Cytogenomics Unveil Possible Transposable Elements Driving Rearrangements in Chromosomes 2 and 4 of Solea senegalensis
Abstract
:1. Introduction
2. Results
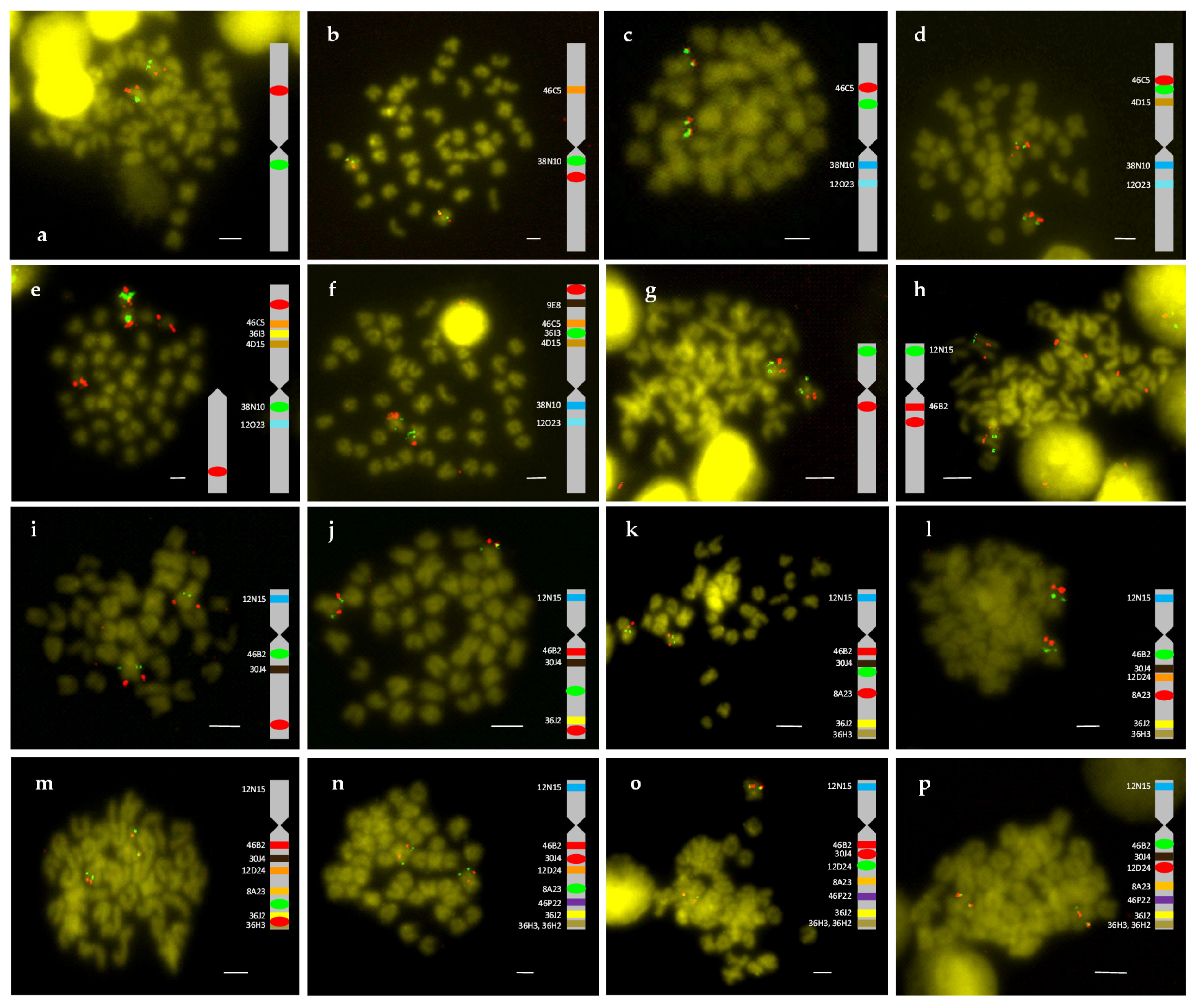
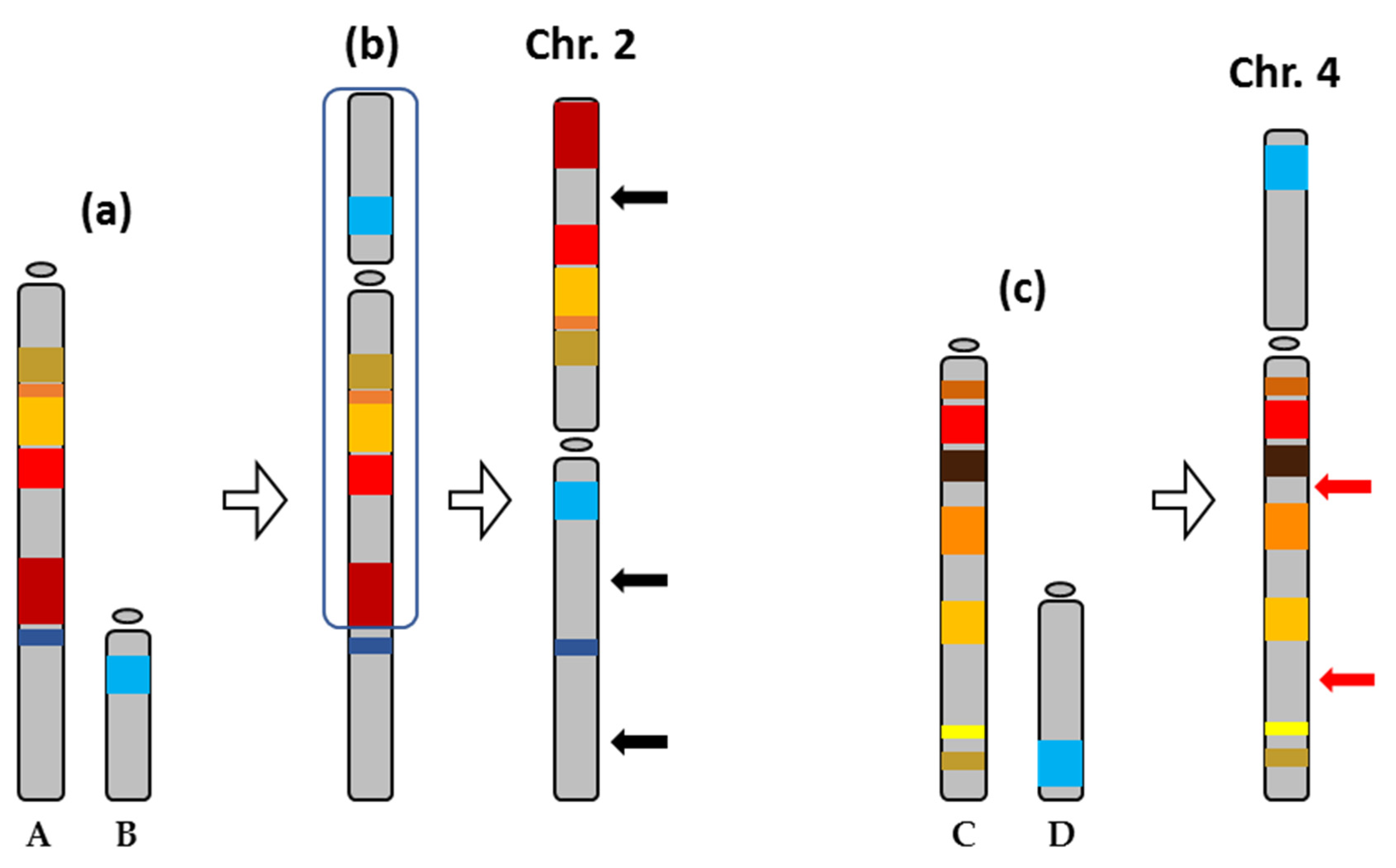
2.1. Description of the Metacentric and Submetacentric Chromosomes 2 and 4
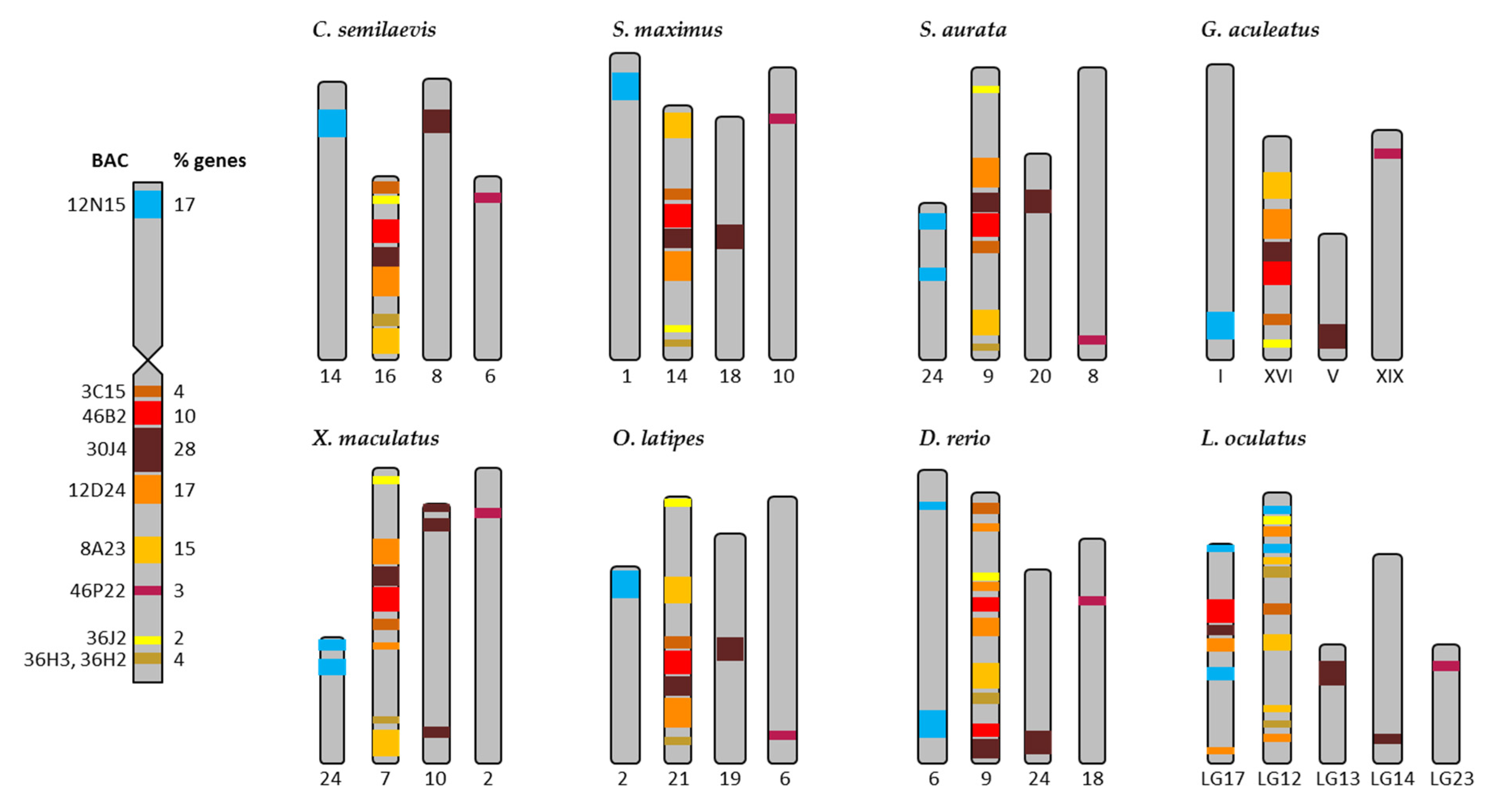
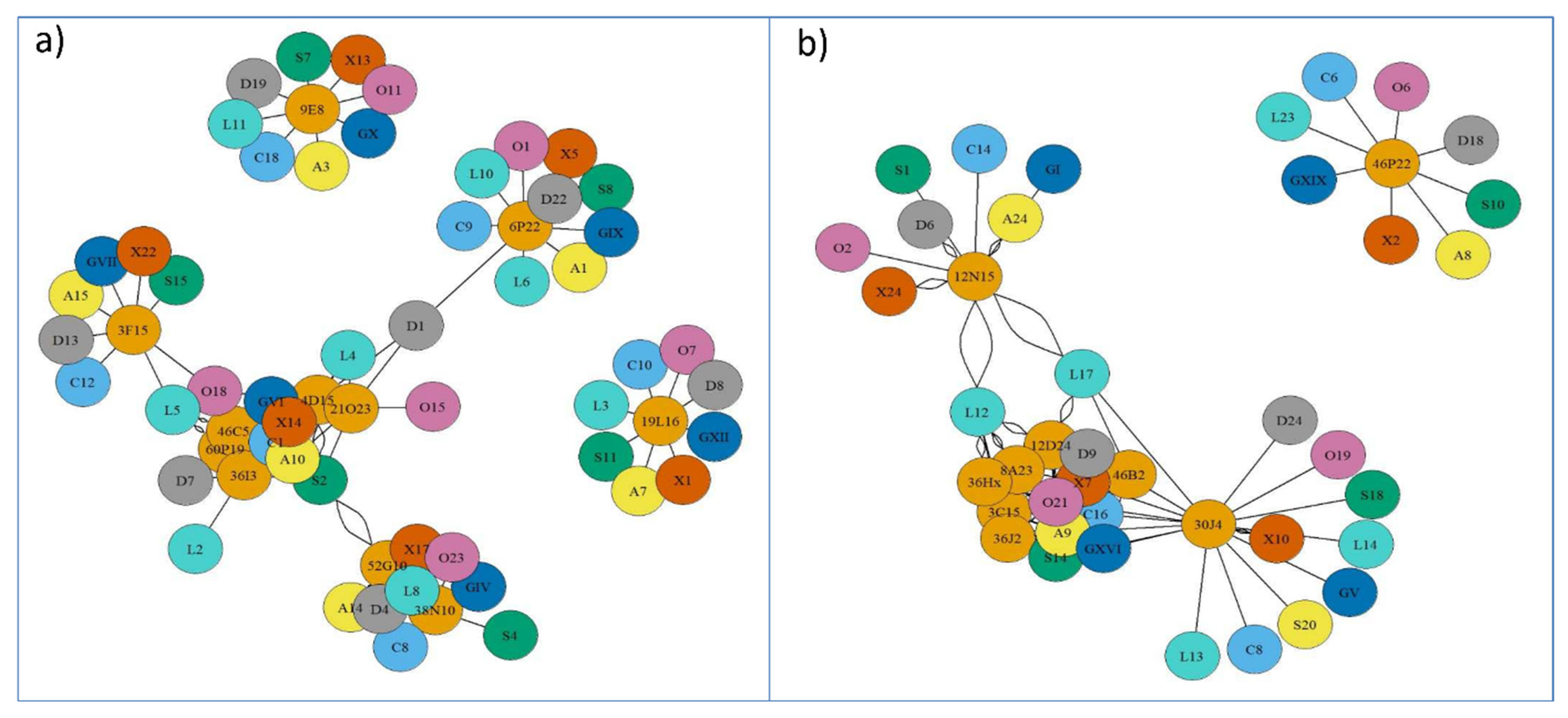
2.2. Synteny Analysis for Metacentric Chromosome 2
2.3. Synteny Analysis for Submetacentric Chromosome 4
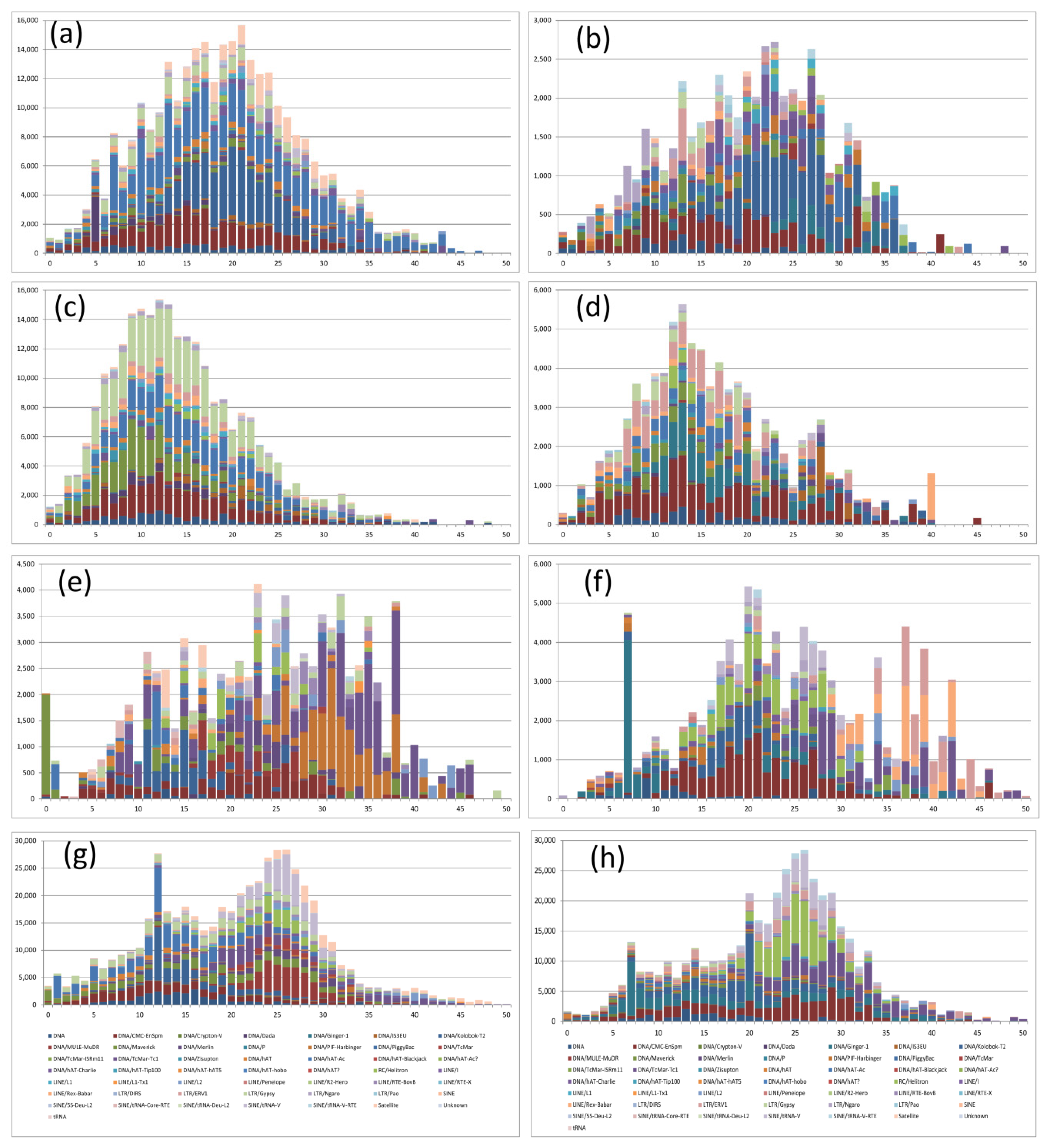
2.4. Distribution of Repeated Sequences
2.5. Analysis of the Transposable Elements Divergence
3. Discussion
4. Materials and Methods
4.1. BAC Clones
4.2. Double FISH-BAC
4.3. Sequencing and Synteny Analysis
Synteny Relationship Net-Graphs
4.4. Repetitive Elements Analysis
4.5. Transposable Elements Divergence
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Symonová, R.; Howell, W.M. Vertebrate Genome Evolution in the Light of Fish Cytogenomics and rDNAomics. Genes 2018, 9, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, M.E.; Molina, B.; Merlo, M.A.; Arias-Pérez, A.; Portela-Bens, S.; García-Angulo, A.; Cross, I.; Liehr, T.; Rebordinos, L. Evolution of the proto sex-chromosome in Solea senegalensis. Int. J. Mol. Sci. 2019, 20, 5111. [Google Scholar] [CrossRef] [Green Version]
- García-Angulo, A.; Merlo, M.A.; Portela-Bens, S.; Rodríguez, M.E.; García, E.; Al-Rikabi, A.B.H.; Liehr, T.; Rebordinos, L. Evidence for a Robertsonian fusion in Solea senegalensis (Kaup, 1858) revealed by zoo-FISH and comparative genome analysis. BMC Genom. 2018, 19, 818. [Google Scholar] [CrossRef] [PubMed]
- García-Angulo, A.; Merlo, M.A.; Rodríguez, M.E.; Portela-Bens, S.; Liehr, T.; Rebordinos, L. Genome and Phylogenetic Analysis of Genes Involved in the Immune System of Solea senegalensis – Potential Applications in Aquaculture. Front. Genet. