Characterization of ALBA Family Expression and Localization in Arabidopsis thaliana Generative Organs
Abstract
1. Introduction
2. Results
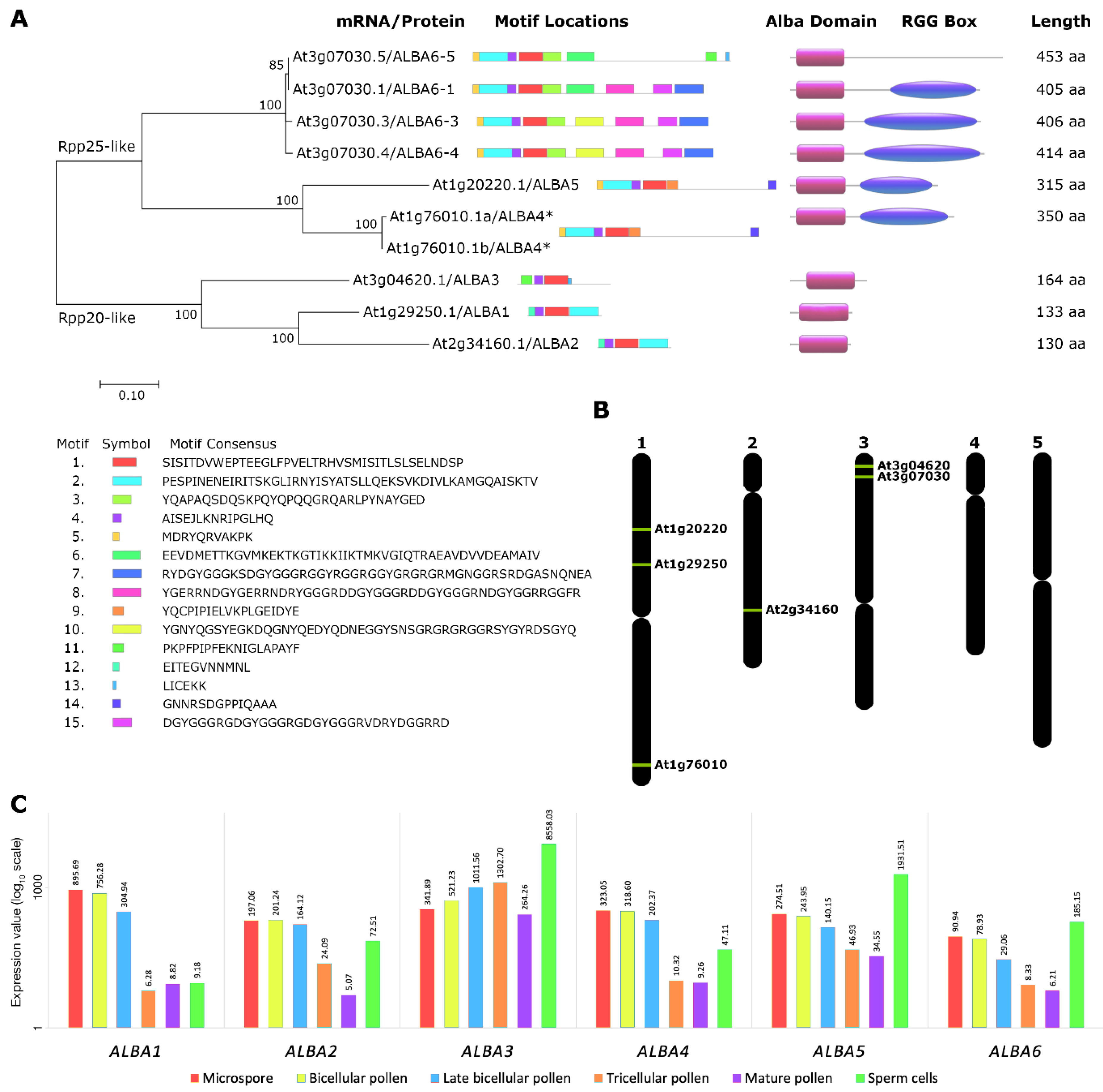
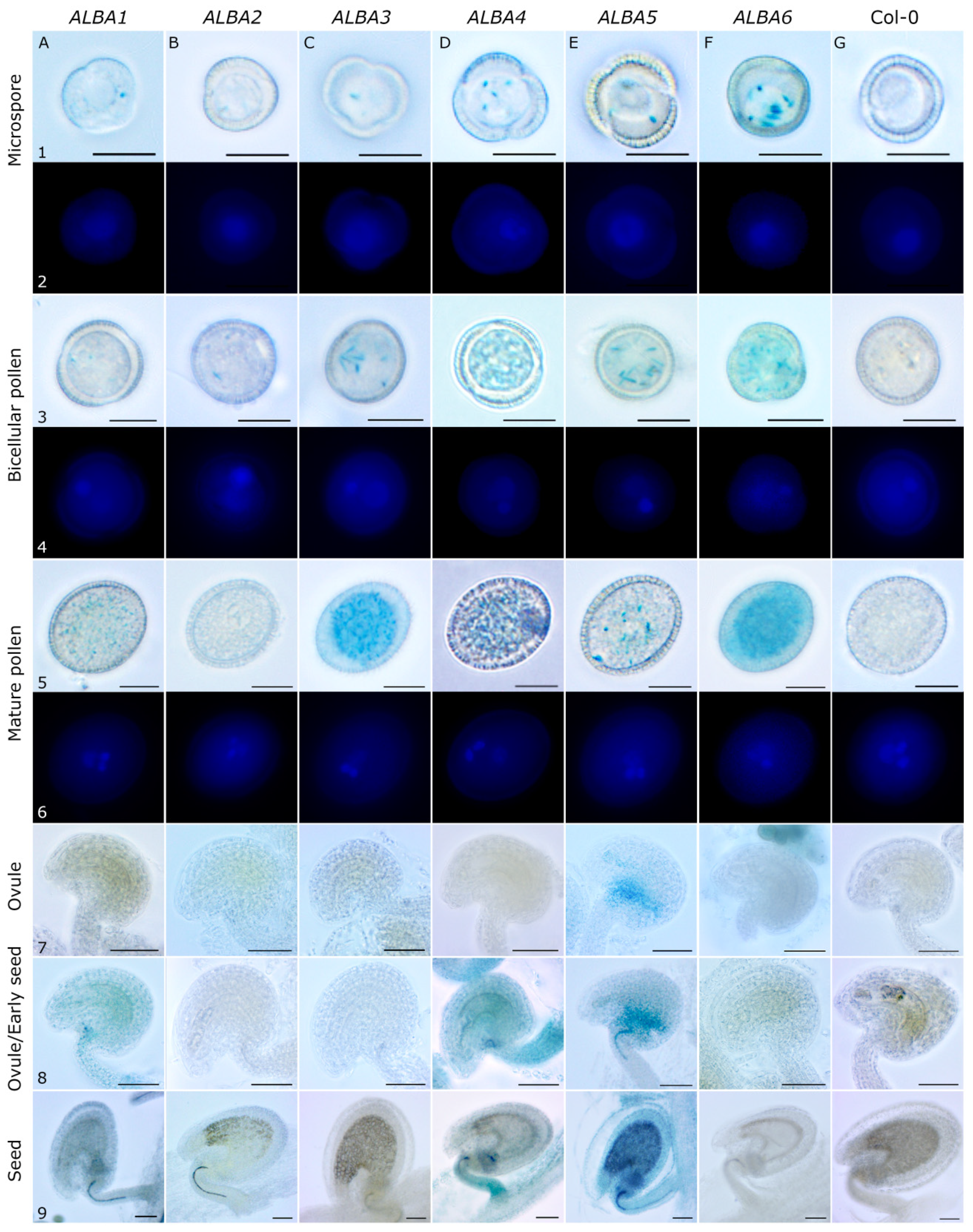
2.1. Expression Analysis of ALBA Genes in Arabidopsis Inflorescences
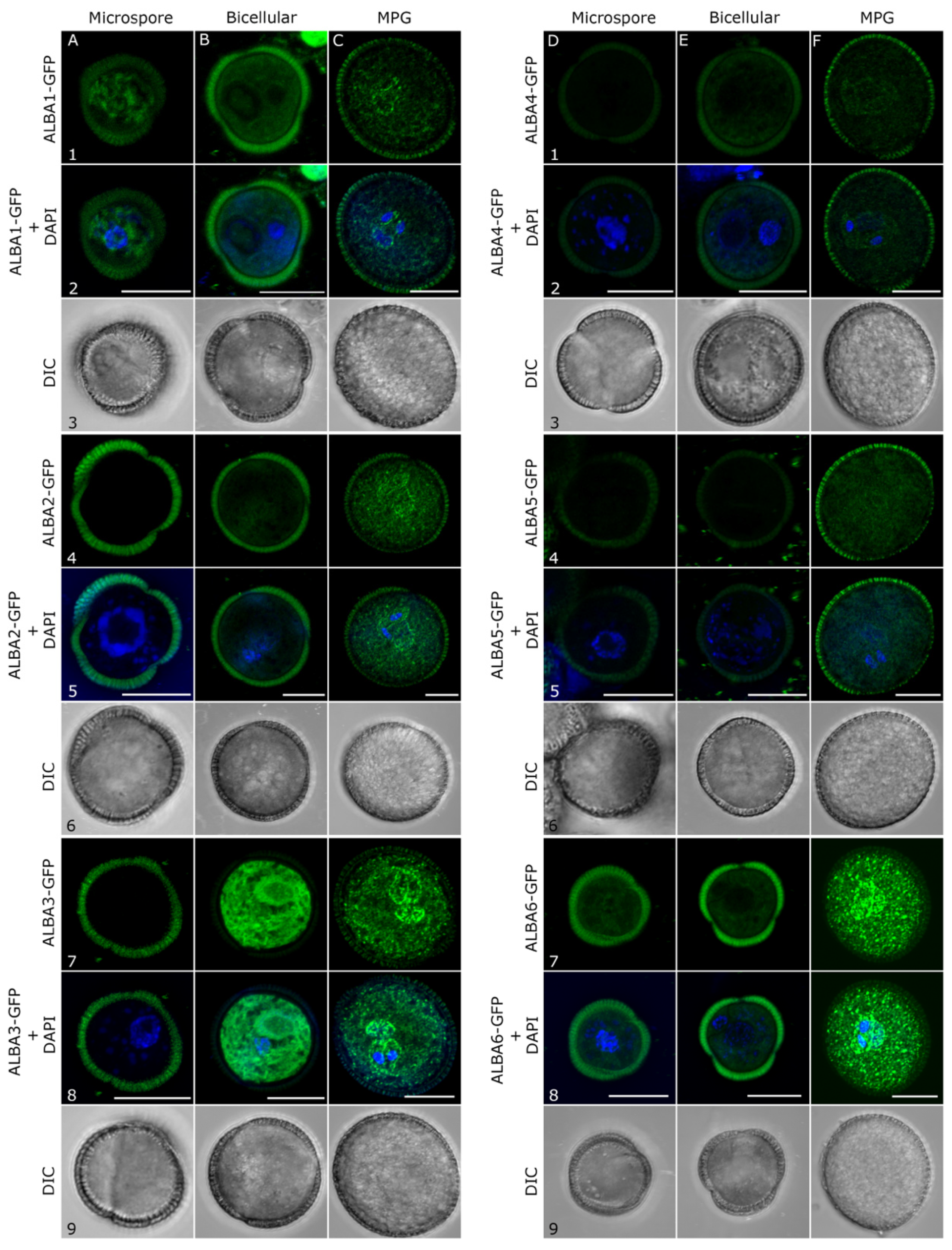
2.2. Subcellular Localization of ALBA Proteins during Pollen Development
2.3. ALBA Genes Expression in Arabidopsis Inflorescences after Heat Stress
2.4. Heat-Stress Induces Aggregation of ALBA Proteins in Pollen
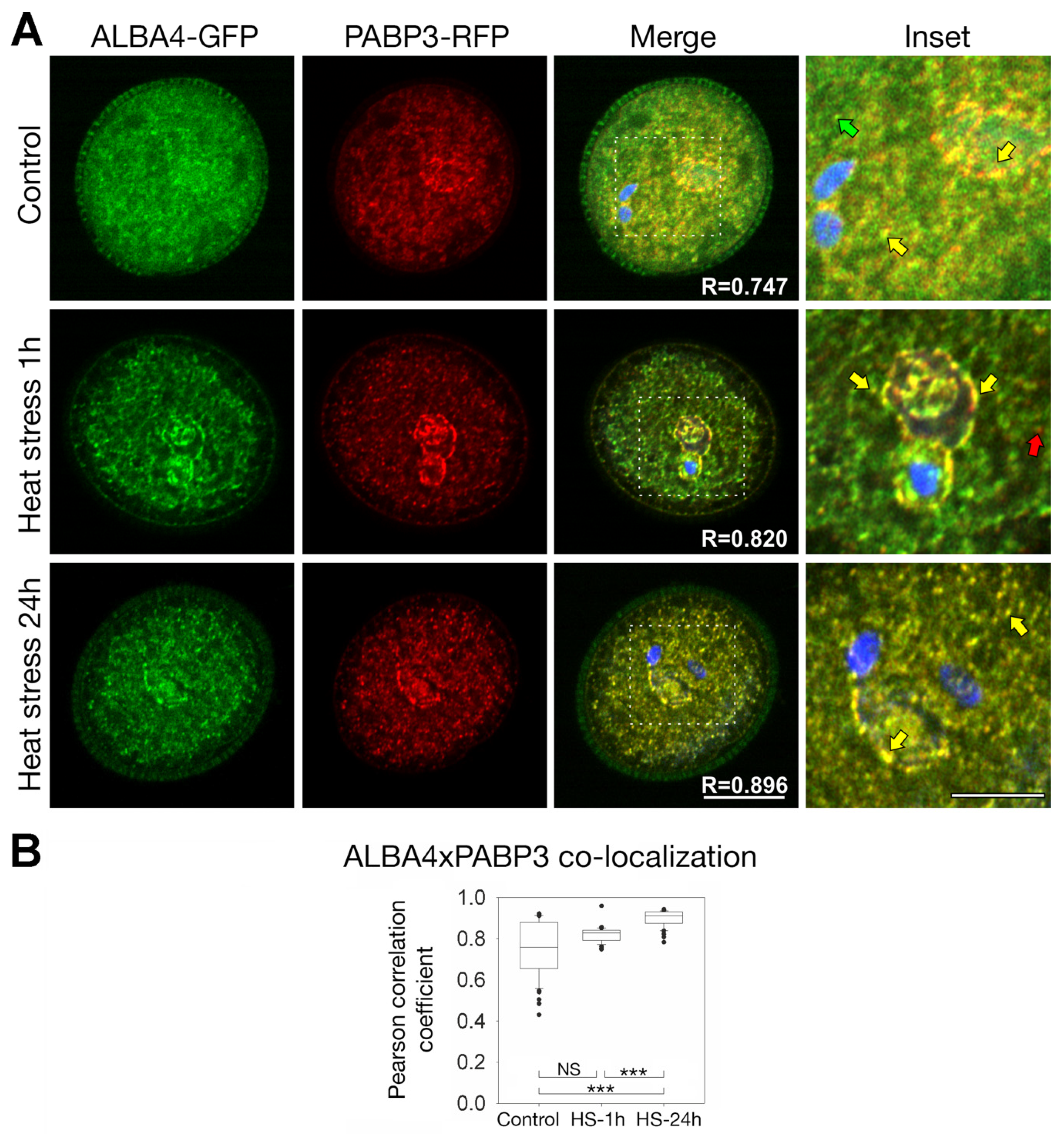
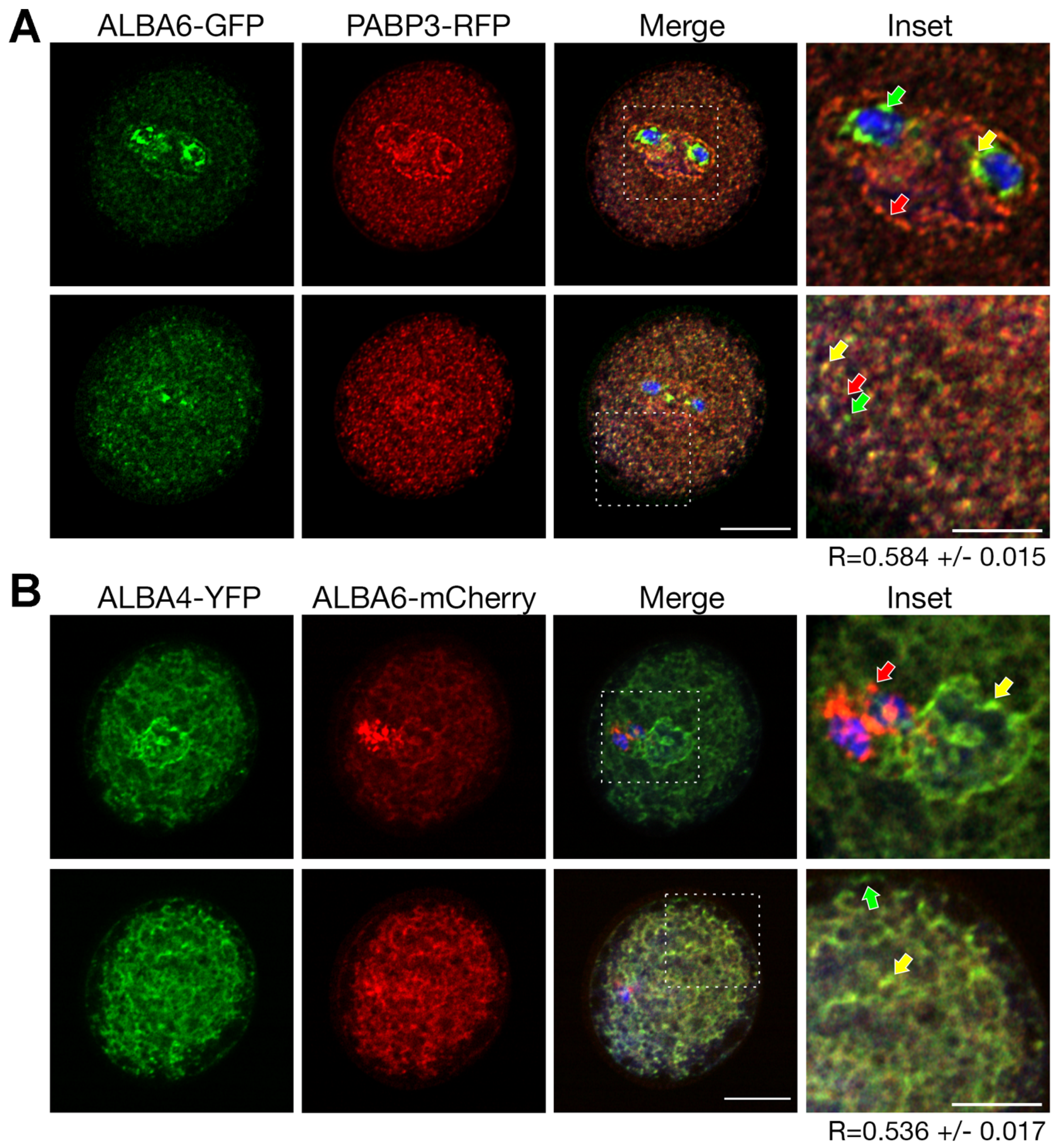
2.5. ALBA4 and ALBA6 Co-Localization with PABP3
3. Discussion
4. Materials and Methods
4.1. Sequence Motif, Phylogenetic, and Transcriptomic Analyses
4.2. Cloning and Plant Transformation
4.3. Plant cultivation and Treatment
4.4. RNA Isolation and RT-qPCR
4.5. GUS Activity
4.6. DAPI Staining
4.7. Light Microscopy
4.8. Microscopy Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALBA | Acetylation lowers binding affinity |
| CV | Coefficient of variation |
| HS | Heat stress |
| GFP | Green fluorescent protein |
| GUS | β-glucuronidase |
| MGU | Male germ unit |
| ML | Maximum likelihood |
| mRNP | Messenger ribonucleoprotein particle |
| PABP | Poly(A)-binding protein |
| PABP3 | Poly(A)-binding protein 3 |
| PCC | Pearson correlation coefficient |
| RFP | Red fluorescent protein |
| Rpp20 | Ribonuclease P protein subunit p20 |
| Rpp25 | Ribonuclease P protein subunit p25 |
| SC | Sperm cell |
| VC | Vegetative cell |
| YFP | Yellow fluorescent protein |
References
