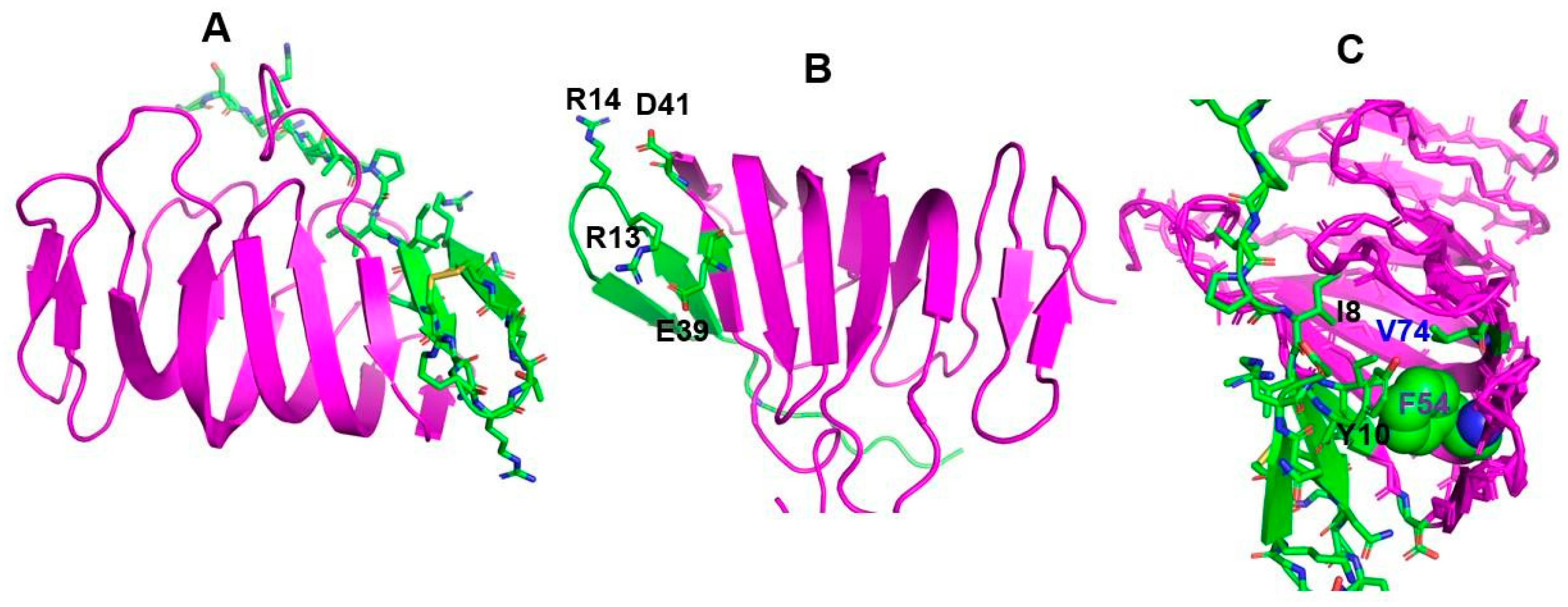
6. Binding of Thanatin with LPS Transport Protein Complex and Interactions with Metallo-β-Lactamase of Gram-Negative Bacteria
7. Mode of Gram-Negative Bacterial Cell Killing by Thanatin
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance (AMR), 2016. Government of the United Kingdom: Wellcome Trust U.K. Available online: https://www.biomerieuxconnection.com/wp-content/uploads/2018/04/Tackling-Drug-Resistant-Infections-Globally_-Final-Report-and-Recommendations.pdf (accessed on 2 February 2021).
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the global burden of disease study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U. Global antimicrobial resistance in gram-negative pathogens and clinical need. Curr. Opin. Microbiol. 2017, 39, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.T.; Leonard, S.P.; DuBois, C.D.; Knauf, G.A.; Cunningham, A.L.; Wilke, C.O.; Trent, M.S.; Davies, B.W. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell 2018, 172, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef]
- World Health Organization. Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-000019-3. [Google Scholar]
- CDC. Antibiotic Resistance Threats in the United States, 2019, US Department of Health and Human Services: Atlanta, GA, USA, December 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf(accessed on 2 February 2021).