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Han, M.; Peng, Z. Evolution and diversity of transposable elements in fish genomes. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Jurka, J.; Kapitonov, V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Manchado, M.; Planas, J.V.; Cousin, X.; Rebordinos, L.; Claros, M.G. Genetic and Genomic Characterization of Soles. Biol. Sole 2019, 6, 375–394. [Google Scholar] [CrossRef]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.-N. Comparative Analysis of Transposable Elements Highlights Mobilome Diversity and Evolution in Vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef]
- García, E.; Cross, I.; Portela-Bens, S.; Rodríguez, M.E.; García-Angulo, A.; Molina, B.; Cuadrado, Á.; Liehr, T.; Rebordinos, L. Integrative genetic map of repetitive DNA in the sole Solea senegalensis genome shows a Rex transposon located in a proto-sex chromosome. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World, 5th ed.; Willey & Sons: Hoboken, NY, USA, 2016; o7030-5774. [Google Scholar]
- Glasauer, S.M.K.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970; ISBN 978-3-642-86661-6. [Google Scholar]
- Brum, M.J.L.; Galetti, P.M., Jr. Teleostei ground plan karyotype. J. Comp. Biol. 1997, 2, 91–102. [Google Scholar]
- Bitencourt, J.A.; Sampaio, I.; Ramos, R.T.; Affonso, P.R. Chromosomal fusion in Brazilian populations of Trinectes inscriptus Gosse, 1851 (Pleuronectiformes; Achiridae) as revealed by internal telomere sequences (ITS). J. Exp. Mar. Biol. Ecol. 2014, 452, 101–104. [Google Scholar] [CrossRef]
- Shi, W.; Chen, S.; Kong, X.; Si, L.; Gong, L.; Zhang, Y.; Yu, H. Flatfish monophyly refereed by the relationship of Psettodes in Carangimorphariae. BMC Genom. 2018, 19, 400. [Google Scholar] [CrossRef]
- Campbell, M.A.; Chen, W.-J.; Lopez, J.A. Are flatfishes (Pleuronectiformes) monophyletic? Mol. Phylogenet. Evol. 2013, 69, 664–673. [Google Scholar] [CrossRef] [Green Version]
