Advances in Understanding of the Copper Homeostasis in Pseudomonas aeruginosa
Abstract
:1. Introduction
P. aeruginosa Origin, Occurrence, Risks, and Pathogenesis
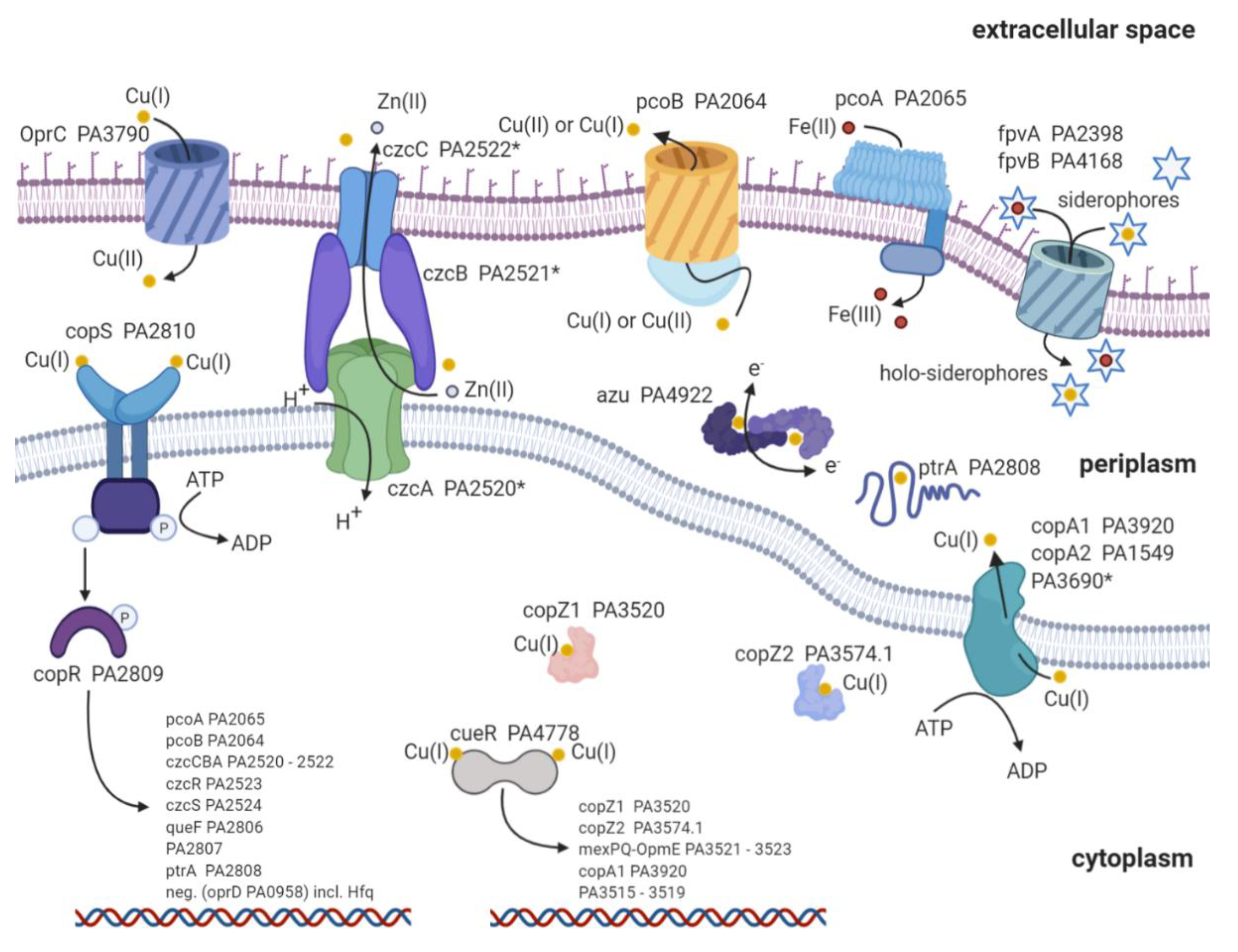
2. Copper Import
3. Copper Defense Mechanisms in P. aeruginosa
| TF | MT | CYTO-C | 2CS | P-Type | RND | Peri C | MCO | Sidero | Others | |
|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa | cueR [86] | MT * [87] | copZ1 copZ2 [86,88] | copRS [89] | copA1 copA2 [90] | czcCBA * | ptrA [91] azurin [92] | pcoA [93] | PVD PCH [82,83,84] | pcoB * [94] |
| E. coli | cueR [95] | N/A | N/A | pcoRS [96] cusRS [97] | copA [93] | cusCFBA [98] | pcoE [99] pcoC [100] cusF [98] | pcoA [101] cueO [102] | Ybt [103] | cut [95] pcoB [104] pcoD [104] porins |
| Transcription Regulator | Locus Tag | Regulated Proteins | Reference |
|---|---|---|---|
| cueR (first responder) PA4778 | PA3515–PA3519 | Five hypothetical proteins potentially involved in glycolysis and fatty acid metabolism | [86,88] |
| PA3520 | copZ1 | [86,105] | |
| PA3521–PA3523 | mexPQ-opmE | [86,88,117] | |
| PA3574.1 | copZ2 | [86,88,123] | |
| PA3920 | copA1 | [86,105] | |
| copRS (second responder) PA2809/PA2810 | PA2065 | pcoA | [88,107] |
| PA2064 | pcoB | [88,107] | |
| * PA2520–PA2522 | czcCBA (in conjunction with cadA activity) | [88,122,124,125] | |
| PA2523 | czcR | [88,122,126] | |
| PA2524 | czcS | [88,122,126] | |
| PA2806 | Hypothetical protein: potentially a NADPH-dependent reductase | [107] | |
| PA2807 | Hypothetical protein: azurin/plastocyanin family | [88,107] | |
| PA2808 | ptrA | [88,91] | |
| PA0958 | oprD (downregulation with cofactor: Hfq) | [88,127] |
4. Structural Insights into Proteins Involved in Copper Homeostasis
5. Copper as Antimicrobial
6. Copper and Antibiotics—The Power Couple
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | Adenosine triphosphate |
| CMT | Cytoplasmic membrane transporter |
| COPD | Severe chronic obstructive pulmonary disease |
| Cryo-EM | Cryogenic electron microscopy |
| CYTO-C | Cytoplasmic copper chaperone |
| CZC | Co(II), Zn(II), and Cd(II) |
| DNA | Deoxyribonucleic acid |
| EPA | Environmental Protection Agency |
| EPR | Electron paramagnetic resonance |
| ExoS | Exoenzyme S |
| ExoT | Exoenzyme T |
| ExoU | Exoenzyme U |
| ExoY | Exoenzyme Y |
| Glu | Glutamic acid |
| HIV | Human immunodeficiency viruses |
| HME | Heavy metal efflux |
| IRVS | Intensive respiratory or vasopressor support |
| iTOL | Interactive Tree of Life |
| KD | Dissociation constant |
| LPS | Lipopolysaccharide |
| MCO | Multicopper oxidase |
| merR | Mercuric resistance operon regulatory protein |
| Met | Methionine |
| MFP | Membrane fusion protein |
| MFS | Major facilitator superfamily |
| MT | Metallothionein |
| OMF | Outer membrane factor |
| PCH | Pyochelin |
| Peri C | Periplasmic copper chaperone |
| PtrA | Pseudomonas type III repressor A |
| P-type | P-type copper ATPase |
| PVD | Pyoverdine |
| QS | Quorum sensing |
| RNA | Ribonucleic acid |
| RND | Resistance nodulation division |
| rpoD | RNA polymerase sigma factor |
| Sidero | Siderophores |
