The In Vivo Selection Method in Breast Cancer Metastasis
Abstract
1. Introduction
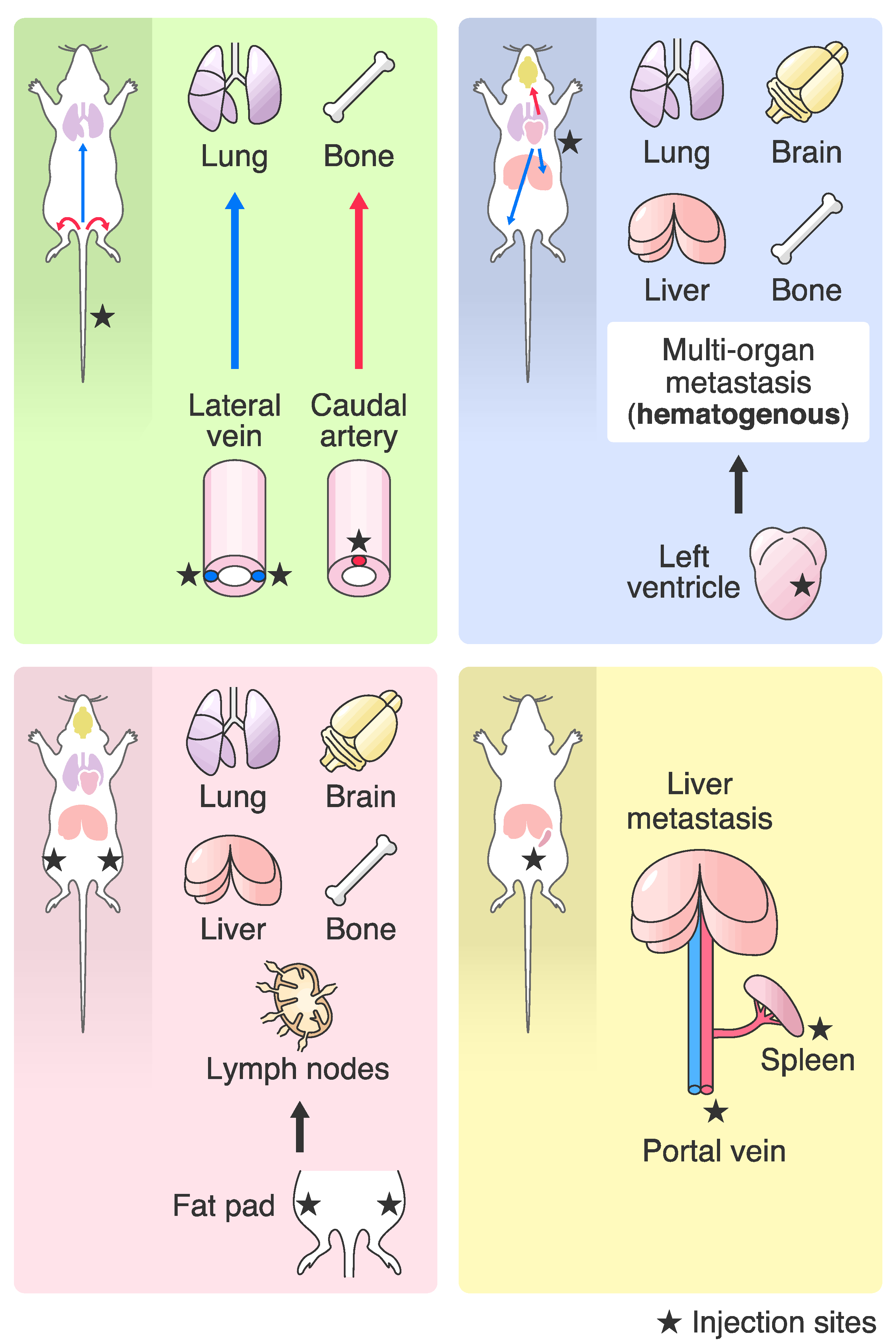
2. Differences in Transplantation Methods for the In Vivo Selection
2.1. Differences in Highly Metastatic Breast Cancer Cell Lines
2.2. Breast Cancer Metastasis in PDX Models
3. Highly Metastatic Breast Cancer Cell Lines
3.1. Introduction of Highly Metastatic Breast Cancer Cell Lines
3.2. Lung Metastasis
3.3. Bone Metastasis
3.4. Brain Metastasis
3.5. Liver Metastasis
3.6. Lymph Node Metastasis
4. Differences in Metastasis Gene Signatures According to the Molecular Subtypes of Breast Cancer
5. Genomic Profiling of Breast Cancer
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SCID | Severe combined immunodeficiency |
| NOD | Non-obese diabetic |
| GEMM | Genetically engineered mouse model |
| PDX | Patient derived xenograft |
| MetMap | Metastasis map |
| QOL | Quality of life |
| BBB | Blood-brain barrier |
| HR | Hormone receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| TNBC | Triple negative breast cancer |
| EMT | Epithelial to mesenchymal transition |
| TCGA | The cancer genome atlas |
| METABRIC | Molecular taxonomy of breast cancer international consortium |
References
- Valastyan, S.; Weinberg, R.A. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Paget, S. Distribution of secondary growths in cancer of the breast. Lancet 1889, 133, 571–573. [Google Scholar] [CrossRef]
- Fidler, I.J. The pathogenesis of cancer metastasis: The ’seed and soil’ hypothesis revisited. Nat. Rev. Cancer 2003, 3. [Google Scholar] [CrossRef]
- Suhail, Y.; Cain, M.P.; Vanaja, K.; Kurywchak, P.A.; Levchenko, A.; Kalluri, R.; Kshitiz. Systems Biology of Cancer Metastasis. Cell Syst. 2019, 9, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Obenauf, A.C.; Massague, J. Surviving at a Distance: Organ-Specific Metastasis. Trends Cancer 2015, 1, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Talmadge, J.E.; Fidler, I.J. AACR Centennial Series: The Biology of Cancer Metastasis: Historical Perspective. Cancer Res. 2010, 70, 5649–5669. [Google Scholar] [CrossRef]
- Takahashi, M. An experimental study of metastasis. J. Pathol. Bacteriol. 1915, 20, 1–13. [Google Scholar] [CrossRef]
- Fidler, I.J. Selection of Successive Tumour Lines for Metastasis. Nat. New Biol. 1973, 242, 148–149. [Google Scholar] [CrossRef]
- Bross, I.D.J.; Viadana, E.; Pickren, J.W. The metastatic spread of myeloma and leukemias in men. Virchows Arch. 1975, 365, 91–101. [Google Scholar] [CrossRef]
- Belizario, J.E. Immunodeficient Mouse Models: An Overview. Open Immunol. J. 2009, 2, 79–85. [Google Scholar] [CrossRef]
- Milsom, C.C.; Lee, C.R.; Hackl, C.; Man, S.; Kerbel, R.S. Differential post-surgical metastasis and survival in SCID, NOD-SCID and NOD-SCID-IL-2Rγ(null) mice with parental and subline variants of human breast cancer: Implications for host defense mechanisms regulating metastasis. PLoS ONE 2013, 8, e71270. [Google Scholar] [CrossRef]
- Bos, P.D.; Nguyen, D.X.; Massagué, J. Modeling metastasis in the mouse. Curr. Opin. Pharmacol. 2010, 10, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cuadrado, L.; Tracey, N.; Ma, R.; Qian, B.; Brunton, V.G. Mouse models of metastasis: Progress and prospects. Dis. Model. Mech. 2017, 10, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Kersten, K.; de Visser, K.E.; van Miltenburg, M.H.; Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 2017, 9, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Fantozzi, A.; Christofori, G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006, 8, 212. [Google Scholar] [CrossRef]
- Tian, X.; Gu, T.; Patel, S.; Bode, A.M.; Lee, M.-H.; Dong, Z. CRISPR/Cas9—An evolving biological tool kit for cancer biology and oncology. NPJ Precis. Oncol. 2019, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Boire, A.; Zou, Y.; Shieh, J.; Macalinao, D.G.; Pentsova, E.; Massagué, J. Complement Component 3 Adapts the Cerebrospinal Fluid for Leptomeningeal Metastasis. Cell 2017, 168, 1101–1113. [Google Scholar] [CrossRef] [PubMed]
- Er, E.E.; Valiente, M.; Ganesh, K.; Zou, Y.; Agrawal, S.; Hu, J.; Griscom, B.; Rosenblum, M.; Boire, A.; Brogi, E.; et al. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat. Cell Biol. 2018, 20, 966–978. [Google Scholar] [CrossRef]
- Rodrigues, G.; Hoshino, A.; Kenific, C.M.; Matei, I.R.; Steiner, L.; Freitas, D.; Kim, H.S.; Oxley, P.R.; Scandariato, I.; Casanova-Salas, I.; et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef]