- Campbell, M.A.; Chen, W.-J.; López, J.A. Molecular data do not provide unambiguous support for the monophyly of flatfishes (Pleuronectiformes): A reply to Betancur-R and Ortí. Mol. Phylogenetics Evol. 2014, 75, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; López, J.A.; Satoh, T.P.; Chen, W.-J.; Miya, M. Mitochondrial genomic investigation of flatfish monophyly. Gene 2014, 551, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Harrington, R.C.; Faircloth, B.C.; Eytan, R.I.; Smith, W.L.; Near, T.J.; Alfaro, M.E.; Friedman, M. Phylogenomic analysis of carangimorph fishes reveals flatfish asymmetry arose in a blink of the evolutionary eye. BMC Evol. Biol. 2016, 16, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinegardner, R. Evolution of Cellular DNA Content in Teleost Fishes. Am. Nat. 1968, 102, 517–523. [Google Scholar] [CrossRef]
- Azevedo, M.F.C.; Oliveira, C.; Pardo, B.G.; Martínez, P.; Foresti, F. Cytogenetic characterization of six species of flatfishes with comments to karyotype differentiation patterns in Pleuronectiformes (Teleostei). J. Fish Biol. 2007, 70, 1–15. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.-N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardie, D.C.; Hebert, P.D. Genome-size evolution in fishes. Can. J. Fish. Aquat. Sci. 2004, 61, 1636–1646. [Google Scholar] [CrossRef]
- Pardo, B.G.; Bouza, C.; Castro, J.; Martínez, P.; Sánchez, L. Localization of ribosomal genes in Pleuronectiformes using Ag-, CMA3-banding and in situ hybridization. Heredity 2001, 86, 531–536. [Google Scholar] [CrossRef]
- Robledo, D.; Hermida, M.; Rubiolo, J.A.; Fernández, C.; Blanco, A.; Bouza, C.; Martínez, P. Integrating genomic resources offlatfish (Pleuronectiformes) to boost aquaculture production. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 21, 41–55. [Google Scholar]
- Vega, L.; Díaz, E.; Cross, I.; Rebordinos, L. Caracterizaciones citogenética e isoenzimática del lengauado Solea senegalensis Kaup, 1858. Bol. Inst. Esp. Oceanogr. 2002, 18, 245–250. [Google Scholar]