- Aravind, L.; Iyer, L.M.; Anantharaman, V. The two faces of Alba: The evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003, 4, R64. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.D.; Botting, C.H.; Wardleworth, B.N.; Jackson, S.P.; White, M.F. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 2002, 296, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Eder, P.S.; Gopalan, V.; Altman, S. Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA 2002, 8, 290–295. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Welting, T.J.; Peters, F.M.; Hensen, S.M.; van Doorn, N.L.; Kikkert, B.J.; Raats, J.M.; van Venrooij, W.J.; Pruijn, G.J. Heterodimerization regulates RNase MRP/RNase P association, localization, and expression of Rpp20 and Rpp25. RNA 2007, 13, 65–75. [Google Scholar] [CrossRef]
- Welting, T.J.; van Venrooij, W.J.; Pruijn, G.J. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004, 32, 2138–2146. [Google Scholar] [CrossRef]
- Dupé, A.; Dumas, C.; Papadopoulou, B. Differential Subcellular Localization of Leishmania Alba-Domain Proteins throughout the Parasite Development. PLoS ONE 2015, 10, e0137243. [Google Scholar] [CrossRef]
- Mair, G.R.; Lasonder, E.; Garver, L.S.; Franke-Fayard, B.M.; Carret, C.K.; Wiegant, J.C.; Dirks, R.W.; Dimopoulos, G.; Janse, C.J.; Waters, A.P. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010, 6, e1000767. [Google Scholar] [CrossRef]