- Pein, M.; Insua-Rodríguez, J.; Hongu, T.; Riedel, A.; Meier, J.; Wiedmann, L.; Decker, K.; Essers, M.A.G.; Sinn, H.-P.; Spaich, S.; et al. Metastasis-initiating cells induce and exploit a fibroblast niche to fuel malignant colonization of the lungs. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 1–17. [Google Scholar] [CrossRef]
- Ross, J.B.; Huh, D.; Noble, L.B.; Tavazoie, S.F. Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nat. Cell Biol. 2015, 17, 651–664. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.-A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef]
- Tavora, B.; Mederer, T.; Wessel, K.J.; Ruffing, S.; Sadjadi, M.; Missmahl, M.; Ostendorf, B.N.; Liu, X.; Kim, J.-Y.; Olsen, O.; et al. Tumoural activation of TLR3–SLIT2 axis in endothelium drives metastasis. Nature 2020, 586, 1–6. [Google Scholar] [CrossRef]
- Minn, A.J.; Gupta, G.P.; Siegel, P.M.; Bos, P.D.; Shu, W.; Giri, D.D.; Viale, A.; Olshen, A.B.; Gerald, W.L.; Massagué, J. Genes that mediate breast cancer metastasis to lung. Nature 2005, 436, 518–524. [Google Scholar] [CrossRef]
- Nakayama, J.; Ito, E.; Fujimoto, J.; Watanabe, S.; Semba, K. Comparative analysis of gene regulatory networks of highly metastatic breast cancer cells established by orthotopic transplantation and intra-circulation injection. Int. J. Oncol. 2017, 50, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Pillar, N.; Polsky, A.L.; Weissglas-Volkov, D.; Shomron, N. Comparison of breast cancer metastasis models reveals a possible mechanism of tumor aggressiveness. Cell Death Dis. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Ross, C.; Szczepanek, K.; Lee, M.; Yang, H.; Peer, C.J.; Kindrick, J.; Shankarappa, P.; Lin, Z.W.; Sanford, J.D.; Figg, W.D.; et al. Metastasis-Specific Gene Expression in Autochthonous and Allograft Mouse Mammary Tumor Models: Stratification and Identification of Targetable Signatures. Mol. Cancer Res. 2020, 18, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.-P.; Varma, S.; Gottesman, M.M. The Clinical Relevance of Cancer Cell Lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef]
- Sulaiman, A.; Wang, L. Bridging the divide: Preclinical research discrepancies between triple-negative breast cancer cell lines and patient tumors. Oncotarget 2017, 8, 113269–113281. [Google Scholar] [CrossRef]
- Shi, J.; Li, Y.; Jia, R.; Fan, X. The fidelity of cancer cells in PDX models: Characteristics, mechanism and clinical significance. Int. J. Cancer 2020, 146, 2078–2088. [Google Scholar] [CrossRef]
- Murayama, T.; Gotoh, N. Patient-Derived Xenograft Models of Breast Cancer and Their Application. Cells 2019, 8, 621. [Google Scholar] [CrossRef]
- Lee, M.W.; Miljanic, M.; Triplett, T.; Ramirez, C.; Aung, K.L.; Eckhardt, S.G.; Capasso, A. Current methods in translational cancer research. Cancer Metastasis Rev. 2020, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M. Patient-derived orthotopic xenografts: Better mimic of metastasis than subcutaneous xenografts. Nat. Rev. Cancer 2015, 15, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Claerhout, S.; Pratt, A.; Dobrolecki, L.E.; Petrovic, I.; Lai, Q.; Landis, M.D.; Wiechmann, L.; Schiff, R.; Giuliano, M.; et al. A Renewable Tissue Resource of Phenotypically Stable, Biologically and Ethnically Diverse, Patient-Derived Human Breast Cancer Xenograft Models. Cancer Res. 2013, 73, 4885–4897. [Google Scholar] [CrossRef] [PubMed]
- González-González, A.; Muñoz-Muela, E.; Marchal, J.A.; Cara, F.E.; Molina, M.P.; Cruz-Lozano, M.; Jiménez, G.; Verma, A.; Ramírez, A.; Qian, W.; et al. Activating Transcription Factor 4 Modulates TGFβ-Induced Aggressiveness in Triple-Negative Breast Cancer via SMAD2/3/4 and mTORC2 Signaling. Clin. Cancer Res. 2018, 24, 5697–5709. [Google Scholar] [CrossRef] [PubMed]
- Paez-Ribes, M.; Man, S.; Xu, P.; Kerbel, R.S. Development of Patient Derived Xenograft Models of Overt Spontaneous Breast Cancer Metastasis: A Cautionary Note. PLoS ONE 2016, 11, e0158034. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Okano, M.; Maiti, A.; Rashid, O.M.; Saito, K.; Kono, K.; Matsuyama, R.; Endo, I.; Takabe, K. Novel Breast Cancer Brain Metastasis Patient-Derived Orthotopic Xenograft Model for Preclinical Studies. Cancers 2020, 12, 444. [Google Scholar] [CrossRef]