- Cross, I.; Merlo, A.; Manchado, M.; Infante, C.; Cañavate, J.-P.; Rebordinos, L.; Torres, M.A.M. Cytogenetic characterization of the sole Solea senegalensis (Teleostei: Pleuronectiformes: Soleidae): Ag-NOR, (GATA) n, (TTAGGG) n and ribosomal genes by one-color and two-color FISH. Genetics 2006, 128, 253–259. [Google Scholar] [CrossRef]
- Portela-Bens, S.; Merlo, M.A.; Rodríguez, M.E.; Cross, I.; Manchado, M.; Kosyakova, N.; Liehr, T.; Rebordinos, L. Integrated gene mapping and synteny studies give insights into the evolution of a sex proto-chromosome in Solea senegalensis. Chromosom 2016, 126, 261–277. [Google Scholar] [CrossRef]
- Cegarra, A.M.G.; Merlo, M.; Ponce, M.; Portela-Bens, S.; Cross, I.; Manchado, M.; Rebordinos, L. A Preliminary Genetic Map inSolea senegalensis(Pleuronectiformes, Soleidae) Using BAC-FISH and Next-Generation Sequencing. Cytogenet. Genome Res. 2013, 141, 227–240. [Google Scholar] [CrossRef]
- Merlo, M.A.; Iziga, R.; Portela-Bens, S.; Cross, I.; Kosyakova, N.; Liehr, T.; Manchado, M.; Rebordinos, L. Analysis of the histone cluster in Senegalese sole (Solea senegalensis): Evidence for a divergent evolution of two canonical histone clusters. Genome 2017, 60, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Da Motta-Neto, C.C.; Cioffi, M.D.B.; Da Costa, G.W.W.F.; Amorim, K.D.J.; Bertollo, L.A.C.; Artoni, R.F.; Molina, W.F. Overview on Karyotype Stasis in Atlantic Grunts (Eupercaria, Haemulidae) and the Evolutionary Extensions for Other Marine Fish Groups. Front. Mar. Sci. 2019, 6, 628. [Google Scholar] [CrossRef] [Green Version]
- Amores, A.; Catchen, J.; Nanda, I.; Warren, W.; Walter, R.; Schartl, M.; Postlethwait, J.H. A RAD-Tag Genetic Map for the Platyfish (Xiphophorus maculatus) Reveals Mechanisms of Karyotype Evolution Among Teleost Fish. Genetics 2014, 197, 625–641. [Google Scholar] [CrossRef] [Green Version]