- Bhattacharjya, S.; Straus, S.K. Design, engineering and discovery of novel α-helical and β-boomerang antimicrobial peptides against drug resistant bacteria. Int. J. Mol. Sci. 2020, 21, 5773. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility. In Vivo Biomol. 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Wang, Z.; Han, H.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Wang, X.; Wang, J. Dual antibacterial activities and biofilm eradication of a marine peptide-N6NH2 and its analogs against multidrug-resistant aeromonas veronii. Int. J. Mol. Sci. 2020, 21, 9637. [Google Scholar] [CrossRef]
- Małuch, I.; Stachurski, O.; Kosikowska-Adamus, P.; Makowska, M.; Bauer, M.; Wyrzykowski, D.; Hać, A.; Kamysz, W.; Deptuła, M.; Pikuła, M.; et al. Double-headed cationic lipopeptides: An EMERGING class of antimicrobials. Int. J. Mol. Sci. 2020, 21, 8944. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Wang, X.; Li, Z.; Wang, X.; Wang, J. Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia Coli. Commun. Biol. 2020, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.-L.; Chih, Y.-H.; Peng, K.-L.; Wu, C.-L.; Yu, H.-Y.; Cheng, D.; Chou, Y.-T.; Cheng, J.-W. Antimicrobial peptides with enhanced salt resistance and antiendotoxin properties. Int. J. Mol. Sci. 2020, 21, 6810. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, A.T.Y.; Gellatly, S.L.; Hancock, R.E.W. Multifunctional cationic host defence peptides and their clinical applications. Cell. Mol. Life. Sci. 2011, 68, 2161–2176. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Reassessing the host defense peptide landscape. Front. Chem. 2019, 7, 43. [Google Scholar] [CrossRef] [Green Version]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K.; et al. integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019, 10, 4538. [Google Scholar] [CrossRef]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B.; et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef] [Green Version]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, 111, e24122. [Google Scholar] [CrossRef]
- Haney, E.F.; Hancock, R.E.W. Peptide design for antimicrobial and immunomodulatory applications. Pept. Sci. 2013, 100, 572–583. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, M.Y.; Hong, S.Y.; Lee, K.H. Structure-activity analysis of brevinin 1E amide, an antimicrobial peptide from rana esculenta. Biochim. Biophys. Acta 1998, 1387, 239–248. [Google Scholar] [CrossRef]
- Sinha, S.; Ng, W.J.; Bhattacharjya, S. NMR Structure and localization of the host defense antimicrobial peptide thanatin in zwitterionic dodecylphosphocholine micelle: Implications in antimicrobial activity. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183432. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Zheng, L.; Mu, Y.; Ng, W.J.; Bhattacharjya, S. Structure and interactions of a host defense antimicrobial peptide thanatin in lipopolysaccharide micelles reveal mechanism of bacterial cell agglutination. Sci. Rep. 2017, 7, 17795. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.; Niu, C.; Zhou, Y.; Xue, X.; Meng, J.; Luo, X.; Hou, Z. The disulfide bond of the peptide thanatin is dispensible for its antimicrobial activity In Vivo and In Vitro. Antimicrob. Agents. Chemother. 2016, 60, 4283–4289. [Google Scholar] [CrossRef] [Green Version]
