Complexation with Random Methyl-β-Cyclodextrin and (2-Hidroxypropyl)-β-Cyclodextrin Enhances In Vivo Anti-Fibrotic and Anti-Inflammatory Effects of Chrysin via the Inhibition of NF-κB and TGF-β1/Smad Signaling Pathways and Modulation of Hepatic Pro/Anti-Fibrotic miRNA
Abstract
:1. Introduction
2. Results
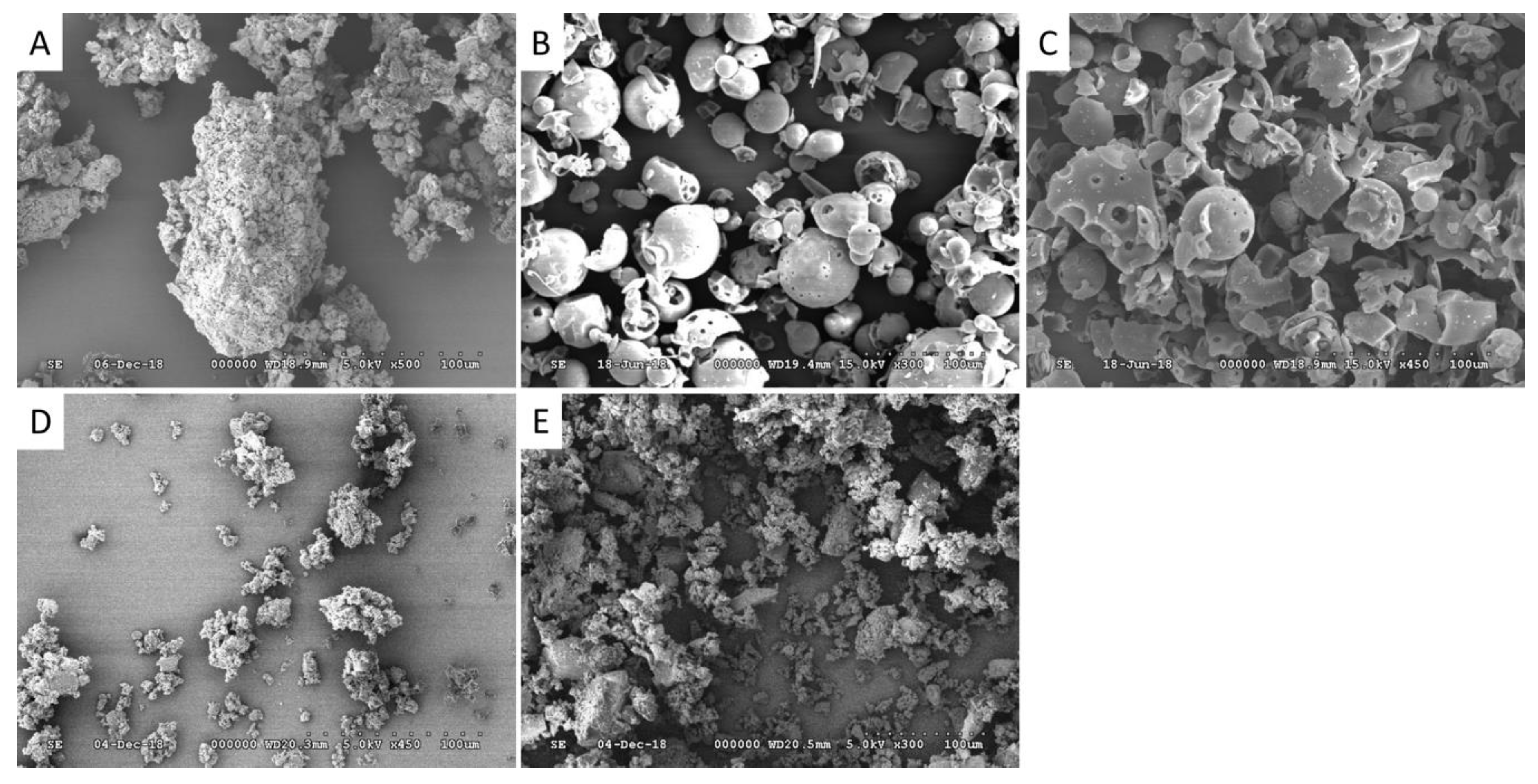
2.1. Characterization of CHR-HPBCD and CHR-RAMEB Nanocomplexes
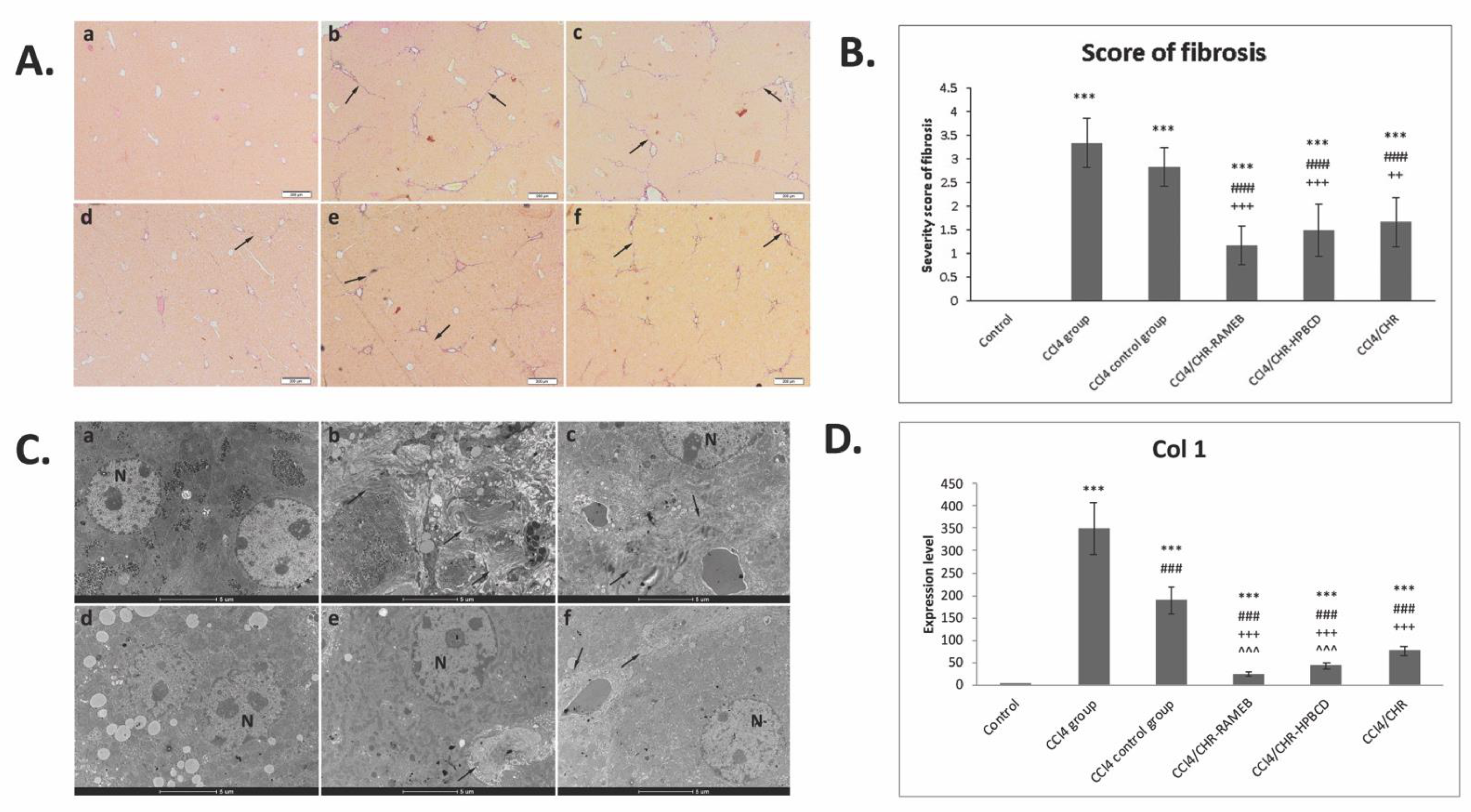