- Mani, J.; Guttinger, A.; Schimanski, B.; Heller, M.; Acosta-Serrano, A.; Pescher, P.; Spath, G.; Roditi, I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS ONE 2011, 6, e22463. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Verma, J.K.; Wardhan, V.; Singh, D.; Chakraborty, S.; Chakraborty, N. Genome-Wide Identification of the Alba Gene Family in Plants and Stress-Responsive Expression of the Rice Alba Genes. Genes (Basel) 2018, 9, 183. [Google Scholar] [CrossRef]
- Verma, J.K.; Gayali, S.; Dass, S.; Kumar, A.; Parveen, S.; Chakraborty, S.; Chakraborty, N. OsAlba1, a dehydration-responsive nuclear protein of rice (Oryza sativa L. ssp. indica), participates in stress adaptation. Phytochemistry 2014, 100, 16–25. [Google Scholar] [CrossRef]
- Wang, N.; Jalajakumari, M.; Miller, T.; Asadi, M.; Millar, A.A. The ALBA RNA-binding proteins function redundantly to promote growth and flowering in Arabidopsis. BioRxiv 2019. [Google Scholar] [CrossRef]
- Borg, M.; Brownfield, L.; Khatab, H.; Sidorova, A.; Lingaya, M.; Twell, D. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 2011, 23, 534–549. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Kirungu, J.N.; Lu, P.; Cai, X.; Xu, Y.; Wang, X.; Zhou, Z.; Hou, Y.; Agong, S.G.; Wang, K.; et al. Knockdown of ghAlba_4 and ghAlba_5 Proteins in Cotton Inhibits Root Growth and Increases Sensitivity to Drought and Salt Stresses. Front. Plant Sci. 2019, 10, 1292. [Google Scholar] [CrossRef]
- Reichel, M.; Liao, Y.; Rettel, M.; Ragan, C.; Evers, M.; Alleaume, A.M.; Horos, R.; Hentze, M.W.; Preiss, T.; Millar, A.A. In Planta Determination of the mRNA-Binding Proteome of Arabidopsis Etiolated Seedlings. Plant Cell 2016, 28, 2435–2452. [Google Scholar] [CrossRef]