| TF | Transcription factor |
| T3SS | Type III secretion system |
| T6SS | The type VI secretion system |
| WHO | World Health Organization |
| 2CS | Copper-sensing two-component |
References
- Talon, D.; Pseudomonas, B.X.M. Etymologia: Pseudomonas. Emerg. Infect. Dis. 2012, 18, 1241. [Google Scholar]
- Shooter, R.A.; Gaya, H.; Cooke, E.M.; Kumar, P.; Patel, N.; Parker, M.T.; Thom, B.T.; France, D.R. Food and medicaments as possible sources of hospital strains of Pseudomonas aeruginosa. Lancet 1969, 1, 1227–1229. [Google Scholar] [CrossRef]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The environmental occurrence of Pseudomonas aeruginosa. Apmis 2020, 128, 220–231. [Google Scholar] [CrossRef]
- Chatterjee, P.; Davis, E.; Yu, F.; James, S.; Wildschutte, J.H.; Wiegmann, D.D.; Sherman, D.H.; McKay, R.M.; LiPuma, J.J.; Wildschutte, H. Environmental pseudomonads inhibit cystic fibrosis patient-derived Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2017, 83, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ringen, L.M.; Drake, C.H. A study of the incidence of Pseudomonas aeruginosa from various natural sources. J. Bacteriol. 1952, 64, 841–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamidimarri, S.; Young, T.; Shanmugam, M.; Soderholm, S.; Basle, A.; Belzunces, B.; Skylaris, C.; Bumann, D.; Khalid, S.; van den Berg, B. Acquisition of ionic copper by a bacterial outer membrane protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Teitzel, G.M.; Parsek, M.R. Heavy Metal Resistance of Biofilm and Planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2003, 69, 2313–2320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitondo-Silva, A.; Gonçalves, G.B.; Stehling, E.G. Heavy metal resistance and virulence profile in Pseudomonas aeruginosa isolated from Brazilian soils. Apmis 2016, 124, 681–688. [Google Scholar] [CrossRef]
- Hobman, J.L.; Crossman, L.C. Bacterial antimicrobial metal ion resistance. J. Med. Microbiol. 2015, 64, 471–497. [Google Scholar] [CrossRef]
- Redfield, R.R. Antibiotic resistance threats in the United States. Cent. Dis. Control Prev. 2019. [Google Scholar] [CrossRef] [Green Version]
- Gransden, W.R.; Leibovici, L.; Eykyn, S.J.; Pitlik, S.D.; Samra, Z.; Konisberger, H.; Drucker, M.; Phillips, I. Risk factors and a clinical index for diagnosis of Pseudomonas aeruginosa bacteremia. Clin. Microbiol. Infect. 1995, 1, 119–123. [Google Scholar] [CrossRef] [Green Version]
- Restrepo, M.I.; Babu, B.L.; Reyes, L.F.; Chalmers, J.D.; Soni, N.J.; Sibila, O.; Faverio, P.; Cilloniz, C.; Rodriguez-Cintron, W.; Aliberti, S.; et al. Burden and risk factors for Pseudomonas aeruginosa community-acquired pneumonia: A multinational point prevalence study of hospitalised patients. Eur. Respir. J. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, S.; Georget, A.; Asselineau, J.; Venier, A.G.; Leroyer, C.; Rogues, A.M.; Thiébaut, R. Risk factors for colonization and infection by Pseudomonas aeruginosa in patients hospitalized in intensive care units in France. PLoS ONE 2018, 13, e0193300. [Google Scholar] [CrossRef]
