Characterization of the FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 Homolog SlFKF1 in Tomato as a Model for Plants with Fleshy Fruit
Abstract
:1. Introduction
2. Results
2.1. SlFKF cDNA Sequence
2.2. Expression Analysis of SlFKF1
2.3. Effect of Light Quality on Lycopene Concentration
2.4. Analysis of SlFKF1 Promoter Sequences
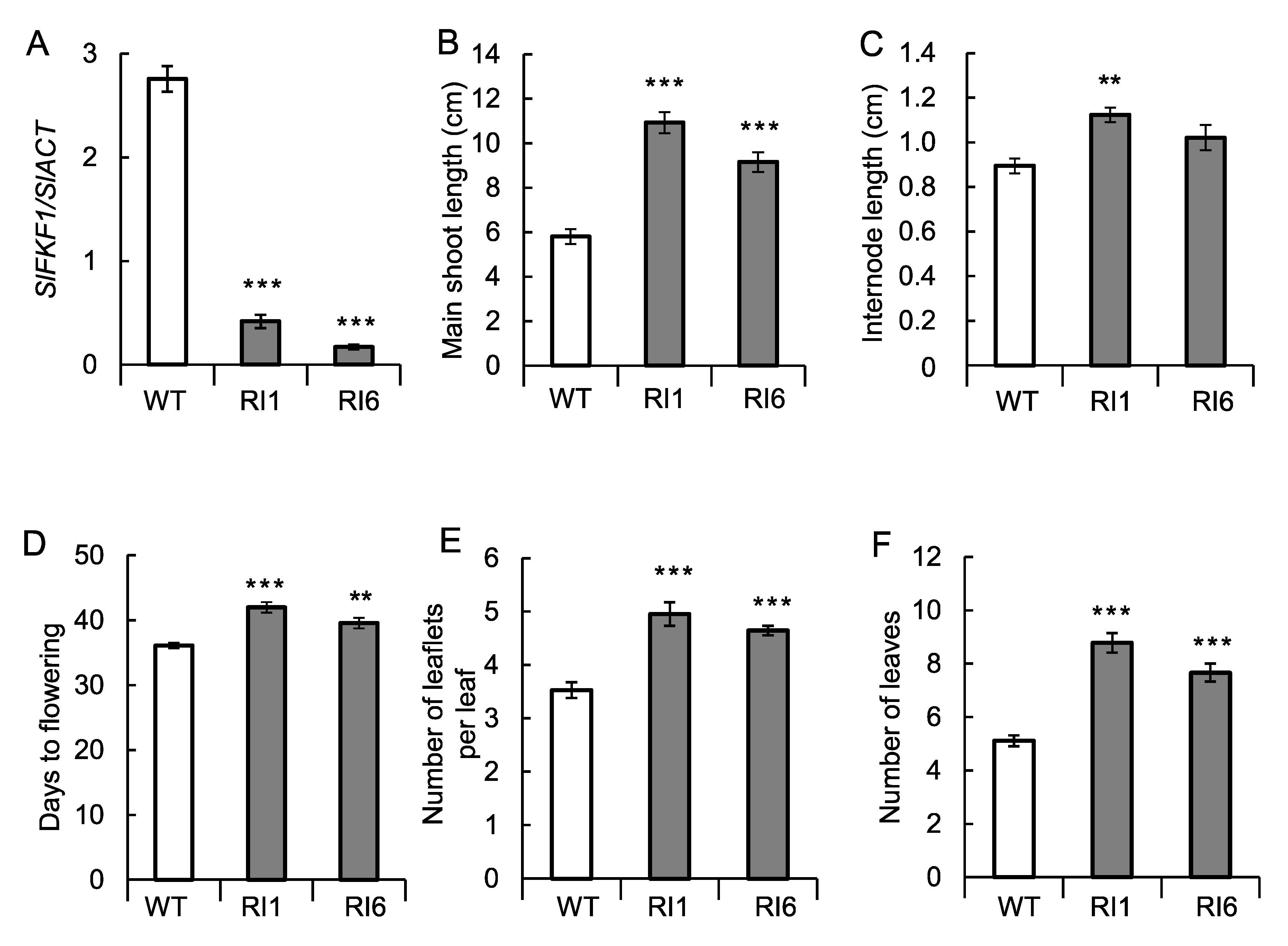
2.5. Transformation Experiments
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Sequence Analysis of Tomato FKF1 Homolog SlFKF1 cDNA
4.3. Expression Analysis
4.4. Alignment, Phylogenetic Analysis, and Promoter Analysis
4.5. Determination of Lycopene Concentration
4.6. Transformation Experiment
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shibuya, T.; Kanayama, Y. Flowering response to blue light and its molecular mechanisms in Arabidopsis and horticultural plants. Adv. Hort. Sci. 2014, 28, 179–183. [Google Scholar]
- Ahmad, M.; Cashmore, A.R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Yang, H.; Mockler, T.C.; Lin, C. Regulation of flowering time by Arabidopsis photoreceptors. Science 1998, 279, 1360–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Assal, S.; Alonso-Blanco, C.; Peeters, A.; Raz, V.; Koornneef, M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 2001, 29, 435–440. [Google Scholar] [CrossRef]
- Kagawa, T.; Wada, M. Blue light-induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol. 2000, 41, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Sakai, T.; Kagawa, T.; Kasahara, M.; Swartz, T.E.; Christie, J.M.; Briggs, W.R.; Wada, M. Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6969–6974. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, T.; Doi, M.; Suetsugu, N.; Kagawa, T.; Wada, M.; Shimazaki, K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 2001, 414, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Wada, M.; Kagawa, T.; Sato, Y. Chloroplast movement. Annu. Rev. Plant Biol. 2003, 54, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Ishii, Y.; Ezura, H.; Olsen, J.E. Effects of light quality under red and blue light emitting diodes on growth and expression of FBP28 in petunia. Acta Hort. 2011, 907, 361–366. [Google Scholar] [CrossRef]
- Shibuya, T.; Takahashi, T.; Hashimoto, S.; Nishiyama, M.; Kanayama, Y. Effects of overnight radiation with monochromatic far-red and blue light on flower budding and expression of flowering-related and light quality-responsive genes in Eustoma grandiflorum. J. Agric. Meteorol. 2019, 75, 160–165. [Google Scholar] [CrossRef] [Green Version]
- Hwang, D.Y.; Park, S.; Lee, S.; Lee, S.S.; Imaizumi, T.; Song, Y.H. GIGANTEA regulates the timing stabilization of CONSTANS by altering the interaction between FKF1 and ZEITLUPE. Mol. Cells 2019, 42, 693–701. [Google Scholar]