- Lefley, D.; Howard, F.; Arshad, F.; Bradbury, S.; Brown, H.; Tulotta, C.; Eyre, R.; Alférez, D.; Wilkinson, J.M.; Holen, I.; et al. Development of clinically relevant in vivo metastasis models using human bone discs and breast cancer patient-derived xenografts. Breast Cancer Res. 2019, 21, 130. [Google Scholar] [CrossRef]
- Fujii, E.; Kato, A.; Suzuki, M. Patient-derived xenograft (PDX) models: Characteristics and points to consider for the process of establishment. J. Toxicol. Pathol. 2020, 33, 153–160. [Google Scholar] [CrossRef]
- Van ‘t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart AA, M.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Van de Vijver, M.J.; He, Y.D.; van ‘t Veer, L.J.; Dai, H.; Hart AA, M.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordón-Cardo, C.; Guise, T.A.; Massagué, J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003, 3, 537–549. [Google Scholar] [CrossRef]
- Oskarsson, T.; Batlle, E.; Massagué, J. Metastatic Stem Cells: Sources, Niches, and Vital Pathways. Cell Stem Cell 2014, 14, 306–321. [Google Scholar] [CrossRef]
- Vanharanta, S.; Massagué, J. Origins of Metastatic Traits. Cancer Cell 2013, 24, 410–421. [Google Scholar] [CrossRef]
- Gupta, G.P.; Massagué, J. Cancer Metastasis: Building a Framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Neophytou, C.; Boutsikos, P.; Papageorgis, P. Molecular Mechanisms and Emerging Therapeutic Targets of Triple-Negative Breast Cancer Metastasis. Front. Oncol. 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D.E.; Hornig, Y.S.; Oei, Y.; Dusich, J.; Purchio, T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res. 2005, 7, R444–R454. [Google Scholar] [CrossRef] [PubMed]
- Kuchimaru, T.; Kataoka, N.; Nakagawa, K.; Isozaki, T.; Miyabara, H.; Minegishi, M.; Kadonosono, T.; Kizaka-Kondoh, S. A reliable murine model of bone metastasis by injecting cancer cells through caudal arteries. Nat. Commun. 2018, 9, 1–7. [Google Scholar] [CrossRef]
- Goddard, E.T.; Fischer, J.; Schedin, P. A Portal Vein Injection Model to Study Liver Metastasis of Breast Cancer. J. Vis. Exp. 2016, 118, e54903. [Google Scholar] [CrossRef]
- Zhang, C.; Lowery, F.J.; Yu, D. Intracarotid Cancer Cell Injection to Produce Mouse Models of Brain Metastasis. J. Vis. Exp. 2017, 120, 55085. [Google Scholar] [CrossRef]
- Ozawa, T.; James, C.D. Establishing Intracranial Brain Tumor Xenografts with Subsequent Analysis of Tumor Growth and Response to Therapy using Bioluminescence Imaging. J. Vis. Exp. 2010, 41, e1986. [Google Scholar] [CrossRef]
- Haley, H.R.; Shen, N.; Qyli, T.; Buschhaus, J.M.; Pirone, M.; Luker, K.E.; Luker, G.D. Enhanced Bone Metastases in Skeletally Immature Mice. Tomography 2018, 4, 84–93. [Google Scholar] [CrossRef]
- Yu, J.; Zhou, Y.; Hua, X.; Zhu, Z.; Yu, X.-Y. In Situ Characterization of Hydrated Proteins in Water by SALVI and ToF-SIMS. J. Vis. Exp. 2016, 108, e53708. [Google Scholar] [CrossRef]
- Li, X.; Sterling, J.A.; Fan, K.H.; Vessella, R.L.; Shyr, Y.; Hayward, S.W.; Matrisian, L.M.; Bhowmick, N.A. Loss of TGF-β responsiveness in prostate stromal cells alters chemokine levels and facilitates the development of mixed osteoblastic/osteolytic bone lesions. Mol. Cancer Res. 2012, 10, 494–503. [Google Scholar] [CrossRef]
- Valiente, M.; Van Swearingen, A.E.; Anders, C.K.; Bairoch, A.; Boire, A.; Bos, P.D.; Cittelly, D.M.; Erez, N.; Ferraro, G.B.; Fukumura, D.; et al. Brain Metastasis Cell Lines Panel: A Public Resource of Organotropic Cell Lines. Cancer Res. 2020, 80, 4314–4323. [Google Scholar] [CrossRef]
- Jin, X.; DeMere, Z.; Nair, K.; Ali, A.; Ferraro, G.B.; Natoli, T.; Deik, A.; Petronio, L.; A Tang, A.; Zhu, C.; et al. A metastasis map of human cancer cell lines. Nature 2020, 588, 331–336. [Google Scholar] [CrossRef]
- Kuroiwa, Y.; Nakayama, J.; Adachi, C.; Inoue, T.; Watanabe, S.; Semba, K. Proliferative Classification of Intracranially Injected HER2-positive Breast Cancer Cell Lines. Cancers 2020, 12, 1811. [Google Scholar] [CrossRef] [PubMed]
- Tabaries, S.; Dong, Z.; Annis, M.G.; Omeroglu, A.; Pepin, F.; Ouellet, V.; Russo, C.; Hassanain, M.; Metrakos, P.; Diaz, Z.H.; et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene 2011, 30, 1318–1328. [Google Scholar] [CrossRef]