- Hedhly, A. Sensitivity of flowering plant gametophytes to temperature fluctuations. Environ. Exp. Bot. 2011, 74, 9–16. [Google Scholar] [CrossRef]
- Hafidh, S.; Fila, J.; Honys, D. Male gametophyte development and function in angiosperms: A general concept. Plant Reprod. 2016, 29, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Young, L.W.; Wilen, R.W.; Bonham-Smith, P.C. High temperature stress of Brassica napus during flowering reduces micro- and megagametophyte fertility, induces fruit abortion, and disrupts seed production. J. Exp. Bot. 2004, 55, 485–495. [Google Scholar] [CrossRef]
- Zinn, K.E.; Tunc-Ozdemir, M.; Harper, J.F. Temperature stress and plant sexual reproduction: Uncovering the weakest links. J. Exp. Bot. 2010, 61, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Benitez, M.; Corvera-Poire, A.; Chaos Cador, A.; de Folter, S.; Gamboa de Buen, A.; Garay-Arroyo, A.; Garcia-Ponce, B.; Jaimes-Miranda, F.; Perez-Ruiz, R.V.; et al. Flower development. Arab. Book 2010, 8, e0127. [Google Scholar] [CrossRef]
- Gray, N.K.; Hrabalkova, L.; Scanlon, J.P.; Smith, R.W. Poly(A)-binding proteins and mRNA localization: Who rules the roost? Biochem. Soc. Trans. 2015, 43, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Belostotsky, D.A. Unexpected Complexity of Poly(A)-Binding Protein Gene Families in Flowering Plants: Three Conserved Lineages That Are at Least 200 Million Years Old and Possible Auto- and Cross-Regulation. Genetics 2003, 163, 311–319. [Google Scholar] [PubMed]
- Belostotsky, D.A.; Meagher, R.B. Differential organ-specific expression of three poly(A)-binding protein genes from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1993, 90, 6686–6690. [Google Scholar] [CrossRef]