- Úbeda-Manzanaro, M.; Merlo, M.A.; Palazón, J.L.; Cross, I.; Sarasquete, C.; Rebordinos, L. Chromosomal mapping of the major and minor ribosomal genes, (GATA)n and U2 snRNA gene by double-colour FISH in species of the Batrachoididae family. Genetics 2010, 138, 787–794. [Google Scholar] [CrossRef]
- Sun, A.; Chen, S.-L.; Gao, F.-T.; Li, H.-L.; Liu, X.-F.; Wang, N.; Sha, Z.-X. Establishment and characterization of a gonad cell line from half-smooth tongue sole Cynoglossus semilaevis pseudomale. Fish Physiol. Biochem. 2015, 41, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Taboada, X.; Pansonato-Alves, J.C.; Foresti, F.; Martínez, P.; Viñas, A.; Pardo, B.G.; Bouza, C. Consolidation of the genetic and cytogenetic maps of turbot (Scophthalmus maximus) using FISH with BAC clones. Chromosoma 2014, 123, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, D.; Volff, J.-N. Analysis of the spotted gar genome suggests absence of causative link between ancestral genome duplication and transposable element diversification in teleost fish. J. Exp. Zool. Part B Mol. Dev. Evol. 2017, 328, 629–637. [Google Scholar] [CrossRef]
- Gao, B.; Shen, D.; Xue, S.; Chen, C.; Cui, H.; Song, C. The contribution of transposable elements to size variations between four teleost genomes. Mob. DNA 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.; Liu, S.; Zhou, T.; Tian, C.; Bao, L.; Dunham, R.A.; Liu, Z. Comparative genome analysis of 52 fish species suggests differential associations of repetitive elements with their living aquatic environments. BMC Genom. 2018, 19, 1–10. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Hellemans, B.; Volckaert, F.A. Microsatellites and their genomic distribution, evolution, function and applications: A review with special reference to fish genetics. Aquaculture 2006, 255, 1–29. [Google Scholar] [CrossRef]