- Sasaki, S.; Baba, T.; Nishimura, T.; Hayakawa, Y.; Hashimoto, S.-I.; Gotoh, N.; Mukaida, N. Essential roles of the interaction between cancer cell-derived chemokine, CCL4, and intra-bone CCR5-expressing fibroblasts in breast cancer bone metastasis. Cancer Lett. 2016, 378, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Furihata, M.; Akimitsu, N.; Watanabe, M.; Kaul, S.; Yumoto, N.; Okada, T. A highly bone marrow metastatic murine breast cancer model established through in vivo selection exhibits enhanced anchorage-independent growth and cell migration mediated by ICAM-1. Clin. Exp. Metastasis 2008, 25, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Erin, N.; Wang, N.; Xin, P.; Bui, V.; Weisz, J.; Barkan, G.A.; Zhao, W.; Shearer, D.; Clawson, G.A. Altered gene expression in breast cancer liver metastases. Int. J. Cancer 2009, 124, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Lala, P.K.; Parhar, R.S. Eradication of spontaneous and experimental adenocarcinoma metastases with chronic indomethacin and intermittent IL-2 therapy. Int. J. Cancer 1993, 54, 677–684. [Google Scholar] [CrossRef]
- Tavazoie, S.F.; Alarcón, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massagué, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar] [CrossRef]
- Han, Y.; Nakayama, J.; Hayashi, Y.; Jeong, S.; Futakuchi, M.; Ito, E.; Watanabe, S.; Semba, K. Establishment and characterization of highly osteolytic luminal breast cancer cell lines by intracaudal arterial injection. Genes Cells 2020, 25, 111–123. [Google Scholar] [CrossRef]
- Jiang, H.-L.; Sun, H.-F.; Gao, S.-P.; Li, L.-D.; Hu, X.; Wu, J.; Jin, W. Loss of RAB1B promotes triple-negative breast cancer metastasis by activating TGF-β/SMAD signaling. Oncotarget 2015, 6, 16352–16365. [Google Scholar] [CrossRef][Green Version]
- Fietz, E.R.; Keenan, C.R.; López-Campos, G.; Tu, Y.; Johnstone, C.N.; Harris, T.; Stewart, A.G. Glucocorticoid resistance of migration and gene expression in a daughter MDA-MB-231 breast tumour cell line selected for high metastatic potential. Sci. Rep. 2017, 7, 43774. [Google Scholar] [CrossRef]
- Chang, X.-Z.; Li, D.-Q.; Hou, Y.-F.; Wu, J.; Lu, J.-S.; Di, G.-H.; Jin, W.; Ou, Z.-L.; Shen, Z.-Z.; Shao, Z.-M. Identification of the functional role of AF1Q in the progression of breast cancer. Breast Cancer Res. Treat. 2008, 111, 65–78. [Google Scholar] [CrossRef]
- Li, D.-Q.; Wang, L.; Fei, F.; Hou, Y.-F.; Luo, J.-M.; Chen, W.; Zeng, R.; Wu, J.; Lu, J.-S.; Di, G.-H.; et al. Identification of breast cancer metastasis-associated proteins in an isogenic tumor metastasis model using two-dimensional gel electrophoresis and liquid chromatography-ion trap-mass spectrometry. Proteomics 2006, 6, 3352–3368. [Google Scholar] [CrossRef]
- Vantyghem, S.A.; Allan, A.L.; Postenka, C.O.; Al-Katib, W.; Keeney, M.; Tuck, A.B.; Chambers, A.F. A new model for lymphatic metastasis: Development of a variant of the MDA-MB-468 human breast cancer cell line that aggressively metastasizes to lymph nodes. Clin. Exp. Metastasis 2005, 22, 351–361. [Google Scholar] [CrossRef]
- Kim, H.; Kim, B.; Kim, S.I.; Kim, H.J.; Ryu, B.Y.; Chung, J.; Lee, Z.H.; Kim, H.-H. S100A4 released from highly bone-metastatic breast cancer cells plays a critical role in osteolysis. Bone Res. 2019, 7, 1–13. [Google Scholar] [CrossRef]
- Esposito, M.; Mondal, N.; Greco, T.M.; Wei, Y.; Spadazzi, C.; Lin, S.C.; Zheng, H.; Cheung, C.; Magnani, J.L.; Lin, S.H.; et al. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat. Cell Biol. 2019, 21, 627–639. [Google Scholar] [CrossRef]
- Johnstone, C.N.; Pattison, A.D.; Gorringe, K.L.; Harrison, P.F.; Powell, D.R.; Lock, P.; Baloyan, D.; Ernst, M.; Stewart, A.G.; Beilharz, T.H.; et al. Functional and genomic characterisation of a xenograft model system for the study of metastasis in triple-negative breast cancer. Dis. Models Mech. 2018, 11, dmm032250. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, T.; Acharyya, S.; Zhang, X.H.-F.; Vanharanta, S.; Tavazoie, S.F.; Morris, P.G.; Downey, R.J.; Manova-Todorova, K.; Brogi, E.; Massagué, J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 2011, 17, 867–874. [Google Scholar] [CrossRef]
- Song, K.-H.; Park, M.S.; Nandu, T.S.; Gadad, S.; Kim, S.-C.; Kim, M.-Y. GALNT14 promotes lung-specific breast cancer metastasis by modulating self-renewal and interaction with the lung microenvironment. Nat. Commun. 2016, 7, 13796. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yin, B.; Yi, Z.; Liu, X.; Hu, Z.; Gao, W.; Yu, H.; Li, Q. Breast cancer stem cells characterized by CD70 expression preferentially metastasize to the lungs. Breast Cancer 2018, 25, 706–716. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Z.; Chen, C.; Hu, Q.; Fu, Z.; Chen, J.; Wang, Z.; Wang, Q.; Li, A.; Marks, J.R.; et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018, 430, 179–192. [Google Scholar] [CrossRef]