- Twell, D. Male gametogenesis and germline specification in flowering plants. Sex. Plant Reprod. 2011, 24, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Hafidh, S.; Potesil, D.; Muller, K.; Fila, J.; Michailidis, C.; Herrmannova, A.; Fecikova, J.; Ischebeck, T.; Valasek, L.S.; Zdrahal, Z.; et al. Dynamics of the Pollen Sequestrome Defined by Subcellular Coupled Omics. Plant Physiol. 2018, 178, 258–282. [Google Scholar] [CrossRef] [PubMed]
- Honys, D.; Twell, D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004, 5, R85. [Google Scholar] [CrossRef]
- Hamada, T.; Yako, M.; Minegishi, M.; Sato, M.; Kamei, Y.; Yanagawa, Y.; Toyooka, K.; Watanabe, Y.; Hara-Nishimura, I. Stress granule formation is induced by a threshold temperature rather than a temperature difference in Arabidopsis. J. Cell Sci. 2018, 131, jcs216051. [Google Scholar] [CrossRef] [PubMed]
- Honys, D.; Renak, D.; Fecikova, J.; Jedelsky, P.L.; Nebesarova, J.; Dobrev, P.; Capkova, V. Cytoskeleton-associated large RNP complexes in tobacco male gametophyte (EPPs) are associated with ribosomes and are involved in protein synthesis, processing, and localization. J. Proteome Res. 2009, 8, 2015–2031. [Google Scholar] [CrossRef]
- Kosmacz, M.; Gorka, M.; Schmidt, S.; Luzarowski, M.; Moreno, J.C.; Szlachetko, J.; Leniak, E.; Sokolowska, E.M.; Sofroni, K.; Schnittger, A.; et al. Protein and metabolite composition of Arabidopsis stress granules. New Phytol. 2019, 222, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Scarpin, M.R.; Sigaut, L.; Temprana, S.G.; Boccaccio, G.L.; Pietrasanta, L.I.; Muschietti, J.P. Two Arabidopsis late pollen transcripts are detected in cytoplasmic granules. Plant Direct 2017, 1, e00012. [Google Scholar] [CrossRef]