2.2. CHR–HPBCD and CHR–RAMEB Nanocomplexes Alleviate CCl4-Induced Liver Fibrosis Collagen Deposition, and Ultrastructural Changes
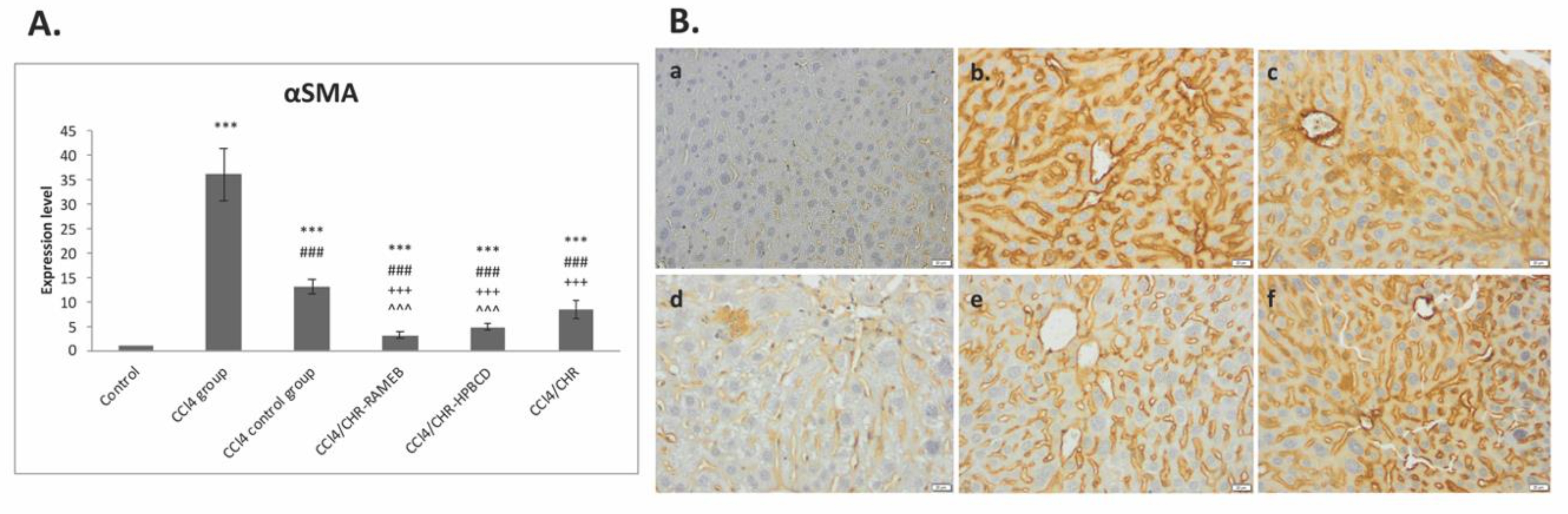
2.3. CHR–HPBCD and CHR–RAMEB Nanocomplexes Inhibit the Activation of Hepatic Stellate Cells
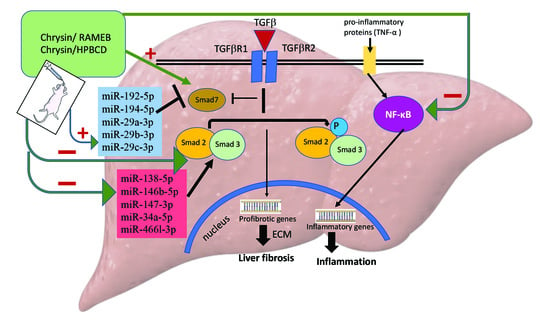
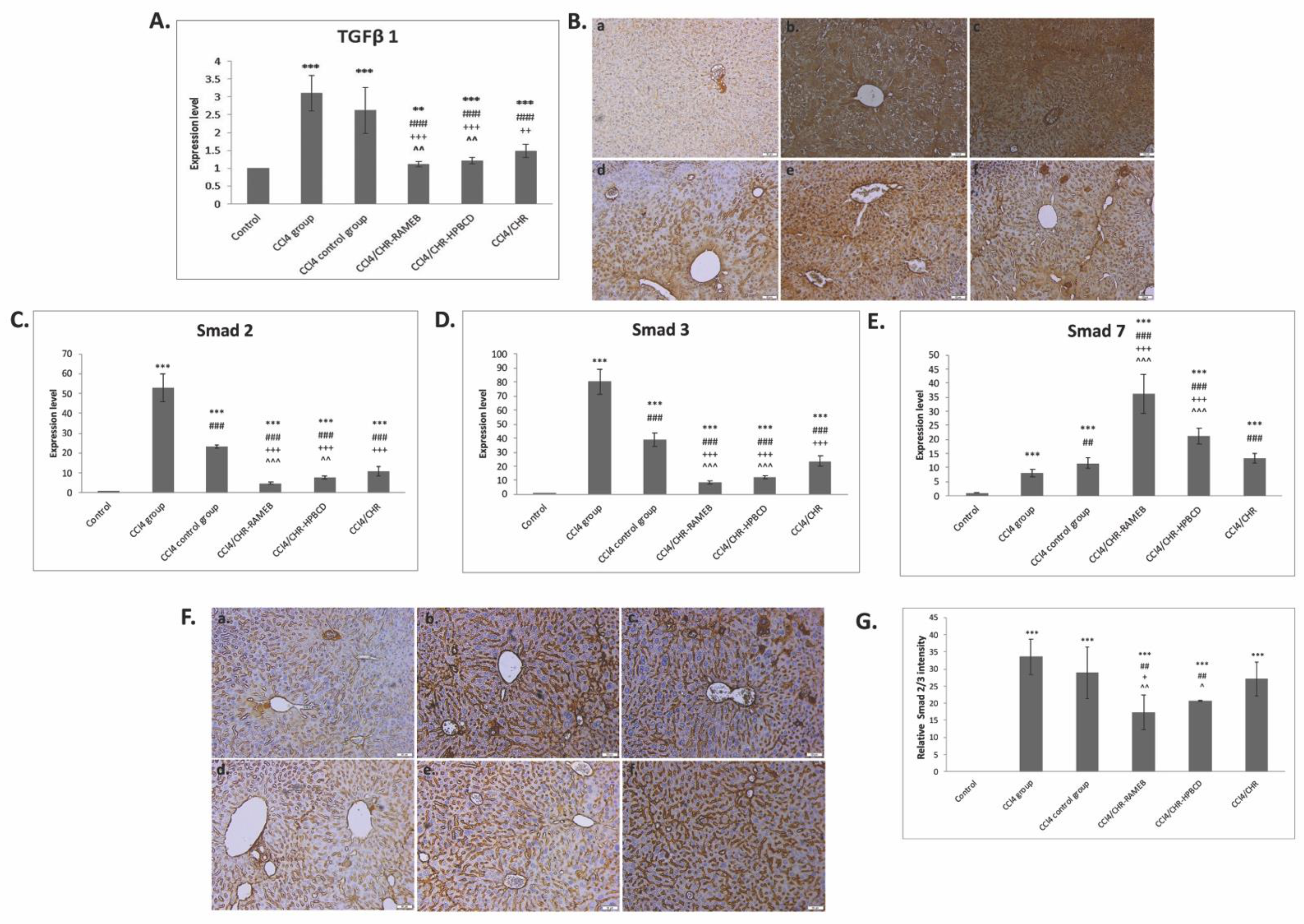
2.4. CHR–HPBCD and CHR–RAMEB Complexes Downregulate the TGF-β1/Smad Signaling Pathway
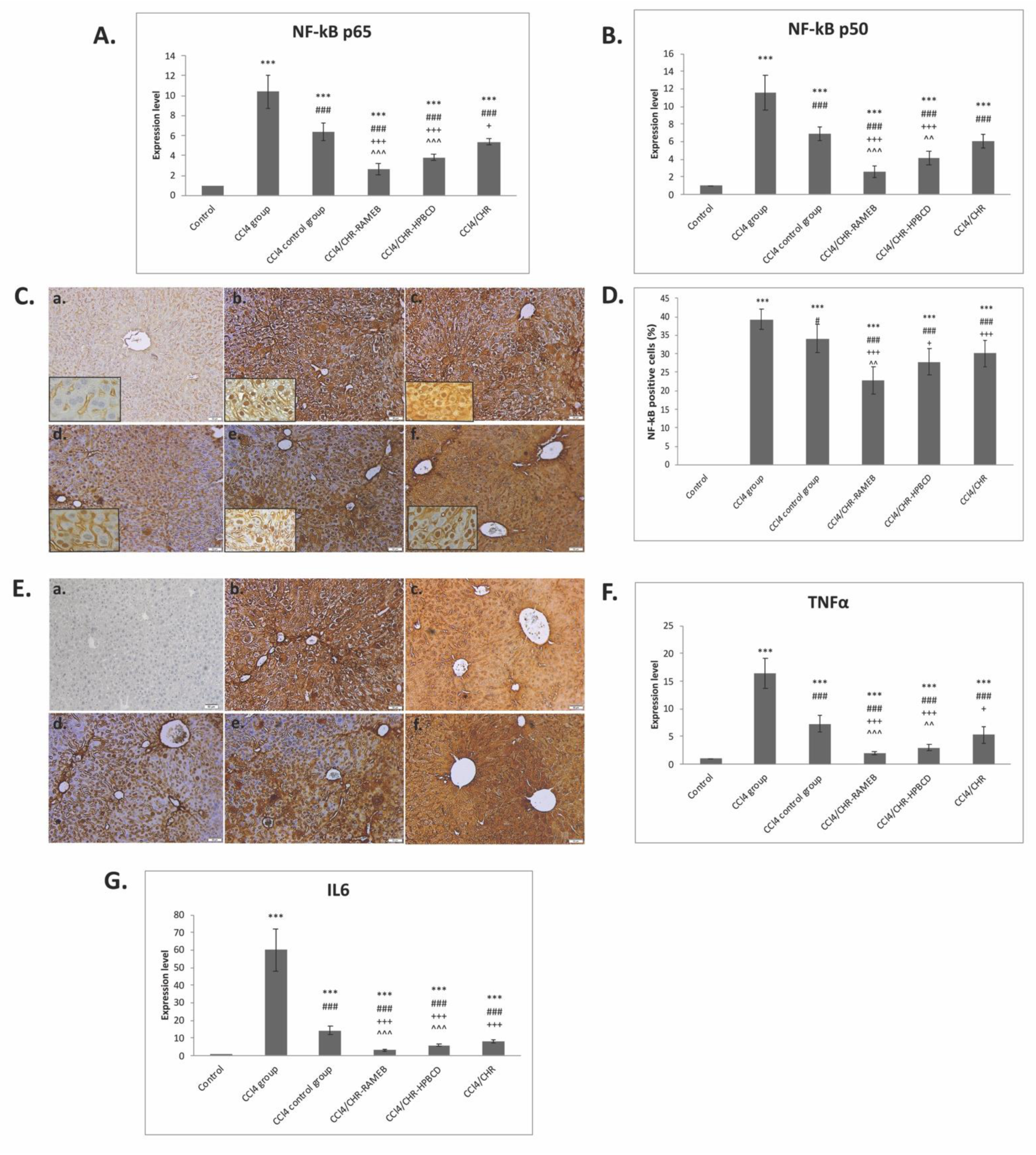