- Edwards, Y.J.; Elgar, G.; Clark, M.S.; Bishop, M.J. The identification and characterization of microsatellites in the compact genome of the japanese pufferfish, Fugu rubripes: Perspectives in functional and comparative genomic analyses. J. Mol. Biol. 1998, 278, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Underwood, C.J.; Choi, K. Heterogeneous transposable elements as silencers, enhancers and targets of meiotic recombination. Chromosoma 2019, 128, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Carducci, F.; Barucca, M.; Canapa, A.; Carotti, E.; Biscotti, M.A. Mobile Elements in Ray-Finned Fish Genomes. Life 2020, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, S.; Abe, I.; Kudoh, Y.; Kishi, N.; Wang, Y.; Kubota, R.; Kudoh, J.; Kawasaki, K.; Minoshima, S.; Shimizu, N. Human BAC library: Construction and rapid screening. Gene 1997, 191, 69–79. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Csardi, G.; Nepusz, T. The igraph software package from complex network research. Int. J. Complex Syst. 2006, 1695. Available online: http://igraph.org (accessed on 28 August 2020).
- Fruchterman, T.M.J.; Reingold, E.M. Graph drawing by force-directed placement. Software Pr. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-3.0 (1996–2010). Available online: http://www.repeatmasker.org/ (accessed on 10 April 2013).
- Quinian, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]



| Chromosome | BAC | Gene Annotation | References |
|---|---|---|---|
| Chr. 2 | 52G10 | lamb1, ssbp1, creb3l2, dgki, calu, opn1sw, tnpo3, irf5, atp6v1f, dennd6b, gcc1, arf5, lta4h, mical3a | [3,9] |
| 6P22 | mri1, rfx1, dcaf15, khsrp, slc25a24, rgs5, c19orf53, gng10, ssx2ip, nanos3, cc2d1a, umod, slc26a11, sgsh, npb, sirt7, pcyt2, gnao1, mafg, map2k5, skor1b | [29] | |
| 9E8 | thrβ, nr1d2, ube2e2 | [4,30] | |
| 60P19 | trim39, c1qtnf3, casq1b, pea15, mrpl48, supt16h, trim25, fam234a, mrc2 | [3,4] | |
| 46C5 | ndrg2, hnrnpc, fhod1, klhl33, c1ql2, parp14, mrc1, slc12a3, ltb4r, myadm, arhgef11a, apol6 | [3] | |
| 36I3 | chd3, tnfsf12 | [3] | |
| 4D15 | ppme1, rnf150, inpp4b, hmgb2, dlg4 | [3,9] | |