- Hiraga, T. Bone metastasis: Interaction between cancer cells and bone microenvironment. J. Oral Biosci. 2019, 61, 95–98. [Google Scholar] [CrossRef]
- Esposito, M.; Kang, Y. Targeting tumor–stromal interactions in bone metastasis. Pharmacol. Ther. 2014, 141, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Mundy, G.R. Metastasis to bone: Causes, consequences and therapeutic opportunities. Nat. Rev. Cancer 2002, 2, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Khanna, C.; Hunter, K. Modeling metastasis in vivo. Carcinogenesis 2005, 26, 513–523. [Google Scholar] [CrossRef]
- Simmons, J.K.; Hildreth, B.E., 3rd; Supsavhad, W.; Elshafae, S.M.; Hassan, B.B.; Dirksen, W.P.; Toribio, R.E.; Rosol, T.J. Animal Models of Bone Metastasis. Vet. Pathol. 2015, 52, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Allinen, M.; Beroukhim, R.; Cai, L.; Brennan, C.; Lahti-Domenici, J.; Huang, H.; Porter, D.; Hu, M.; Chin, L.; Richardson, A.; et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 2004, 6, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Eck, S.M.; Hoopes, P.J.; Petrella, B.L.; Coon, C.I.; Brinckerhoff, C.E. Matrix metalloproteinase-1 promotes breast cancer angiogenesis and osteolysis in a novel in vivo model. Breast Cancer Res. Treat. 2009, 116, 79–90. [Google Scholar] [CrossRef]
- Zhuang, X.; Zhang, H.; Li, X.; Li, X.; Cong, M.; Peng, F.; Yu, J.; Zhang, X.; Yang, Q.; Hu, G. Differential effects on lung and bone metastasis of breast cancer by Wnt signalling inhibitor DKK1. Nat. Cell Biol. 2017, 19, 1274–1285. [Google Scholar] [CrossRef]
- Nayak, L.; Lee, E.Q.; Wen, P.Y. Epidemiology of Brain Metastases. Curr. Oncol. Rep. 2012, 14, 48–54. [Google Scholar] [CrossRef]
- Lim, M.; Puttick, S.; Houston, Z.H.; Thurecht, K.J.; Croft, P.K.-D.; Mahler, S.; Rose, S.E.; Jeffree, R.L.; Mazzieri, R.; Dolcetti, R.; et al. Innovative Therapeutic Strategies for Effective Treatment of Brain Metastases. Int. J. Mol. Sci. 2019, 20, 1280. [Google Scholar] [CrossRef]
- Lin, X.; DeAngelis, L.M. Treatment of Brain Metastases. J. Clin. Oncol. 2015, 33, 3475–3484. [Google Scholar] [CrossRef]
- Bos, P.D.; Zhang, X.H.-F.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; Van De Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Kabraji, S.; Xie, S.; Pan, P.; Liu, Z.; Ni, J.; Zhao, J.J. Improving orthotopic mouse models of patient-derived breast cancer brain metastases by a modified intracarotid injection method. Sci. Rep. 2019, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Priego, N.; Zhu, L.; Monteiro, C.; Mulders, M.; Wasilewski, D.; Bindeman, W.; Doglio, L.; Martínez-Saez, E.; Cajal, S.R.Y.; Fustero-Torre, C.; et al. Abstract 2746: Stat3 labels a subpopulation of reactive astrocytes required for brain metastasis. Tumor Biol. 2019, 24, 1024–1035. [Google Scholar] [CrossRef]
- Wyatt, E.A.; Davis, M.E. Method of establishing breast cancer brain metastases affects brain uptake and efficacy of targeted, therapeutic nanoparticles. Bioeng. Transl. Med. 2018, 4, 30–37. [Google Scholar] [CrossRef]
- Sevenich, L.; Bowman, R.L.; Mason, S.D.; Quail, D.F.; Rapaport, F.; Elie, B.T.; Brogi, E.; Brastianos, P.K.; Hahn, W.C.; Holsinger, L.J.; et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat. Cell Biol. 2014, 16, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Valiente, M.; Obenauf, A.C.; Jin, X.; Chen, Q.; Zhang, X.H.-F.; Lee, D.J.; Chaft, J.E.; Kris, M.G.; Huse, J.T.; Brogi, E.; et al. Serpins Promote Cancer Cell Survival and Vascular Co-Option in Brain Metastasis. Cell 2014, 156, 1002–1016. [Google Scholar] [CrossRef]
- Zeng, Q.; Michael, I.P.; Zhang, P.; Saghafinia, S.; Knott, G.; Jiao, W.; McCabe, B.D.; Galván, J.A.; Robinson, H.P.C.; Zlobec, I.; et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 2019, 573, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.S.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma–astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016, 533, 493–498. [Google Scholar] [CrossRef]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lotvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015, 6, 6716. [Google Scholar] [CrossRef]