2.5. CHR–HPBCD and CHR–RAMEB Nanocomplexes Downregulate the NF-kB-Mediated Inflammatory Pathway
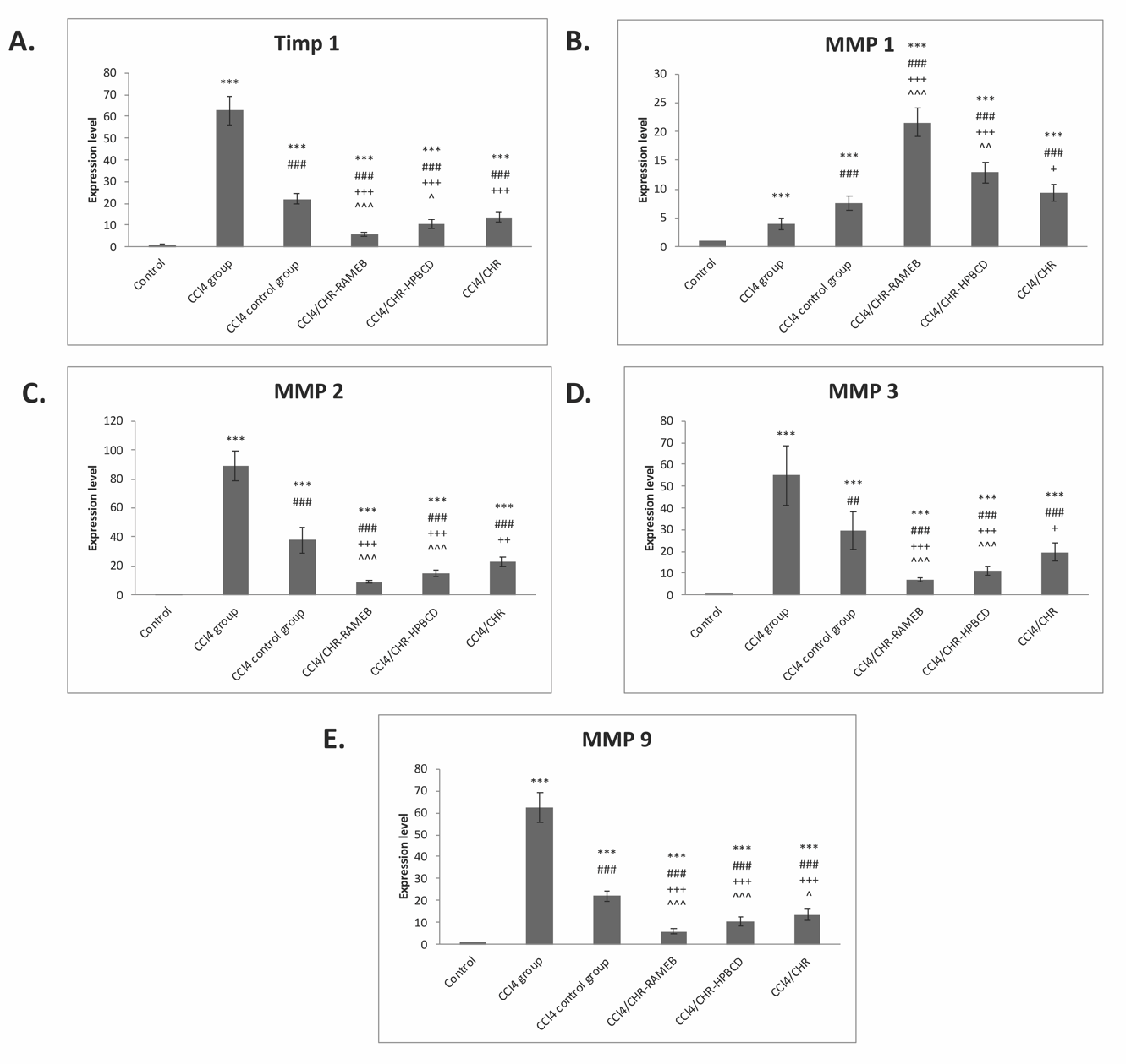
2.6. CHR–HPBCD and CHR–RAMEB Nanocomplexes Modulate ECM by TIMP-1/MMPs Balance
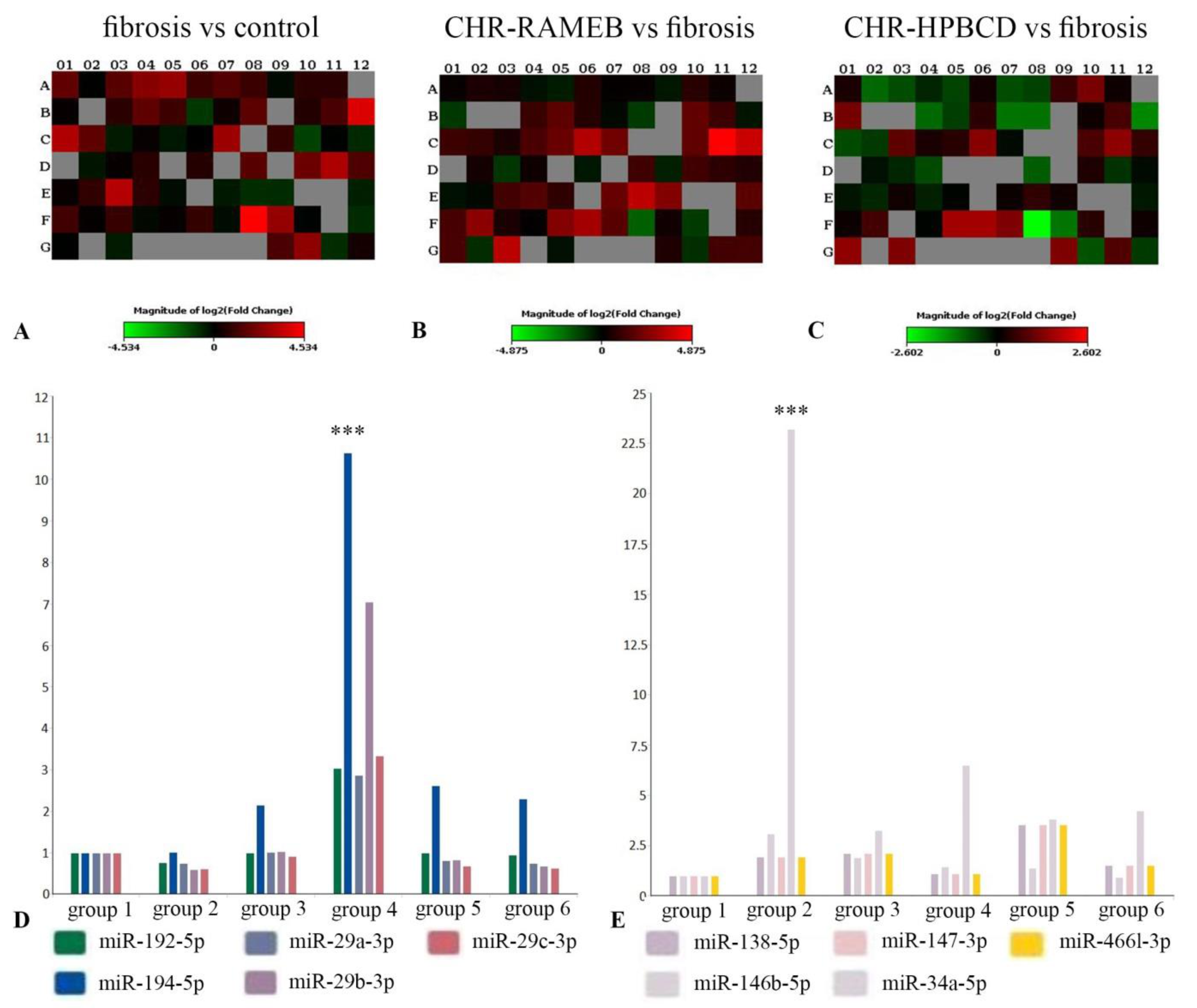
2.7. CHR–HPBCD and CHR–RAMEB Nanocomplexes Modulate Profibrotic and Antifibrotic miRNA Expression
3. Discussion
4. Materials and Methods
4.1. Preparation of Chrysin-Loaded HPBCD/RAMEB Nanocomplexes
4.2. Scanning Electron Microscopy
4.3. In Vivo Experimental Design
- -
- Group 1 (control group) orally received a saline solution for 7 weeks and 0.7% carboxymethyl cellulose (CMC) for the next 2 weeks;
- -
- Group 2 (CCl4 group) i.p. injected with CCl4 solution (20% v/v, 2 mL/kg b.w.) 2 times a week for 7 weeks and euthanatized for liver fibrosis confirmation;
- -
- Group 3 (CCl4 control group) received CCl4 chemical induction of liver fibrosis for 7 weeks, followed by 2 weeks of de novo self-recovery (spontaneous fibrosis resolution);
- -
- Group 4 (CCl4/CHR–RAMEB group) received CCl4 chemical induction of liver fibrosis for 7 weeks, followed by oral administration of 50 mg/kg CHR–RAMEB for 2 weeks;
- -
- Group 5 (CCl4/CHR–HPBCD group) received CCl4 chemical induction of liver fibrosis for 7 weeks, followed by oral administration of 50 mg/kg CHR–HPBCD for 2 weeks;
- -
- Group 6 (CCl4/CHR group) received CCl4 chemical induction of liver fibrosis for 7 weeks, followed by oral administration of 50 mg/kg free CHR for 2 weeks.