| 38N10 | tmbim4, b2m, gdi2, asb13, net1, hspa14, manf, irak3, prl | [3] | |
| 3F15 | slc18a3a, rgr, lrit1 | This work | |
| 21O23 | fabp2, dmrt4, adcyap1r1 | [29] | |
| 19L16 | kiaa1324, c1orf194, smc5, fam107b | This work | |
| Chr. 4 | 12N15 | tuba1c, taar13c, atp1b1b, ube3a, gpd2, hsf2bp, cnga3, ddx4, ccdc14, ankrd10, dpp10, nr4a2 | [3,29] |
| 3C15 | pde11a, cycsb, osbpl6 | [3] | |
| 46B2 | uggt2, dnajc3, cldn10, abcc4, dct, sox21, gpc6 | [3] | |
| 30J4 | nlrc3, wdr90, rhot2, H1.0B, rhbdl1, wdr24, anks3, c8orf33, H3.3, gcgr, pcdh8, ednrb, cog3, mid1, arhgap6, tlr7, tlr8, tyb12, egfl6 | [3,30,31] | |
| 12D24 | igsf3, rpe, ackr3, nqo1, nhlh2, vangl1, casq2, kcne2, slc5a3, dop1b, morc3, ptprn | [3,4] | |
| 8A23 | hibch, c2orf88, mstn, pms1, ormdl1, adat3, alkbh6, osgepl1, fkbp7, gls | [3] | |
| 46P22 | zfpm1, trhr2 | [4] | |
| 36J2 | crygs | [3] | |
| 36H3, 36H2 | ccdc141, tchh, ttn | [3] |
| NL/Mb | Coverage (%) | |||
|---|---|---|---|---|
| Class/Family | Chr.2 | Chr.4 | Chr.2 | Chr.4 |
| Retroelements | 114.209 | 124.649 | 1.12 | 1.68 |
| SINEs: | 34.686 | 22.879 | 0.34 | 0.23 |
| Penelope | 1.692 | 3.156 | 0.01 | 0.03 |
| LINEs: | 47.376 | 44.968 | 0.57 | 0.81 |
| L2/CR1/Rex | 27.072 | 29.190 | 0.32 | 0.58 |
| R1/LOA/Jockey | 3.384 | 3.156 | 0.02 | 0.03 |
| R2/R4/NeSL | 0.000 | 0.789 | 0 | 0.11 |
| RTE/Bov-B | 9.306 | 0.789 | 0.14 | 0.02 |
| L1/CIN4 | 2.538 | 4.734 | 0.02 | 0.03 |
| LTR elements | 32.148 | 56.802 | 0.21 | 0.64 |
| BEL/Pao | 5.922 | 4.734 | 0.04 | 0.03 |
| Ty1/Copia | 0.000 | 0.000 | 0 | 0 |
| Gypsy/DIRS1 | 13.536 | 22.090 | 0.08 | 0.29 |
| Retroviral | 3.384 | 20.512 | 0.02 | 0.24 |
| DNA transposons | 317.247 | 268.232 | 2.97 | 2.81 |
| hobo-Activator | 141.281 | 85.203 | 1.15 | 0.69 |
| Tc1-IS630-Pogo | 45.684 | 20.512 | 0.83 | 0.43 |
| PiggyBac | 1.692 | 1.578 | 0.02 | 0.04 |
| Tourist/Harbinger | 9.306 | 7.100 | 0.16 | 0.07 |
| Other | 0.846 | 0.000 | 0 | 0 |