- Hou, Z.; Lu, J.; Fang, C.; Zhou, Y.; Bai, H.; Zhang, X.; Xue, X.; Chen, Y.; Luo, X. Underlying mechanism of In Vivo and In Vitro activity of C-terminal-amidated thanatin against clinical isolates of extended-spectrum beta-lactamase-producing Escherichia Coli. J. Infect. Dis. 2011, 203, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Fan, X.; Li, L.; Wang, H.; Ding, J.; Hongbin, W.; Zhao, R.; Gou, L.; Shen, Z.; Xi, T. Interaction of antimicrobial peptide S-thanatin with lipopolysaccharide In Vitro and in an experimental mouse model of septic shock caused by a multidrug-resistant clinical isolate of Escherichia Coli. Int. J. Antimicrob. Agents 2010, 35, 250–254. [Google Scholar] [CrossRef]
- Wu, G.; Li, X.; Fan, X.; Wu, H.; Wang, S.; Shen, Z.; Xi, T. The activity of antimicrobial peptide S-thanatin is independent on multidrug-resistant spectrum of bacteria. Peptides 2011, 32, 1139–1145. [Google Scholar] [CrossRef]
- Wu, G.; Wu, P.; Xue, X.; Yan, X.; Liu, S.; Zhang, C.; Shen, Z.; Xi, T. Application of S-thanatin, an antimicrobial peptide derived from thanatin, in mouse model of klebsiella pneumoniae infection. Peptides 2013, 45, 73–77. [Google Scholar] [CrossRef]
- Mah, T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef] [PubMed]
- De Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; de Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; van der Heijde, T.; Boekema, B.K.; et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10, 4044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayana, J.L.; Mishra, B.; Lushnikova, T.; Golla, R.M.; Wang, G. Modulation of antimicrobial potency of human cathelicidin peptides against the ESKAPE pathogens and In Vivo efficacy in a murine catheter-associated biofilm model. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1592–1602. [Google Scholar] [CrossRef] [PubMed]
- Woodburn, K.W.; Jaynes, J.; Clemens, L.E. Designed antimicrobial peptides for topical treatment of antibiotic resistant acne vulgaris. Antibiotics 2020, 9, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, F.; Teixeira, C.; Gomes, P.; Martins, M.C.L. Clinical Application of AMPs. Adv. Exp. Med. Biol. 2019, 1117, 281–298. [Google Scholar] [CrossRef]
- Luong, H.X.; Thanh, T.T.; Tran, T.H. Antimicrobial peptides—Advances in development of therapeutic applications. Life. Sci. 2020, 260, 118407. [Google Scholar] [CrossRef]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.T.; Cooper, M.A. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef]
- Dimarcq, J.-L.; Bulet, P.; Hetru, C.; Hoffmann, J. Cysteine-rich antimicrobial peptides in invertebrates. Pept. Sci. 1998, 47, 465–477. [Google Scholar] [CrossRef]
- Pagès, J.-M.; Dimarcq, J.-L.; Quenin, S.; Hetru, C. Thanatin activity on multidrug resistant clinical isolates of enterobacter aerogenes and klebsiella pneumoniae. Int. J. Antimicrob. Agents 2003, 22, 265–269. [Google Scholar] [CrossRef]
- Andrès, E. Cationic antimicrobial peptides in clinical development, with special focus on thanatin and heliomicin. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Cha, L.; Lee, S.-H.; Hahm, K.-S. Role of amino acid residues within the disulfide loop of thanatin, a potent antibiotic peptide. J. Biochem. Mol. Biol. 2002, 35, 291–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandard, N.; Sodano, P.; Labbe, H.; Bonmatin, J.-M.; Bulet, P.; Hetru, C.; Ptak, M.; Vovelle, F. Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. Eur. J. Biochem. 1998, 256, 404–410. [Google Scholar] [CrossRef]