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imaizumi, T.; Tran, H.G.; Swartz, T.E.; Briggs, W.R.; Kay, S.A. FKF1 is essential for photoperiodic-specific light signaling in Arabidopsis. Nature 2003, 426, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnu, J. Molecular mechanisms suppressing COP1/SPA E3 ubiquitin ligase activity in blue light. Physiol. Plant. 2020, 169, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-D.; Cha, J.-Y.; Kim, M.R.; Shin, G.-I.; Paek, N.-C.; Kim, W.-Y. Light-dependent suppression of COP1 multimeric complex formation is determined by the blue-light receptor FKF1 in Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 508, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-D.; Kim, M.R.; Kang, M.-Y.; Cha, J.-Y.; Han, S.-H.; Nawkar, G.M.; Sakuraba, Y.; Lee, S.Y.; Imaizumi, T.; McClung, C.R.; et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat. Commun. 2017, 8, 2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibuya, T.; Murakawa, Y.; Nishidate, K.; Nishiyama, M.; Kanayama, Y. Characterization of flowering-related genes and flowering response in relation to blue light in Gypsophila paniculata. Hort. J. 2017, 86, 94–104. [Google Scholar] [CrossRef] [Green Version]
- Hori, Y.; Nishidate, K.; Nishiyama, M.; Kanahama, K.; Kanayama, Y. Flowering and expression of flowering-related genes under long-day conditions with light-emitting diodes. Planta 2011, 234, 321–330. [Google Scholar] [CrossRef]
- Han, S.-H.; Yoo, S.-C.; Lee, B.-D.; An, G.; Paek, N.-C. Rice FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (OsFKF1) promotes flowering independent of photoperiod. Plant Cell Environ. 2015, 38, 2527–2540. [Google Scholar] [CrossRef] [PubMed]
- Kubota, A.; Kita, S.; Ishizaki, K.; Nishihama, R.; Yamato, K.T.; Kohchi, T. Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat. Commun. 2014, 5, 3668. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-M.; Feke, A.; Li, M.-W.; Adamchek, C.; Webb, K.; Pruneda-Paz, J.; Bennett, E.J.; Kay, S.A.; Gendron, J.M. Decoys untangle complicated redundancy and reveal targets of circadian clock F-BOX proteins. Plant Physiol. 2018, 177, 1170–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, N.; Balasubramanian, V.K.; Chopra, R.; Mendu, V. The photoperiodic flowering time regulator fkf1 negatively regulates cellulose biosynthesis. Plant Physiol. 2019, 180, 2240–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, D.C.; Lasswell, J.; Rogg, L.; Cohen, M.A.; Bartel, B. FKF1, a Clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 2000, 101, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Obulareddy, N.; Panchal, S.; Melotto, M. Guard cell purification and RNA isolation suitable for high-throughput transcriptional analysis of cell-type responses to biotic stresses. Mol. Plant Microbe Interact. 2013, 26, 844–849. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Zhang, X.; Hu, R.; Wu, F.; Ma, J.; Meng, Y.; Fu, Y. Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS ONE 2013, 8, e79036. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.; Mo, Y.; Chen, J.; Li, C.; Zhao, L.; Liu, Y. Expression and interaction proteins analysis of BjuFKF1 in stem mustard. Sci. Hortic. 2020, 269, 109430. [Google Scholar] [CrossRef]
- Huang, F.; Fu, Z.; Zeng, L.; Morley-Bunker, M. Isolation and characterization of GI and FKF1 homologous genes in the subtropical fruit tree. Dimocarpus longan. Mol. Breed. 2017, 37, 90. [Google Scholar] [CrossRef]