- Niwińska, A.; Murawska, M.; Pogoda, K. Breast cancer brain metastases: Differences in survival depending on biological subtype, RPA RTOG prognostic class and systemic treatment after whole-brain radiotherapy (WBRT). Ann. Oncol. 2010, 21, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Gril, B.; Palmieri, D.; Bronder, J.L.; Herring, J.M.; Vega-Valle, E.; Feigenbaum, L.; Liewehr, D.J.; Steinberg, S.M.; Merino, M.J.; Rubin, S.D.; et al. Effect of Lapatinib on the Outgrowth of Metastatic Breast Cancer Cells to the Brain. J. Natl. Cancer Inst. 2008, 100, 1092–1103. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Bronder, J.L.; Herring, J.M.; Yoneda, T.; Weil, R.J.; Stark, A.M.; Kurek, R.; Vega-Valle, E.; Feigenbaum, L.; Halverson, D.; et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 2007, 67, 4190–4198. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, T.; Zhang, H.; Gou, X.; Han, C.; Wang, J.; Chen, A.T.; Ma, J.; Liu, J.; Chen, Z.; et al. LRRC31 inhibits DNA repair and sensitizes breast cancer brain metastasis to radiation therapy. Nat. Cell Biol. 2020, 22, 1276–1285. [Google Scholar] [CrossRef]
- Gril, B.; Palmieri, D.; Qian, Y.; Smart, D.; Ileva, L.; Liewehr, D.J.; Steinberg, S.M.; Steeg, P.S. Pazopanib Reveals a Role for Tumor Cell B-Raf in the Prevention of HER2+ Breast Cancer Brain Metastasis. Clin. Cancer Res. 2011, 17, 142–153. [Google Scholar] [CrossRef]
- Beyer, I.; Li, Z.; Persson, J.; Liu, Y.; Van Rensburg, R.; Yumul, R.; Zhang, X.-B.; Hung, M.-C.; Lieber, A. Controlled Extracellular Matrix Degradation in Breast Cancer Tumors Improves Therapy by Trastuzumab. Mol. Ther. 2011, 19, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Dhillon, J.; Wang, M.Y.; Gao, Y.; Hu, K.; Park, E.; Astanehe, A.; Hung, M.-C.; Eirew, P.; Eaves, C.J.; et al. Targeting YB-1 in HER-2 Overexpressing Breast Cancer Cells Induces Apoptosis via the mTOR/STAT3 Pathway and Suppresses Tumor Growth in Mice. Cancer Res. 2008, 68, 8661–8666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, W.-C.; Zhang, L.; Zhang, C.; Lowery, F.J.; Ding, Z.; Guo, H.; Wang, H.; Huang, S.; Sahin, A.A.; et al. Src Family Kinases as Novel Therapeutic Targets to Treat Breast Cancer Brain Metastases. Cancer Res. 2013, 73, 5764–5774. [Google Scholar] [CrossRef] [PubMed]
- Sirkisoon, S.R.; Carpenter, R.L.; Rimkus, T.; Doheny, D.; Zhu, D.; Aguayo, N.R.; Xing, F.; Chan, M.; Ruiz, J.; Metheny-Barlow, L.J.; et al. TGLI1 transcription factor mediates breast cancer brain metastasis via activating metastasis-initiating cancer stem cells and astrocytes in the tumor microenvironment. Oncogene 2020, 39, 64–78. [Google Scholar] [CrossRef]
- Xing, F.; Liu, Y.; Sharma, S.; Wu, K.; Chan, M.D.; Lo, H.-W.; Carpenter, R.L.; Metheny-Barlow, L.J.; Zhou, X.; Qasem, S.A.; et al. Activation of the c-Met Pathway Mobilizes an Inflammatory Network in the Brain Microenvironment to Promote Brain Metastasis of Breast Cancer. Cancer Res. 2016, 76, 4970–4980. [Google Scholar] [CrossRef]
- Lyle, L.T.; Lockman, P.R.; Adkins, C.E.; Mohammad, A.S.; Sechrest, E.; Hua, E.; Palmieri, D.; Liewehr, D.J.; Steinberg, S.M.; Kloc, W.; et al. Alterations in Pericyte Subpopulations Are Associated with Elevated Blood–Tumor Barrier Permeability in Experimental Brain Metastasis of Breast Cancer. Clin. Cancer Res. 2016, 22, 5287–5299. [Google Scholar] [CrossRef]
- Murrell, D.H.; Hamilton, A.M.; Mallett, C.L.; Van Gorkum, R.; Chambers, A.F.; Foster, P.J. Understanding Heterogeneity and Permeability of Brain Metastases in Murine Models of HER2-Positive Breast Cancer Through Magnetic Resonance Imaging: Implications for Detection and Therapy. Transl. Oncol. 2015, 8, 176–184. [Google Scholar] [CrossRef][Green Version]
- Palmieri, D.; Duchnowska, R.; Woditschka, S.; Hua, E.; Qian, Y.; Biernat, W.; Sosińska-Mielcarek, K.; Gril, B.; Stark, A.M.; Hewitt, S.M.; et al. Profound Prevention of Experimental Brain Metastases of Breast Cancer by Temozolomide in an MGMT-Dependent Manner. Clin. Cancer Res. 2014, 20, 2727–2739. [Google Scholar] [CrossRef]
- Tanaka, Y.; Hirata, M.; Shinonome, S.; Torii, M.; Nezasa, K.-I.; Tanaka, H. Distribution analysis of epertinib in brain metastasis of HER2-positive breast cancer by imaging mass spectrometry and prospect for antitumor activity. Sci. Rep. 2018, 8, 343. [Google Scholar] [CrossRef]
- Erin, N.; Kale, S.; Tanrıöver, G.; Köksoy, S.; Duymuş, O.; Korcum, A.F. Differential characteristics of heart, liver, and brain metastatic subsets of murine breast carcinoma. Breast Cancer Res. Treat. 2013, 139, 677–689. [Google Scholar] [CrossRef]
- Gao, H.; Chakraborty, G.; Zhang, Z.; Akalay, I.; Gadiya, M.; Gao, Y.; Sinha, S.; Hu, J.; Jiang, C.; Akram, M.; et al. Multi-organ Site Metastatic Reactivation Mediated by Non-canonical Discoidin Domain Receptor 1 Signaling. Cell 2016, 166, 47–62. [Google Scholar] [CrossRef]