- Mohanram, H.; Bhattacharjya, S. Cysteine deleted protegrin-1 (CDP-1): Anti-bacterial activity, outer-membrane disruption and selectivity. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 3006–3016. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, R.; Mohanram, H.; Joshi, M.; Domadia, P.N.; Torres, J.; Ruedl, C.; Bhattacharjya, S. Structure, activity and interactions of the cysteine deleted analog of tachyplesin-1 with lipopolysaccharide micelle: Mechanistic insights into outer-membrane permeabilization and endotoxin neutralization. Biochim. Biophys. Acta 2013, 1818, 1613–1624. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorthy, A.; Thennarasu, S.; Tan, A.; Gottipati, K.; Sreekumar, S.; Heyl, D.L.; An, F.Y.P.; Shelburne, C.E. Deletion of all cysteines in tachyplesin I abolishes hemolytic activity and retains antimicrobial activity and lipopolysaccharide selective binding. Biochemistry 2006, 45, 6529–6540. [Google Scholar] [CrossRef] [Green Version]
- Imamura, T.; Yamamoto, T.; Tamura, A.; Murabayashi, S.; Hashimoto, S.; Shimada, H.; Taguchi, S. NMR based structure–activity relationship analysis of an antimicrobial peptide thanatin, engineered by site-specific chemical modification: Activity improvement and spectrum alteration. Biochem. Biophys. Res. Commun. 2008, 369, 609–615. [Google Scholar] [CrossRef]
- Taguchi, S.; Kuwasako, K.; Suenaga, A.; Okada, M.; Momose, H. Functional mapping against escherichia coli for the broad-spectrum antimicrobial peptide, thanatin, based on an In Vivo monitoring assay system. J. Biochem. 2000, 128, 745–754. [Google Scholar] [CrossRef]
- Jiang, X.; Qian, K.; Liu, G.; Sun, L.; Zhou, G.; Li, J.; Fang, X.; Ge, H.; Lv, Z. Design and activity study of a melittin-thanatin hybrid peptide. AMB Express. 2019, 9, 14. [Google Scholar] [CrossRef]
- Tian, L.; Zhang, D.; Su, P.; Wei, Y.; Wang, Z.; Wang, P.X.; Dai, C.J.; Gong, G.L. Design, recombinant expression, and antibacterial activity of a novel hybrid magainin-thanatin antimicrobial peptide. Prep. Biochem. Biotechnol. 2019, 49, 427–434. [Google Scholar] [CrossRef]
- Hongbiao, W.; Baolong, N.; Mengkui, X.; Lihua, H.; Weifeng, S.; Zhiqi, M. Biological activities of cecropin B-thanatin hybrid peptides. J. Pept. Res. 2005, 66, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.N.; Yu, B.; Han, G.Q.; He, J.; Chen, D.W. Design, expression and characterization of recombinant hybrid peptide attacin-thanatin in Escherichia Coli. Mol. Biol. Rep. 2010, 37, 3495–3501. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Tang, D.; Chen, W.; Huang, H.; Wang, R.; Chen, Y. Expression of antimicrobial peptides thanatin(S) in transgenic arabidopsis enhanced resistance to phytopathogenic fungi and bacteria. Gene 2013, 527, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Yasuda, M.; Kusano, H.; Nakashita, H.; Ohno, Y.; Kamakura, T.; Taguchi, S.; Shimada, H. Acquired resistance to the rice blast in transgenic rice accumulating the antimicrobial peptide thanatin. Transgenic. Res. 2010, 19, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Houdelet, M.; Kogel, K.-H.; Fischer, R.; Schillberg, S.; Nölke, G. Thanatin confers partial resistance against aflatoxigenic fungi in maize (zea mays). Transgenic. Res. 2015, 24, 885–895. [Google Scholar] [CrossRef]
- Wu, G.; Wu, H.; Li, L.; Fan, X.; Ding, J.; Li, X.; Xi, T.; Shen, Z. Membrane aggregation and perturbation induced by antimicrobial peptide of s-thanatin. Biochem. Biophys. Res. Commun. 2010, 395, 31–35. [Google Scholar] [CrossRef]