- He, Y.; Li, R.; Lin, F.; Xiong, Y.; Wang, L.; Wang, B.; Guo, J.; Hu, C. Transcriptome changes induced by different potassium levels in banana roots. Plants 2020, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Mason, M.; Risseeuw, E.P.; Crosby, W.L.; Somers, D.E. Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J. 2004, 40, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, P.; Mizzotti, C.; Colombo, M.; Masiero, S. Genetic regulation and structural changes during tomato fruit development and ripening. Front. Plant Sci. 2014, 5, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lifschitz, E.; Eviatar, T.; Rozman, A.; Shalit, A.; Goldshmidt, A.; Amsellem, Z.; Alvarez, J.; Eshed, Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 2006, 103, 6398–6403. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; How-Kit, A.; Stammittia, L.; Teyssier, E.; Rolin, D.; Mortain-Bertrand, A.; Halle, S.; Liu, M.; Kong, J.; Wu, C.; et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 2015, 112, 10804–10809. [Google Scholar] [CrossRef] [Green Version]
- Más, P.; Kim, W.-Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 02163. [Google Scholar] [CrossRef]
- Kiba, T.; Henriques, R.; Sakakibara, H.; Chua, N.-H. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 2007, 19, 2516–2530. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, S.; Wang, L.; Han, L.; Suh, S.-S.; Salomé, P.A.; McClung, R.C.; Somers, D.E. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008, 283, 23073–23083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, S.B.; Shen, Y.; Chang, H.-C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef]
- Gnesutta, N.; Kumimoto, R.W.; Swain, S.; Chiara, M.; Siriwardana, C.S.; Horner, D.; Ben, F.; Holt, B.F.; Mantovania, R. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell 2017, 29, 1516–1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef]
- Shalit, A.; Rozman, A.; Goldshmidt, A.; Alvarez, J.P.; Bowman, J.; Eshed, Y.; Lifschitz, E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 2009, 106, 8392–8397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinero-Rosales, N.; Latorre, A.; Jamilena, M.; Lozano, R. SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 2004, 218, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Li, X.; Zeng, B.; Zhong, M.; Yang, J.; Yang, P.; Li, X.; He, C.; Lin, J.; Liu, X.; et al. FKF1 F-box protein promotes flowering in part by negatively regulating DELLA protein stability under long-day photoperiod in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1717–1740. [Google Scholar] [CrossRef]
- Silva, G.F.F.; Silva, E.M.; Correa, J.P.O.; Vicente, M.H.; Jiang, N.; Notini, M.M.; Junior, A.C.; De Jesus, F.A.; Castilho, P.; Carrera, E.; et al. Tomato floral induction and flower development are orchestrated by the interplay between gibberellin and two unrelated microRNA-controlled modules. New Phytol. 2019, 221, 1328–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, B.-X.; Wei, J.-J.; Zhang, Y.-T.; Song, S.-W.; Su, W.; Sun, G.-W.; Hao, Y.-W.; Liu, H.-C. Supplemental blue and red light promote lycopene synthesis in tomato fruits. J. Integr. Agric. 2019, 18, 590–598. [Google Scholar] [CrossRef] [Green Version]