- Weber, C.; Nover, L.; Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008, 56, 517–530. [Google Scholar] [CrossRef]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef]
- Chekanova, J.A.; Belostotsky, D.A. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA 2003, 9, 1476–1490. [Google Scholar] [CrossRef]
- Subota, I.; Rotureau, B.; Blisnick, T.; Ngwabyt, S.; Durand-Dubief, M.; Engstler, M.; Bastin, P. ALBA proteins are stage regulated during trypanosome development in the tsetse fly and participate in differentiation. Mol. Biol. Cell 2011, 22, 4205–4219. [Google Scholar] [CrossRef]
- Gissot, M.; Walker, R.; Delhaye, S.; Alayi, T.D.; Huot, L.; Hot, D.; Callebaut, I.; Schaeffer-Reiss, C.; Dorsselaer, A.V.; Tomavo, S. Toxoplasma gondii Alba proteins are involved in translational control of gene expression. J. Mol. Biol. 2013, 425, 1287–1301. [Google Scholar] [CrossRef]
- Keller, M.; Simm, S. The coupling of transcriptome and proteome adaptation during development and heat stress response of tomato pollen. BMC Genom. 2018, 19, 447. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Neron, B.; Menager, H.; Maufrais, C.; Joly, N.; Maupetit, J.; Letort, S.; Carrere, S.; Tuffery, P.; Letondal, C. Mobyle: A new full web bioinformatics framework. Bioinformatics 2009, 25, 3005–3011. [Google Scholar] [CrossRef] [PubMed]