| Chromosome of S. senegalensis. | Spp. | Main Divergence Peak (K-Value) | Families |
|---|---|---|---|
| 2 | C. semilaevis | 16–21 | DNA/CMC/Emspm, DNA/Kolobok and DNA/hAT-Ac elements |
| S. maximus | 9–13 | DNA/CMC-En, DNA/Maverick, DNA/hAT-Ac and LTR/Gypsy | |
| O. latipes | 23–38 | DNA/PIF-Harburger and DNA/Tcmar-TC1 | |
| D. rerio | 24–27 | DNA/Kolobok-T2 | |
| 4 | C. semilaevis | 22–27 | DNA/Kolobok and DNA/hAt-Carlie |
| S. maximus | 12–14 | DNA/CMC-EnSpm, DNA/Ginger-1 and LTR/ERV-1 | |
| O. latipes | 20–21 | DNA/CMC-EnSpm and RC Helitron | |
| D. rerio | 24–27 | RC/Helitron and SINE/tRNA-V |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, M.E.; Cross, I.; Arias-Pérez, A.; Portela-Bens, S.; Merlo, M.A.; Liehr, T.; Rebordinos, L. Cytogenomics Unveil Possible Transposable Elements Driving Rearrangements in Chromosomes 2 and 4 of Solea senegalensis. Int. J. Mol. Sci. 2021, 22, 1614. https://doi.org/10.3390/ijms22041614
Rodríguez ME, Cross I, Arias-Pérez A, Portela-Bens S, Merlo MA, Liehr T, Rebordinos L. Cytogenomics Unveil Possible Transposable Elements Driving Rearrangements in Chromosomes 2 and 4 of Solea senegalensis. International Journal of Molecular Sciences. 2021; 22(4):1614. https://doi.org/10.3390/ijms22041614
Chicago/Turabian StyleRodríguez, María Esther, Ismael Cross, Alberto Arias-Pérez, Silvia Portela-Bens, Manuel Alejandro Merlo, Thomas Liehr, and Laureana Rebordinos. 2021. "Cytogenomics Unveil Possible Transposable Elements Driving Rearrangements in Chromosomes 2 and 4 of Solea senegalensis" International Journal of Molecular Sciences 22, no. 4: 1614. https://doi.org/10.3390/ijms22041614
APA StyleRodríguez, M. E., Cross, I., Arias-Pérez, A., Portela-Bens, S., Merlo, M. A., Liehr, T., & Rebordinos, L. (2021). Cytogenomics Unveil Possible Transposable Elements Driving Rearrangements in Chromosomes 2 and 4 of Solea senegalensis. International Journal of Molecular Sciences, 22(4), 1614. https://doi.org/10.3390/ijms22041614