- Dhakal, R.; Baek, K.-H. Short period irradiation of single blue wavelength light extends the storage period of mature green tomatoes. Postharvest Biol. Technol. 2014, 90, 73–77. [Google Scholar] [CrossRef]
- Lin, C.; Yang, H.; Guo, H.; Mockler, T.; Chen, J.; Cashmore, A.R. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 1998, 95, 2686–2690. [Google Scholar] [CrossRef] [Green Version]
- Novák, A.; Boldizsár, Á.; Ádám, É.; Kozma-Bognár, L.; Majláth, I.; Båga, M.; Tóth, B.; Chibbar, R.; Galiba, G. Light-quality and temperature-dependent CBF14 gene expression modulates freezing tolerance in cereals. J. Exp. Bot. 2016, 67, 1285–1295. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.-J.; Kim, P.; Chen, Y.; Chen, B.; Yang, J.; Liu, X.; Kawabata, S.; Wang, Y.; Li, Y. Blue and UV-B light synergistically induce anthocyanin accumulation by co-activating nitrate reductase gene expression in Anthocyanin fruit (Aft) tomato. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Giliberto, L.; Perrotta, G.; Pallara, P.; Weller, J.; Fraser, P.D.; Bramley, P.M.; Fiore, A.; Tavazza, M.; Giuliano, G. Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 2005, 137, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-C.; Ahammed, G.J.; Wang, G.-T.; Xu, C.-J.; Chen, K.-S.; Zhou, Y.-H.; Yu, J.-Q. Tomato CRY1a plays a critical role in the regulation of phytohormone homeostasis, plant development, and carotenoid metabolism in fruits. Plant Cell Environ. 2018, 41, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Nanya, K.; Ishigami, Y.; Hikosaka, S.; Goto, E. Effects of blue and red light on stem elongation and flowering of tomato seedlings. Acta Hort. 2012, 956, 261–266. [Google Scholar] [CrossRef]
- Kim, H.M.; Hwang, S.J. The growth and development of ‘Mini Chal’ tomato plug seedlings grown under monochromatic or combined red and blue light-emitting diodes. Hort. Sci. Technol. 2019, 37, 190–205. [Google Scholar]
- Yoshida, H.; Mizuta, D.; Fukuda, N.; Hikosaka, S.; Goto, E. Effects of varying light quality from single-peak blue and red light-emitting diodes during nursery period on flowering, photosynthesis, growth, and fruit yield of everbearing strawberry. Plant Biotechnol. 2016, 33, 267–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanayama, Y. Sugar metabolism and fruit development in tomato. Hort. J. 2017, 86, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Mawphlang, O.I.L.; Kharshiing, E.V. Photoreceptor mediated plant growth responses: Implications for photoreceptor engineering toward improved performance in crops. Front. Plant Sci. 2017, 8, 1181. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, S.; Miyamoto, K.; Li, Y. cDNA microarray analysis of differential gene expression in tomato fruits exposed to blue, UV-A, and UV-A+UV-B. Acta Hort. 2011, 907, 371–374. [Google Scholar] [CrossRef]