- Selzner, M.; Morse, M.A.; Vredenburgh, J.J.; Meyers, W.C.; Clavien, P.-A. Liver metastases from breast cancer: Long-term survival after curative resection. Surgery 2000, 127, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.; Aloia, T.; Krissat, J.; Bralet, M.-P.; Paule, B.; Giacchetti, S.; Delvart, V.; Azoulay, D.; Bismuth, H.; Castaing, D. Is Liver Resection Justified for Patients with Hepatic Metastases from Breast Cancer? Ann. Surg. 2006, 244, 897–908. [Google Scholar] [CrossRef]
- Ma, R.; Feng, Y.; Lin, S.; Chen, J.; Lin, H.; Liang, X.; Zheng, H.; Cai, X. Mechanisms involved in breast cancer liver metastasis. J. Transl. Med. 2015, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.Z.; Li, D.Q.; Hou, Y.F.; Wu, J.; Lu, J.S.; Di, G.H.; Jin, W.; Ou, Z.L.; Shen, Z.Z.; Shao, Z.M. Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res. 2007, 9, R76. [Google Scholar] [CrossRef]
- Tu, Y.; Johnstone, C.N.; Ryall, J.G.; López-Campos, G.H.; Keenan, C.R.; Stewart, A.G. Altered energy metabolism and metabolic gene expression associated with increased metastatic capacity identified in MDA-MB-231 cell line variants. J. Cancer Metastasis Treat. 2018, 2018, 58. [Google Scholar] [CrossRef]
- Petrova, T.V.; Koh, G.Y. Biological functions of lymphatic vessels. Science 2020, 369, eaax4063. [Google Scholar] [CrossRef]
- Yano, S.; Takehara, K.; Miwa, S.; Kishimoto, H.; Tazawa, H.; Urata, Y.; Kagawa, S.; Bouvet, M.; Fujiwara, T.; Hoffman, R.M. In Vivo Isolation of a Highly-aggressive Variant of Triple-negative Human Breast Cancer MDA-MB-231 Using Serial Orthotopic Transplantation. Anticancer Res. 2016, 36, 3817–3820. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J.; Panel, M. Strategies for subtypes--dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef]
- Chen, W.; Hoffmann, A.D.; Liu, H.; Liu, X. Organotropism: New insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2018, 2, 4. [Google Scholar] [CrossRef]
- Buonomo, O.C.; Caredda, E.; Portarena, I.; Vanni, G.; Orlandi, A.; Bagni, C.; Petrella, G.; Palombi, L.; Orsaria, P. New insights into the metastatic behavior after breast cancer surgery, according to well-established clinicopathological variables and molecular subtypes. PLoS ONE 2017, 12, e0184680. [Google Scholar] [CrossRef]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.U.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic Behavior of Breast Cancer Subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Xiao, W.; Zheng, S.; Yang, A.; Zhang, X.; Zou, Y.; Tang, H.; Xie, X. Breast cancer subtypes and the risk of distant metastasis at initial diagnosis: A population-based study. Cancer Manag. Res. 2018, 10, 5329–5338. [Google Scholar] [CrossRef]
- Smid, M.; Wang, Y.; Zhang, Y.; Sieuwerts, A.M.; Yu, J.; Klijn, J.G.M.; Foekens, J.A.; Martens, J.W.M. Subtypes of Breast Cancer Show Preferential Site of Relapse. Cancer Res. 2008, 68, 3108–3114. [Google Scholar] [CrossRef]
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast cancer subtypes predict the preferential site of distant metastases: A SEER based study. Oncotarget 2017, 8, 27990–27996. [Google Scholar] [CrossRef]
- Gong, Y.; Liu, Y.-R.; Ji, P.; Hu, X.; Shao, Z.-M. Impact of molecular subtypes on metastatic breast cancer patients: A SEER population-based study. Sci. Rep. 2017, 7, 45411. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Rose, D.P. Differential expression and regulation of cyclooxygenase-1 and -2 in two human breast cancer cell lines. Cancer Res. 1996, 56, 5125–5127. [Google Scholar] [PubMed]
- Tan, C.-C.; Li, G.-X.; Tan, L.-D.; Du, X.; Li, X.-Q.; He, R.; Wang, Q.-S.; Feng, Y.-M. Breast cancer cells obtain an osteomimetic feature via epithelial-mesenchymal transition that have undergone BMP2/RUNX2 signaling pathway induction. Oncotarget 2016, 7, 79688–79705. [Google Scholar] [CrossRef]
- Comşa, Ş.; Cimpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar] [PubMed]
- Gerratana, L.; Fanotto, V.; Bonotto, M.; Bolzonello, S.; Minisini, A.M.; Fasola, G.; Puglisi, F. Pattern of metastasis and outcome in patients with breast cancer. Clin. Exp. Metastasis 2015, 32, 125–133. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Curtis, C.; METABRIC Group; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; A Samarajiwa, S.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.A.; Russell, R.; Sammut, S.-J.; et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Ali, H.R.; Team, C.I.G.C.; Jackson, H.W.; Zanotelli, V.R.T.; Danenberg, E.; Fischer, J.R.; Bardwell, H.; Provenzano, E.; Rueda, O.M.; Chin, S.-F.; et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Rev. Cancer 2020, 1, 163–175. [Google Scholar] [CrossRef]