- Robert, É.; Lefèvre, T.; Fillion, M.; Martial, B.; Dionne, J.; Auger, M. Mimicking and understanding the agglutination effect of the antimicrobial peptide thanatin using model phospholipid vesicles. Biochemistry 2015, 54, 3932–3941. [Google Scholar] [CrossRef]
- Bhattacharjya, S. NMR structures and interactions of antimicrobial peptides with lipopolysaccharide: Connecting structures to functions. Curr. Top. Med. Chem. 2016, 16, 4–15. [Google Scholar] [CrossRef]
- Domadia, P.N.; Bhunia, A.; Ramamoorthy, A.; Bhattacharjya, S. Structure, interactions, and antibacterial activities of MSI-594 derived mutant peptide MSI-594F5A in lipopolysaccharide micelles: Role of the helical hairpin conformation in outer-membrane permeabilization. J. Am. Chem. Soc. 2010, 132, 18417–18428. [Google Scholar] [CrossRef]
- Ilyas, H.; Kim, J.; Lee, D.; Malmsten, M.; Bhunia, A. Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism. J. Biol. Chem. 2019, 294, 14615–14633. [Google Scholar] [CrossRef]
- Saravanan, R.; Holdbrook, D.A.; Petrlova, J.; Singh, S.; Berglund, N.A.; Choong, Y.K.; Kjellström, S.; Bond, P.J.; Malmsten, M.; Schmidtchen, A. Structural basis for endotoxin neutralisation and anti-inflammatory activity of thrombin-derived C-terminal peptides. Nat. Commun. 2018, 9, 2762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhunia, A.; Bhattacharjya, S. Mapping residue-specific contacts of polymyxin B with lipopolysaccharide by saturation transfer difference NMR: Insights into outer-membrane disruption and endotoxin neutralization. Biopolymers 2011, 96, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.; Saravanan, R.; Mohanram, H.; Mangoni, M.L.; Bhattacharjya, S. NMR structures and interactions of temporin-1Tl and temporin-1Tb with lipopolysaccharide micelles: Mechanistic insights into outer membrane permeabilization and synergistic activity. J. Biol. Chem. 2011, 286, 24394–24406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.-Y.; Chen, Y.-A.; Yip, B.-S.; Wang, S.-Y.; Wei, H.-J.; Chih, Y.-H.; Chen, K.-H.; Cheng, J.-W. Role of β-naphthylalanine end-tags in the enhancement of antiendotoxin activities: Solution structure of the antimicrobial peptide S1-Nal-Nal in complex with lipopolysaccharide. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-K.; Lee, E.; Shin, S.; Jeong, K.; Lee, J.-Y.; Bae, S.-Y.; Kim, S.-H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, papilio xuthus. J. Biol. Chem. 2011, 286, 41296–41311. [Google Scholar] [CrossRef] [Green Version]
- Bhunia, A.; Domadia, P.N.; Torres, J.; Hallock, K.J.; Ramamoorthy, A.; Bhattacharjya, S. NMR structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide micelles: Mechanism of outer membrane permeabilization. J. Biol. Chem. 2010, 285, 3883–3895. [Google Scholar] [CrossRef] [Green Version]
- Bhunia, A.; Ramamoorthy, A.; Bhattacharjya, S. Helical hairpin structure of a potent antimicrobial peptide MSI-594 in lipopolysaccharide micelles by NMR spectroscopy. Chemistry 2009, 15, 2036–2040. [Google Scholar] [CrossRef] [Green Version]
- Bhunia, A.; Mohanram, H.; Domadia, P.N.; Torres, J.; Bhattacharjya, S. Designed beta-boomerang antiendotoxic and antimicrobial peptides: Structures and activities in lipopolysaccharide. J. Biol. Chem. 2009, 284, 21991–22004. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjya, S.; Ramamoorthy, A. Multifunctional host defense peptides: Functional and mechanistic insights from NMR structures of potent antimicrobial peptides. FEBS J. 2009, 276, 6465–6473. [Google Scholar] [CrossRef] [Green Version]