4.4. Histology
4.5. Immunohistochemistry
4.6. Transmission Electron Microscopy
4.7. Quantitative Real-Time PCR Analysis
4.8. MiRNA PCR Array Analysis
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lackner, C.; Tiniakos, D. Fibrosis and alcohol-related liver disease. J. Hepatol. 2019, 70, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Reyes, G.; Gutierrez-Ruiz, M.C.; Kershenobich, D. Liver fibrosis and chronic viral hepatitis. Arch. Med. Res. 2007, 38, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Penz-Österreicher, M.; Österreicher, C.H.; Trauner, M. Fibrosis in autoimmune and cholestatic liver disease. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 245–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majo, J.; Klinkhammer, B.M.; Boor, P.; Tiniakos, D. Pathology and natural history of organ fibrosis. Curr. Opin. Pharmacol. 2019, 49, 82–89. [Google Scholar] [CrossRef]
- Crawford, J.M.; Bioulac-Sage, P.; Hytiroglou, P. Structure, Function, and Responses to Injury. In MacSween’s Pathology of the Liver; Burt, A.D., Ferrell, L.D., Hubscher, S.G., Eds.; Elsevier: Philadelphia, PA, USA, 2017; pp. 1–68. [Google Scholar]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Friedman, S.L. Hepatic fibrosis—Overview. Toxicology 2008, 254, 120–129. [Google Scholar] [CrossRef]
- Dhar, D.; Baglieri, J.; Kisseleva, T.; Brenner, D.A. Mechanisms of liver fibrosis and its role in liver cancer. Exp. Biol. Med. 2020, 245, 96–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novo, E.; Cannito, S.; Paternostro, C.; Bocca, C.; Miglietta, A.; Parola, M. Cellular and molecular mechanisms in liver fibrogenesis. Arch. Biochem. Biophys. 2014, 548, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Vallée, A.; Lecarpentier, Y. TGF-β in fibrosis by acting as a conductor for contractile properties of myofibroblasts. Cell Biosci. 2019, 9, 98. [Google Scholar] [CrossRef] [Green Version]
- Oo, Y.H.; Shetty, S.; Adams, D.H. The role of chemokines in the recruitment of lymphocytes to the liver. Dig. Dis. 2010, 28, 31–44. [Google Scholar] [CrossRef]
- Borkham-Kamphorst, E.; Weiskirchen, R. The PDGF system and its antagonists in liver fibrosis. Citokine Growth Factor Rev. 2016, 28, 53–61. [Google Scholar] [CrossRef]
- Dooley, S.; ten Dijke, P. TGF-β in progression of liver disease. Cell Tissue Res. 2012, 347, 245–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabregat, I.; Caballero-Díaz, D. Transforming growth factor-β-induced cell plasticity in liver fibrosis and hepatocarcinogenesis. Front. Oncol. 2018, 8, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte, S.; Baber, J.; Fujii, T.; Coito, A.J. Matrix metalloproteinases in liver injury, repair and fibrosis. Matrix Biol. 2015, 147–156. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.S.; Lee, D.H.; Yook, Y.W.; Na, D.; Jang, Y.J.; Kim, J.H.; Lee, Y.S. MicroRNA signatures associated with thioacetamide-induced liver fibrosis in mice. Biosci. Biotechnol. Biochem. 2017, 81, 1348–1355. [Google Scholar] [CrossRef] [Green Version]
- Coll, M.; El Taghdouini, A.; Perea, L.; Mannaerts, I.; Vila-Casadesús, M.; Blaya, D.; Rodrigo-Torres, D.; Affò, S.; Morales-Ibanez, O.; Graupera, I.; et al. Integrative miRNA and gene expression profiling analysis of human quiescent hepatic stellate cells. Sci. Rep. 2015, 5, 11549. [Google Scholar] [CrossRef] [Green Version]
- Maubach, G. miRNA studies in in vitro and in vivo activated hepatic stellate cells. World J. Gastroenterol. 2011, 17, 2748. [Google Scholar] [CrossRef]
- Guo, C.J.; Pan, Q.; Li, D.G.; Sun, H.; Liu, B.W. MiR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J. Hepatol. 2009, 50, 766–778. [Google Scholar] [CrossRef]
- O’Reilly, S. MicroRNAs in fibrosis: Opportunities and challenges. Arthritis Res. Ther. 2016, 18, 11. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Huang, C.; Zhang, S.; Sun, X.; Long, X.; Li, J. The potential of microRNAs in liver fibrosis. Cell Signal. 2012, 24, 2268–2272. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; He, Y.; Ma, T.T.; Huang, C.; Zhang, L.; Li, J. Participation of miR-200a in TGF-β1-mediated hepatic stellate cell activation. Mol. Cell Biochem. 2014, 388, 11–23. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Xu, K.; Lu, J. miR-34a promotes liver fibrosis in patients with chronic hepatitis via mediating Sirt1/p53 signaling pathway. Pathol. Res. Pract. 2020, 216, 152876. [Google Scholar] [CrossRef] [PubMed]
- Santoro, R.; Mangia, A. Progress in promising anti-fibrotic therapies. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 1145–1152. [Google Scholar] [CrossRef]
- Pingili, R.B.; Pawar, A.K.; Challa, S.R.; Kodali, T.; Koppula, S.; Toleti, V. A comprehensive review on hepatoprotective and nephroprotective activities of chrysin against various drugs and toxic agents. Chem. Biol. Interact. 2019, 308, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wu, W.F.; Sheng, L.; Jiang, B.J.; Li, X.; Cai, K.S. Chrysin ameliorates hepatic steatosis induced by a diet deficient in methionine and choline by inducing the secretion of hepatocyte nuclear factor 4 alpha-dependent very low-density lipoprotein. J. Biochem. Mol. Toxicol. 2020, 34, e22497. [Google Scholar] [CrossRef]