- Merchant, S.; Proudfoot, E.M.; Quadri, H.N.; McElroy, H.J.; Wright, W.R.; Gupta, A.; Sarpong, E.M. Risk factors for Pseudomonas aeruginosa infections in Asia-Pacific and consequences of inappropriate initial antimicrobial therapy: A systematic literature review and meta-analysis. J. Glob. Antimicrob. Resist. 2018, 14, 33–44. [Google Scholar] [CrossRef]
- Ghibu, L.; Miftode, E.; Teodor, A.; Bejan, C.; Dorobăţ, C.M. Risk factors for Pseudomonas aeruginosa infections, resistant to carbapenem. Rev. Med. Chir. Soc. Med. Nat. Iasi. 2010, 114, 1012–1016. [Google Scholar]
- Jones, J.L.; Dargelas, V.; Roberts, J.; Press, C.; Remington, J.S.; Montoya, J.G. Risk Factors for Toxoplasma gondii Infection in the United States. Clin. Infect. Dis. 2009, 49, 878–884. [Google Scholar] [CrossRef] [Green Version]
- Hill, S.; Raviglione, M.; Weyer, K.; Tacconelli, E.; Magrini, N. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Pollack, M. The virulence of Pseudomonas aeruginosa. Rev. Infect. Diseases 1984, 6, S617–S626. [Google Scholar] [CrossRef]
- Peterson, J.W. Chapter 7 Bacterial Pathogenesis. Med. Microbiol. 2013, 1, 1–20. [Google Scholar]
- Hauser, A.R. So Many Virulence Factors, So Little Time. Crit. Care Med. 2012, 39, 2193–2194. [Google Scholar] [CrossRef] [Green Version]
- Le Berre, R.; Nguyen, S.; Nowak, E.; Kipnis, E.; Pierre, M.; Quenee, L.; Ader, F.; Lancel, S.; Courcol, R.; Guery, B.P.; et al. Relative contribution of three main virulence factors in Pseudomonas aeruginosa pneumonia. Crit. Care Med. 2011, 39, 2113–2120. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017. [Google Scholar] [CrossRef] [Green Version]
- Newman, J.W.; Floyd, R.V.; Fothergill, J.L. The contribution of Pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. Fems. Microbiol. Lett. 2017, 364, 1–11. [Google Scholar] [CrossRef]
- Azam, M.W.; Khan, A.U. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. Today 2019, 24, 350–359. [Google Scholar] [CrossRef]
- Juhas, M.; Eberl, L.; Tümmler, B. Quorum sensing: The power of cooperation in the world of Pseudomonas. Environ. Microbiol. 2005, 7, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Cámara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Häussler, S.; Becker, T. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 2008, 4, e1000166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Moustafa, D.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017, 13, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Kiratisin, P.; Tucker, K.D.; Passador, L. LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J. Bacteriol. 2002, 184, 4912–4919. [Google Scholar] [CrossRef] [Green Version]
- Cornelis, P. Putting an end to the Pseudomonas aeruginosa IQS controversy. Microbiologyopen 2020, 9, 2019–2020. [Google Scholar] [CrossRef] [Green Version]