- Soding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Julca, I.; Ferrari, C.; Flores-Tornero, M.; Proost, S.; Lindner, A.-C.; Hackenberg, D.; Steinbachová, L.; Michaelidis, C.; Pereira, S.G.; Misra, C.S.; et al. Comparative transcriptomic analysis reveals conserved transcriptional programs underpinning organogenesis and reproduction in land plants. BioRxiv 2020. [Google Scholar] [CrossRef]
- Liang, X.; Peng, L.; Baek, C.H.; Katzen, F. Single step BP/LR combined Gateway reactions. Biotechniques 2013, 55, 265–268. [Google Scholar] [CrossRef]
- Shimada, T.L.; Shimada, T.; Hara-Nishimura, I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 2010, 61, 519–528. [Google Scholar] [CrossRef]
- Sarrion-Perdigones, A.; Vazquez-Vilar, M.; Palaci, J.; Castelijns, B.; Forment, J.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. GoldenBraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013, 162, 1618–1631. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Vilar, M.; Quijano-Rubio, A.; Fernandez-Del-Carmen, A.; Sarrion-Perdigones, A.; Ochoa-Fernandez, R.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. GB3.0: A platform for plant bio-design that connects functional DNA elements with associated biological data. Nucleic Acids Res. 2017, 45, 2196–2209. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.scirp.org/(S(lz5mqp453edsnp55rrgjct55))/reference/ReferencesPapers.aspx?ReferenceID=2342186 (accessed on 4 February 2021).
- Park, S.K.; Howden, R.; Twell, D. The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 1998, 125, 3789–3799. [Google Scholar] [PubMed]