- Pai, S.A.; Munshi, R.P.; Panchal, F.H.; Gaur, I.S.; Juvekar, A.R. Chrysin ameliorates nonalcoholic fatty liver diseases in rats. Naunyn Schmiedebergs Arch. Pharmacol. 2019, 392, 1617–1628. [Google Scholar] [CrossRef]
- Balta, C.; Herman, H.; Boldura, O.M.; Gasca, I.; Rosu, M.; Ardelean, A.; Hermenean, A. Chrysin attenuates liver fibrosis and hepatic stellate cell activation through TGF-β/Smad signaling pathway. Chem. Biol. Interact. 2015, 240, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Balta, C.; Ciceu, A.; Herman, H.; Rosu, M.; Boldura, O.M.; Hermenean, A. Dose-dependent antifibrotic effect of chrysin on regression of liver fibrosis: The role in extracellular matrix remodeling. Dose-Response 2018, 16, 1559325818789835. [Google Scholar] [CrossRef] [Green Version]
- Fenyvesi, F.; Phuong Nguyen, T.L.; Haimhoffer, Á.; Rusznyák, Á.; Vasvári, G.; Bácskay, I.; Vecsernyés, M.; Ignat, S.R.; Dinescu, S.; Costache, M.; et al. Cyclodextrin complexation improves the solubility and Caco-2 permeability of chrysin. Materials 2020, 13, 3618. [Google Scholar] [CrossRef]
- Ignat, S.R.; Dinescu, S.; Váradi, J.; Fenyvesi, F.; Phuong Nguyen, T.L.; Ciceu, A.; Hermenean, A.; Costache, M. Complexation with random methyl-β-cyclodextrin and (2-hydroxypropyl)-β-cyclodextrin promotes chrysin effect and potential for liver fibrosis therapy. Materials 2020, 13, 5003. [Google Scholar] [CrossRef]
- Caprino, I.; Franchitto, A.; Morini, S.; Corradini, S.G.; Merli, M.; Gaudio, E. Activated hepatic stellate cells in liver cirrhosis. A morphologic and morphometrical study. Ital. J. Anat. Embryol. 2004, 109, 225–238. [Google Scholar]
- Domitrovic, R.; Jakovac, H. Effects of standardized bilberry fruit extract (Mirtoselect) on resolution of CCl4-induced liver fibrosis in mice. Food Chem. Toxicol. 2011, 4, 848–854. [Google Scholar] [CrossRef]
- Tsai, J.H.; Liu, J.Y.; Wu, T.T.; Ho, P.C.; Huang, C.Y.; Shyu, J.C.; Hsieh, Y.S.; Tsai, C.C.; Liu, Y.C. Effects of silymarin on the resolution of liver fibrosis induced by carbon tetrachloride in rats. J. Viral Hepat. 2008, 15, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, W.Y.; Wu, J.J.; Wei, W. TGF-β signaling pathway as a pharmacological target in liver diseases. Pharmacol. Res. 2014, 85, 15–22. [Google Scholar] [CrossRef]
- Luedde, T.; Schwabe, R. NF-κB in the liver—linking injury, fibrosis and hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 108–118. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Ghosh, S. NF-kappaB, an evolutionarily conserved mediator of immune and inflammatory responses. Adv. Exp. Med. Biol. 2005, 560, 41–45. [Google Scholar]
- Bilia, A.R.; Isacchi, B.; Righeschi, C.; Guccione, C.; Bergonzi, M.C. Flavonoids loaded in nanocarriers: An opportunity to increase oral bioavailability and bioefficacy. Food Nutr. Sci. 2014, 5, 1212–1327. [Google Scholar] [CrossRef] [Green Version]
- Weiskirchen, R.; Weiskirchen, S.; Tacke, F. Recent advances in understanding liver fibrosis: Bridging basic science and individualized treatment concepts. F1000Res 2018, 7, F1000 Faculty Rev-921. [Google Scholar] [CrossRef] [Green Version]
- Gharbia, S.; Balta, C.; Herman, H.; Rosu, M.; Varadi, J.; Bacskay, I.; Vecsernyés, M.; Gyöngyösi, S.; Fenyvesi, F.; Voicu, S.N.; et al. Enhancement of silymarin anti-fibrotic effects by complexation with hydroxypropyl (HPBCD) and randomly methylated (RAMEB) β-cyclodextrins in a mouse model of liver fibrosis. Front. Pharmacol. 2018, 9, 883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Wang, J.; Wang, J.; Zhou, Q.; Yang, B.; He, Q.; Weng, Q. Intercellular crosstalk of hepatic stellate cells in liver fibrosis: New insights into therapy. Pharmacol. Res. 2020, 155, 104720. [Google Scholar] [CrossRef]
- Khomich, O.; Ivanov, A.V.; Bartosch, B. Metabolic hallmarks of hepatic stellate cells in liver fibrosis. Cells 2020, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, E.B.; Huang, C.; Wang, H.; Chen, X.X.; Zhang, L.; Lv, X.W.; Li, J. Repression of Smad7 mediated by DNMT1 determines hepatic stellate cell activation and liver fibrosis in rats. Toxicol. Lett. 2014, 224, 175–185. [Google Scholar] [CrossRef]
- Li, X.M.; Peng, J.H.; Sun, Z.L.; Tian, H.J.; Duan, X.H.; Liu, L.; Ma, X.; Feng, Q.; Liu, P.; Hu, Y.Y. Chinese medicine CGA formula ameliorates DMN-induced liver fibrosis in rats via inhibiting MMP2/9, TIMP1/2 and the TGF-beta/Smad signalling pathways. Acta Pharmacol. Sin. 2016, 37, 783–793. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Deng, C.; Peng, Y.; Zheng, J.; Wei, L.; Shi, Y.; Gong, Z.; Hu, G. Dynamic Changes in MMP1 and TIMP1 in the Antifibrotic Process of Dahuang Zhechong Pill in Rats with Liver Fibrosis. Open Chem. 2019, 17, 346–356. [Google Scholar] [CrossRef]