- Tsunoda, Y.; Hano, S.; Imoto, N.; Shibuya, T.; Ikeda, H.; Amagaya, K.; Kato, K.; Shirakawa, H.; Aso, H.; Kanayama, Y. Physiological roles of tryptophan decarboxylase revealed by overexpression of SlTDC1 in tomato. Sci. Hortic. 2021, 275, 109672. [Google Scholar] [CrossRef]
- Ikeda, H.; Shibuya, T.; Imanishi, S.; Aso, H.; Nishiyama, M.; Kanayama, Y. Dynamic metabolic regulation by a chromosome segment from a wild relative during fruit development in a tomato introgression line, IL8-3. Plant Cell Physiol. 2016, 57, 1257–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammed, S.A.; Nishio, S.; Takahashi, H.; Shiratake, K.; Ikeda, H.; Kanahama, K.; Kanayama, Y. Role of vacuolar H+-inorganic pyrophosphatase in tomato fruit development. J. Exp. Bot. 2012, 63, 5613–5621. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Shiba, H.; Ohta, M.; Kang, S.; Sato, A.; Yuasa, T.; Iwava-Inoue, M.; Kamada, H.; Ezura, H. SlICE1 encoding a MYC-type transcription factor controls cold tolerance in tomato, Solanum lycopersicum. Plant Biotechnol. 2012, 29, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Ito, H.; Horie, H. Proper solvent selection for lycopene extraction in tomatoes and application to a rapid determination. Bull. Natl. Inst. Veg. Tea Sci. 2009, 8, 165–173. [Google Scholar]




| Gene | Locus 1 | Putative CONSTANS Responsive Element 2 | Strand | Position of 1st C from ATG |
|---|---|---|---|---|
| SFT | Solyc03g063100 | TTTCCACAAAA | Top | −379 |
| PSY1 | Solyc03g031860 | TTCCCACACTG | Bottom | −554 |
| Solyc03g031860 | AAATGTGGTGT | Bottom | −269 | |
| Solyc03g031860 | GTCTGTGGTCT | Bottom | −186 | |
| PSY2 | Solyc02g081330 | TTGTGTGGTCA | Bottom | −274 |
| PSY3 | Solyc01g005940 | not found | ||
| RIN | Solyc05g012020 | CTACCACAAGG | Top | −1049 |
| Solyc05g012020 | ATGTGTGGCTA | Bottom | −701 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shibuya, T.; Nishiyama, M.; Kato, K.; Kanayama, Y. Characterization of the FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 Homolog SlFKF1 in Tomato as a Model for Plants with Fleshy Fruit. Int. J. Mol. Sci. 2021, 22, 1735. https://doi.org/10.3390/ijms22041735
Shibuya T, Nishiyama M, Kato K, Kanayama Y. Characterization of the FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 Homolog SlFKF1 in Tomato as a Model for Plants with Fleshy Fruit. International Journal of Molecular Sciences. 2021; 22(4):1735. https://doi.org/10.3390/ijms22041735
Chicago/Turabian StyleShibuya, Tomoki, Manabu Nishiyama, Kazuhisa Kato, and Yoshinori Kanayama. 2021. "Characterization of the FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 Homolog SlFKF1 in Tomato as a Model for Plants with Fleshy Fruit" International Journal of Molecular Sciences 22, no. 4: 1735. https://doi.org/10.3390/ijms22041735
APA StyleShibuya, T., Nishiyama, M., Kato, K., & Kanayama, Y. (2021). Characterization of the FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 Homolog SlFKF1 in Tomato as a Model for Plants with Fleshy Fruit. International Journal of Molecular Sciences, 22(4), 1735. https://doi.org/10.3390/ijms22041735