- Retzer, K.; Lacek, J.; Skokan, R.; Del Genio, C.I.; Vosolsobe, S.; Lankova, M.; Malinska, K.; Konstantinova, N.; Zazimalova, E.; Napier, R.M.; et al. Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution. Int. J. Mol. Sci. 2017, 18, 1–20. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Náprstková, A.; Malínská, K.; Záveská Drábková, L.; Billey, E.; Náprstková, D.; Sýkorová, E.; Bousquet-Antonelli, C.; Honys, D. Characterization of ALBA Family Expression and Localization in Arabidopsis thaliana Generative Organs. Int. J. Mol. Sci. 2021, 22, 1652. https://doi.org/10.3390/ijms22041652
Náprstková A, Malínská K, Záveská Drábková L, Billey E, Náprstková D, Sýkorová E, Bousquet-Antonelli C, Honys D. Characterization of ALBA Family Expression and Localization in Arabidopsis thaliana Generative Organs. International Journal of Molecular Sciences. 2021; 22(4):1652. https://doi.org/10.3390/ijms22041652
Chicago/Turabian StyleNáprstková, Alena, Kateřina Malínská, Lenka Záveská Drábková, Elodie Billey, Dagmar Náprstková, Eva Sýkorová, Cécile Bousquet-Antonelli, and David Honys. 2021. "Characterization of ALBA Family Expression and Localization in Arabidopsis thaliana Generative Organs" International Journal of Molecular Sciences 22, no. 4: 1652. https://doi.org/10.3390/ijms22041652
APA StyleNáprstková, A., Malínská, K., Záveská Drábková, L., Billey, E., Náprstková, D., Sýkorová, E., Bousquet-Antonelli, C., & Honys, D. (2021). Characterization of ALBA Family Expression and Localization in Arabidopsis thaliana Generative Organs. International Journal of Molecular Sciences, 22(4), 1652. https://doi.org/10.3390/ijms22041652