- Chu, X.; Wang, H.; Jiang, Y.; Zhang, Y.; Bao, Y.; Zhang, X.; Zhang, J.; Guo, H.; Yang, F.; Luan, Y.; et al. Ameliorative effects of tannic acid on carbon tetrachloride-induced liver fibrosis in vivo and in vitro. J. Pharmacol. Sci. 2016, 130, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Papa, S.; Bubici, C.; Zazzeroni, F.; Franzoso, G. Mechanisms of liver disease: The crosstalk between the NF-κB and JNK pathways. Biol. Chem. 2009, 390, 965–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, S.; Shankar, E.; Fu, P.; MacLennan, G.; Gupta, S. Suppression of NF-ĸB and NF-ĸBRegulated Gene Expression by Apigenin through IĸBα and IKK Pathway in TRAMP Mice. PLoS ONE 2015, 10, e0138710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarrats, N.; Moles, A.; Morales, A.; García-Ruiz, C.; Fernández-Checa, J.C.; Marí, M. Critical role of tumor necrosis factor receptor 1, but not 2, in hepatic stellate cell proliferation, extracellular matrix remodeling, and liver fibrogenesis. Hepatology 2011, 54, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.M.; Seki, E. TNFα in liver fibrosis. Curr. Pathobiol. Rep. 2015, 3, 253–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Lee, C.H.; Lee, S.W. Hepatitis C virus infection stimulates transforming growth factor-β1 expression through up-regulating miR-192. J. Microbiol. 2016, 54, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I. MicroRNA Control of TGF-β Signaling. Int. J. Mol. Sci. 2018, 19, 1901. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Ma, L.; Wei, R.; Ye, T.; Zhou, J.; Wen, M.; Yang, L. Twist1-induced miR-199a-3p promotes liver fibrosis by suppressing caveolin-2 and activating TGF-β pathway. Signal Transduct. Target. Ther. 2020, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, J.; Wang, X.; Zhao, X.; Li, Z.; Niu, J.; Song, G. MicroRNA-378 promotes hepatic inflammation and fibrosis via modulation of the NF-κB-TNFα pathway. J. Hepatol. 2019, 70, 87–96. [Google Scholar] [CrossRef]
- Folk, A.; Cotoraci, C.; Balta, C.; Suciu, M.; Herman, H.; Boldura, O.M.; Dinescu, S.; Paiusan, L.; Ardelean, A.; Hermenean, A. Evaluation of Hepatotoxicity with Treatment Doses of Flucytosine and Amphotericin B for Invasive Fungal Infections. BioMed. Res.Int. 2016, 5, 398730. [Google Scholar]
- Han, Y.M.; Koh, J.; Koh, J.; Lee, C.; Koh, S.; Kim, B.; Lee, K.; Im, J.; Kim, J. NF-kappa B activation correlates with disease phenotype in Crohn’s disease. PLoS ONE 2017, 12, e0182071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Target | Sense | Antisense |
|---|---|---|
| NF-κB 50 | 5′ AGGAAGAAAATGGCGGAGTT 3′ | 5′ GCATAAGCTTCTGGCGTTTC 3′ |
| NF-κB 65 | 5′ CTTGGCAACAGCACAGACC 3′ | 5′ GAGAAGTCCATGTCCGCAAT 3′ |
| TNF-α | 5′CTGTAGCCCACGTCGTAGC3′ | 5′ TTGAGATCCATGCCGTTG 3′ |
| IL-6 | 5′AAAGAGTTGTGCAATGGCAATTCT3′ | 5′AAGTGCATCATCGTTGTTCATACA 3′ |
| TGF-β1 | 5′ TTTGGAGCCTGGACACACAGTAC 3′ | 5′ TGTGTTGGTTGTAGAGGGCAAGGA 3′ |
| α-SMA | 5′ CCGACCGAATGCAGAAG GA 3′ | 5′ ACAGAGTATTTGCGCTCCGAA 3′ |
| Smad 2 | 5′ GTTCCTGCCTTTGCTGAGAC 3′ | 5′ TCTCTTTGCCAGGAATGCTT 3′ |
| Smad 3 | 5′ TGCTGGTGACTGGATAGCAG 3′ | 5′ CTCCTTGGAAGGTGCTGAAG 3′ |
| Smad 7 | 5′ GCTCACGCACTCGGTGCTCA 3′ | 5′CCAGGCTCCAGAAGAAGTTG 3′ |
| Col I | 5′CAGCCGCTTCACCTACAGC 3′ | 5′ TTTTGTATTCAATCACTGTCTTGCC 3′ |
| TIMP-1 | 5′GGTGTGCACAGTGTTTCCCTGTTT 3′ | 5′ TCCGTCCACAAACAGTGAGTGTCA 3′ |
| MMP-1 | 5′ GCAGCGTCAAGTTTAACTGGAA 3′ | 5′ AACTACATTTAGGGGAGAGGTGT 3′ |
| MMP-2 | 5′CAGGGAATGAGTACTGGGTCTATT 3′ | 5′ ACTCCAGTTAAAGGCAGCATCTAC 3′ |
| MMP-3 | 5′ACCAACCTATTCCTGGTTGCTGCT 3′ | 5′ATGGAAACGGGACAAGTCTGTGGA 3′ |
| MMP-9 | 5′ GGACCCGAAGCGGACATTG 3′ | 5′ CGTCGTCGAAATGGGCATCT 3′ |
| GAPDH | 5′CGACTTCAACAGCAACTCCCACTCT3′ | 5′TGGGTGGTCCAGGGTTTCTTACTCCT3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciceu, A.; Balta, C.; Herman, H.; Gharbia, S.; Ignat, S.-R.; Dinescu, S.; Váradi, J.; Fenyvesi, F.; Gyöngyösi, S.; Hermenean, A.; et al. Complexation with Random Methyl-β-Cyclodextrin and (2-Hidroxypropyl)-β-Cyclodextrin Enhances In Vivo Anti-Fibrotic and Anti-Inflammatory Effects of Chrysin via the Inhibition of NF-κB and TGF-β1/Smad Signaling Pathways and Modulation of Hepatic Pro/Anti-Fibrotic miRNA. Int. J. Mol. Sci. 2021, 22, 1869. https://doi.org/10.3390/ijms22041869
Ciceu A, Balta C, Herman H, Gharbia S, Ignat S-R, Dinescu S, Váradi J, Fenyvesi F, Gyöngyösi S, Hermenean A, et al. Complexation with Random Methyl-β-Cyclodextrin and (2-Hidroxypropyl)-β-Cyclodextrin Enhances In Vivo Anti-Fibrotic and Anti-Inflammatory Effects of Chrysin via the Inhibition of NF-κB and TGF-β1/Smad Signaling Pathways and Modulation of Hepatic Pro/Anti-Fibrotic miRNA. International Journal of Molecular Sciences. 2021; 22(4):1869. https://doi.org/10.3390/ijms22041869
Chicago/Turabian StyleCiceu, Alina, Cornel Balta, Hidegard Herman, Sami Gharbia, Simona-Rebeca Ignat, Sorina Dinescu, Judit Váradi, Ferenc Fenyvesi, Szilvia Gyöngyösi, Anca Hermenean, and et al. 2021. "Complexation with Random Methyl-β-Cyclodextrin and (2-Hidroxypropyl)-β-Cyclodextrin Enhances In Vivo Anti-Fibrotic and Anti-Inflammatory Effects of Chrysin via the Inhibition of NF-κB and TGF-β1/Smad Signaling Pathways and Modulation of Hepatic Pro/Anti-Fibrotic miRNA" International Journal of Molecular Sciences 22, no. 4: 1869. https://doi.org/10.3390/ijms22041869
APA StyleCiceu, A., Balta, C., Herman, H., Gharbia, S., Ignat, S.-R., Dinescu, S., Váradi, J., Fenyvesi, F., Gyöngyösi, S., Hermenean, A., & Costache, M. (2021). Complexation with Random Methyl-β-Cyclodextrin and (2-Hidroxypropyl)-β-Cyclodextrin Enhances In Vivo Anti-Fibrotic and Anti-Inflammatory Effects of Chrysin via the Inhibition of NF-κB and TGF-β1/Smad Signaling Pathways and Modulation of Hepatic Pro/Anti-Fibrotic miRNA. International Journal of Molecular Sciences, 22(4), 1869. https://doi.org/10.3390/ijms22041869