- Saravanan, R.; Joshi, M.; Mohanram, H.; Bhunia, A.; Mangoni, M.L.; Bhattacharjya, S. NMR structure of temporin-1Ta in lipopolysaccharide micelles: Mechanistic insight into inactivation by outer membrane. PLoS ONE 2013, 8, e72718. [Google Scholar] [CrossRef]
- Vetterli, S.U.; Zerbe, K.; Müller, M.; Urfer, M.; Mondal, M.; Wang, S.-Y.; Moehle, K.; Zerbe, O.; Vitale, A.; Pessi, G.; et al. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia Coli. Sci. Adv. 2018, 4, eaau2634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, E.C.C.M.; Baeta, T.; Romanelli, A.; Laguri, C.; Martorana, A.M.; Erba, E.; Simorre, J.-P.; Sperandeo, P.; Polissi, A. Thanatin impairs lipopolysaccharide transport complex assembly by targeting LptC-LptA interaction and decreasing LptA stability. Front. Microbiol. 2020, 11, 909. [Google Scholar] [CrossRef] [PubMed]
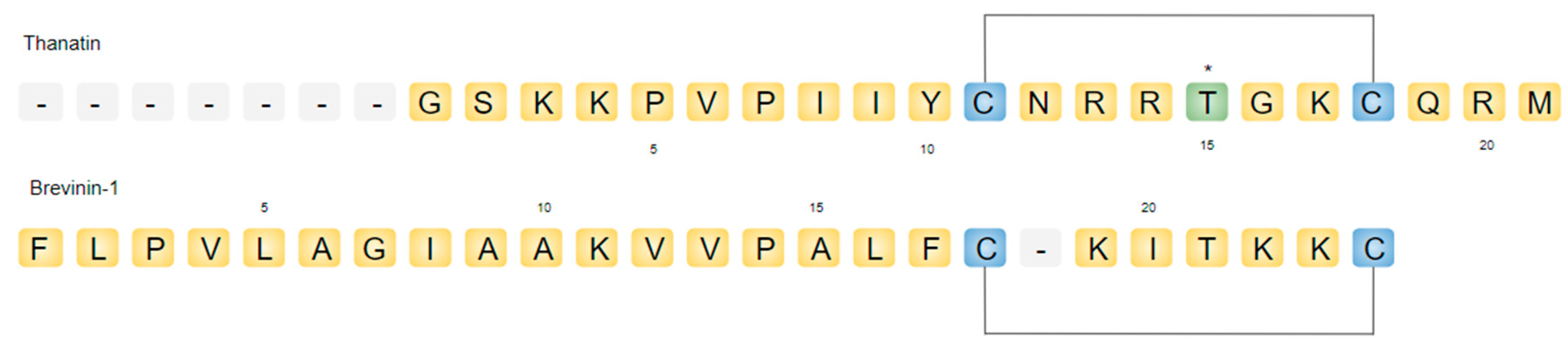
| Microorganism | MIC (µM) |
|---|---|
| Gram-Positive Bacteria | |
| Aerococcus viridans | 0.6–1.2 |
| Micrococcus luteus | 1.2–2.5 |
| Bacillus megaterium | 2.5–5 |
| Bacillus subtilis | 2.5–5 |
| Staphylococcus aureus | No activity |
| Pediococcus acidolactici | 20–40 |
| Gram-Negative Bacteria | |
| Escherichia coli D22 | 0.3–0.6 |
| E. coli D31 | 0.3–0.6 |
| E. coli 1106 | 0.6–1.2 |
| Salmonella typhimurium | 0.6–1.2 |
| Klebsiella pneumoniae | 0.6–1.2 |
| Enterobacter cloacae | 1.2–2.5 |
| Erwinia carotovora | 10–20 |
| Pseudomonas aeruginosa | 20–40 |
| Fungi | |
| Neurospora crassa | 0.6–1.2 |
| Botrytis cinerea | 1.2–2.5 |
| Nectria haematococca | 1.2–2.5 |
| Trichoderma viride | 1.2–2.5 |
| Alternaria brassicola | 2.5–5 |
| Fusarium culmorum | 2.5–5 |
| Ascochyta pisi | 5–10 |
| Fusarium oxysporum | 10–20 |
| Sequence | (G−) Activity | (G+) Activity | Antifungal |
|---|---|---|---|
| GSKKPVPIIYCNRRTGKCQRM (Thanatin) | ++++ | ++++ | ++++ |
| GSKKPVPIIYCNRRTGKCQR (G20R) | + | +++ | +++ |
| GSKKPVPIIYCNRRTGKCQ (G19Q) | − | ++ | +++ |
| GSKKPVPIIYCNRRTGKC (G18C) | − | ++ | +++ |
| KPVPIIYCNRRTGKCQRM (K18M) | ++++ | ++++ | +++ |
| VPIIYCNRRTGKCQRM (V16M) | +++ | +++ | ++ |
| IIYCNRRTGKCQRM (I14M) | + | ++ | + |
| YCNRRTGKCQRM (Y12M) | − | + | − |
| Sequence | (G−) Activity | (G+) Activity |
|---|---|---|
| GSKKPVPIIYCNRRTGKCQRM (Thanatin) | ++++ | ++++ |
| GSKKPVPIIYCNRR-GKCQRM (Del T) | ++++ | +++++ |
| GSKKPVPIIYCNRRT-KCQRM (Del G) | +++ | +++ |
| GSKKPVPIIYCNRR-KCQRM (Del T, G) | +++ | +++ |
| GSKKPVPIIYCNRRATGKCQRM (Ins A) | ++ | ++ |
| GSKKPVPIIYCNRRAATGKCQRM (Ins AA) | ++ | ++ |
| Sequence | (G−) Activity | (G+) Activity |
|---|---|---|
| GSKKPVPIIYCNRRTGKCQRM (Thanatin) | +++ | +++ |
| GSKKPVPIIYCNRRTGKCQRM (L-Thanatin) | +++ | +++ |
| GSKKPVPIIYANRRTGKAQRM (C to A) | − | − |
| GSKKPVPIIYSNRRTGKSQRM (C to S) | − | +++ |
| GSKKPVPIIYXNRRTGKXQRM (C to X, X stands for Cys residues modified with tert-butyl group) | − | +++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dash, R.; Bhattacharjya, S. Thanatin: An Emerging Host Defense Antimicrobial Peptide with Multiple Modes of Action. Int. J. Mol. Sci. 2021, 22, 1522. https://doi.org/10.3390/ijms22041522
Dash R, Bhattacharjya S. Thanatin: An Emerging Host Defense Antimicrobial Peptide with Multiple Modes of Action. International Journal of Molecular Sciences. 2021; 22(4):1522. https://doi.org/10.3390/ijms22041522
Chicago/Turabian StyleDash, Rachita, and Surajit Bhattacharjya. 2021. "Thanatin: An Emerging Host Defense Antimicrobial Peptide with Multiple Modes of Action" International Journal of Molecular Sciences 22, no. 4: 1522. https://doi.org/10.3390/ijms22041522