- Kostylev, M.; Kim, D.Y.; Smalley, N.E.; Salukhe, I.; Peter Greenberg, E.; Dandekar, A.A. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. USA 2019, 116, 7027–7032. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, Y.; Zhang, L.H. The roles of microbial cell-cell chemical communication systems in the modulation of antimicrobial resistance. Antibiotics 2020, 9, 779. [Google Scholar] [CrossRef] [PubMed]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.O.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.Y.Y.; Chua, S.L.; Chen, Y.; Rice, S.A.; Kjelleberg, S.; Nielsen, T.E.; Yang, L.; Givskov, M. Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database. Antimicrob. Agents Chemother. 2013, 57, 5629–5641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, E.; Wuest, W.M.; Wuest, W.M. Virulence attenuating combination therapy: A potential multi-target synergy approach to treat: Pseudomonas aeruginosa infections in cystic fibrosis patients. RSC Med. Chem. 2020, 11, 358–369. [Google Scholar] [CrossRef]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Ulmer, A.J.; Zähringer, U.; Seydel, U.; Di Padova, F.; et al. Bacterial endotoxin: Molecular relationships of structure to activity and function. Faseb J. 1994, 8, 217–225. [Google Scholar] [CrossRef]
- Bystrova, O.V.; Lindner, B.; Moll, H.; Kocharova, N.A.; Knirel, Y.A.; Zahringer, U.; Pier, G.B. Full structure of the lipopolysaccharide of Pseudomonas aeruginosa immunotype 5. Biochemistry 2004, 69, 170–175. [Google Scholar] [CrossRef] [Green Version]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide Endotoxins Endotoxins as Activators of Innate Immunity. Annu. Rev. Biochem. 2008, 71, 635–700. [Google Scholar] [CrossRef] [Green Version]
- Huszczynski, S.M.; Lam, J.S.; Khursigara, C.M. The role of Pseudomonas aeruginosa lipopolysaccharide in bacterial pathogenesis and physiology. Pathogens. 2019, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- Bertani, B.; Ruiz, N. Function and Biogenesis of Lipopolysaccharides. EcoSal. Plus 2018, 8, 139–148. [Google Scholar] [CrossRef]
- Lam, J.S.; Taylor, V.L.; Islam, S.T.; Hao, Y.; Kocíncová, D. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2011, 2, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Pier, G.B. Pseudomonas aeruginosa lipopolysaccharide: A major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 2007, 297, 277–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazgaeen, L.; Gurung, P. Recent advances in lipopolysaccharide recognition systems. Int. J. Mol. Sci. 2020, 21, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, C.J.; Wozniaka, D.J. Psl Produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. MBio 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteban, J.; Pérez-Tanoira, R.; Pérez-Jorge-Peremarch, C.; Gómez-Barrena, E. Bacterial Adherence to Biomaterials Used in Surgical Procedures. Microbiol. Surg. Infect. Diagn. Progn. Treat. 2014, 41–57. [Google Scholar]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [Green Version]