- Yates, L.R.; Knappskog, S.; Wedge, D.; Farmery, J.H.; Gonzalez, S.; Martincorena, I.; Alexandrov, L.B.; Van Loo, P.; Haugland, H.K.; Lilleng, P.K.; et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017, 32, 169–184.e7. [Google Scholar] [CrossRef]
- Chen, R.; Goodison, S.; Sun, Y. Molecular Profiles of Matched Primary and Metastatic Tumor Samples Support a Linear Evolutionary Model of Breast Cancer. Cancer Res. 2020, 80, 170–174. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.-M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymond, V.; et al. Integrative clinical genomics of metastatic cancer. Nature. 2017, 548, 297–303. [Google Scholar] [CrossRef]
- Quinn, J.J.; Jones, M.G.; Okimoto, R.A.; Nanjo, S.; Chan, M.M.; Yosef, N.; Bivona, T.G.; Weissman, J.S. Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 2021, eabc1944. [Google Scholar] [CrossRef] [PubMed]

| PDX Models | Subtype | Method | Metastatic Organs | Reference |
|---|---|---|---|---|
| 3887-LM | Basal-like (TNBC) | Orthotopic | Lung | [36] |
| HCI-002 LM2-1 | TNBC | Orthotopic | Lung | [38] |
| Brain PDX (B0~B3) | TNBC | Intracranially | Brain | [39] |
| MFP PDX (M0~M3) | TNBC | Orthotopic | Brain | [39] |
| Cell Line | Species of Origin | Subtype | Method | Metastatic Organ | References |
|---|---|---|---|---|---|
| 4T1-2776, 2792, 2869 | Mouse | Basal-like (TNBC) | Spleen transplantation | Liver | [61] |
| 4T1.3 | Mouse | Basal-like (TNBC) | Orthotopic | Bone | [62] |
| 4T1E/M3 | Mouse | Basal-like (TNBC) | Intravenous | Bone | [63] |
| 4T1LM | Mouse | Basal-like (TNBC) | Orthotopic | Liver | [64] |
| C3L5 | Mouse | Unknown | Subcutaneous | Lung | [65] |
| CN34-LM1 | Human | Claudin-low (TNBC) | Intravenous | Lung | [66] |
| MCF7-BM02 | Human | Luminal A | Intracaudal arterial | Bone | [67] |
| MDA-MB-231-BM (BoM: 1833) | Human | Claudin-low (TNBC) | Intracardiac | Bone | [45] |
| MDA-MB-231-HM | Human | Claudin-low (TNBC) | Intravenous | Lung | [68] |
| MDA-MB-231-HM.LNm5 | Human | Claudin-low (TNBC) | Orthotopic | Lymph node | [69] |
| MDA-MB-231-LM05 | Human | Claudin-low (TNBC) | Orthotopic | Lung | [26] |
| MDA-MB-231-LM1-2-1 | Human | Claudin-low (TNBC) | Intravenous | Lung | [26] |
| MDA-MB-231-LM2 (4175) | Human | Claudin-low (TNBC) | Intracardiac/Intravenous | Lung | [25] |
| MDA-MB-231-luc-D3H2LN | Human | Claudin-low (TNBC) | Orthotopic | Lymph node | [50] |
| MDA-MB-231HMLNm5 | Human | Claudin-low (TNBC) | Orthotopic | Lung, Liver, Lymph node, Spleen and Paraspinal tissue | [69,70,71] |
| MDA-MB-468LN | Human | Basal-like (TNBC) | Orthotopic | Lymph node | [72] |
| mtMDA | Human | Claudin-low (TNBC) | Intracardiac | Bone | [73] |
| SUM159-M1a | Human | Claudin-low (TNBC) | Intravenous | Lung | [74] |
| Metastasis Organ | Gene Signatures | Reference |
|---|---|---|
| Lung | EREG, CXCL1, MMP1, MMP2, SPARC, VCAM1, COX2 | [25] |
| Lung | CD70, SOX4 | [66] |
| Bone | IL1, CTGF, CXCR4, OPN | [45] |
| Bone | DKK1 | [87] |
| Bone | S100A4 | [73] |
| Brain | COX2, HBEGF, ST6GALNAC5 | [91] |
| Brain | CTSS, L1CAM, GRIN2B, PCDH7, CX43 | [18,95,96,97] |
| Liver | CLDN2 | [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakayama, J.; Han, Y.; Kuroiwa, Y.; Azuma, K.; Yamamoto, Y.; Semba, K. The In Vivo Selection Method in Breast Cancer Metastasis. Int. J. Mol. Sci. 2021, 22, 1886. https://doi.org/10.3390/ijms22041886
Nakayama J, Han Y, Kuroiwa Y, Azuma K, Yamamoto Y, Semba K. The In Vivo Selection Method in Breast Cancer Metastasis. International Journal of Molecular Sciences. 2021; 22(4):1886. https://doi.org/10.3390/ijms22041886
Chicago/Turabian StyleNakayama, Jun, Yuxuan Han, Yuka Kuroiwa, Kazushi Azuma, Yusuke Yamamoto, and Kentaro Semba. 2021. "The In Vivo Selection Method in Breast Cancer Metastasis" International Journal of Molecular Sciences 22, no. 4: 1886. https://doi.org/10.3390/ijms22041886
APA StyleNakayama, J., Han, Y., Kuroiwa, Y., Azuma, K., Yamamoto, Y., & Semba, K. (2021). The In Vivo Selection Method in Breast Cancer Metastasis. International Journal of Molecular Sciences, 22(4), 1886. https://doi.org/10.3390/ijms22041886