- Irie, Y.; Roberts, A.E.L.; Kragh, K.N.; Gordon, V.D.; Hutchison, J.; Allen, R.J.; Melaugh, G.; Bjarnsholt, T.; West, S.A.; Diggle, S.P. The Pseudomonas aeruginosa PSL polysaccharide is a social but noncheatable trait in biofilms. MBio 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Passos da Silva, D.; Matwichuk, M.L.; Townsend, D.O.; Reichhardt, C.; Lamba, D.; Wozniak, D.J.; Parsek, M.R. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Grishin, A.V.; Krivozubov, M.S.; Karyagina, A.S.; Gintsburg, A.L. Pseudomonas Aeruginosa lectins as targets for novel antibacterials. Acta Naturae 2015, 7, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Kolomiets, E.; Johansson, E.M.V.; Renaudet, O.; Darbre, T.; Reymond, J.L. Neoglycopeptide dendrimer libraries as a source of lectin binding ligands. Org. Lett. 2007, 9, 1465–1468. [Google Scholar] [CrossRef]
- Reymond, J.L.; Bergmann, M.; Darbrea, T. Glycopeptide dendrimers as pseudomonas aeruginosa biofilm inhibitors. Chem. Soc. Rev. 2013, 42, 4814–4822. [Google Scholar] [CrossRef] [Green Version]
- Chemani, C.; Imberty, A.; De Bentzmann, S.; Pierre, M.; Wimmerová, M.; Guery, B.P.; Faure, K. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect. Immun. 2009, 77, 2065–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Rhman, S.H. Role of Pseudomonas aeruginosa lipopolysaccharides in modulation of biofilm and virulence factors of Enterobacteriaceae. Ann. Microbiol. 2019, 69, 299–305. [Google Scholar] [CrossRef]
- Lombardi, C.; Tolchard, J.; Bouillot, S.; Signor, L.; Gebus, C.; Liebl, D.; Fenel, D.; Teulon, J.M.; Brock, J.; Habenstein, B.; et al. Structural and functional characterization of the type three secretion system (T3SS) needle of pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 573. [Google Scholar] [CrossRef] [Green Version]
- Galle, M.; Carpentier, I.; Beyaert, R. Structure and Function of the Type III Secretion System of Pseudomonas aeruginosa. Curr. Protein Pept. Sci. 2013, 13, 831–842. [Google Scholar] [CrossRef] [Green Version]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 2009, 7, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Yahr, T.L.; Wolfgang, M.C. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 2006, 62, 631–640. [Google Scholar] [CrossRef]
- McShan, A.C.; De Guzman, R.N. The Bacterial Type III Secretion System as a Target for Developing New Antibiotics. Chem. Biol. Drug Des. 2015, 85, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anantharajah, A.; Mingeot-Leclercq, M.P.; Van Bambeke, F. Targeting the Type Three Secretion System in Pseudomonas aeruginosa. Trends Pharm. Sci. 2016, 37, 734–749. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A.; Szabo, D. Mechanisms of Multidrug Resistance in Acinetobacter Species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006, 43, S49–S56. [Google Scholar] [CrossRef] [Green Version]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa—Mechanisms, epidemiology and evolution. Drug Resist. Updat. 2019, 44, 100640. [Google Scholar] [CrossRef]
- Hwang, W.; Yoon, S.S. Virulence Characteristics and an Action Mode of Antibiotic Resistance in Multidrug-Resistant Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [Green Version]
- Hancock, R.E.W.; Speert, D.P. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and impact on treatment. Drug Resist. Updat. 2000, 3, 247–255. [Google Scholar] [CrossRef] [Green Version]
- van den Akker, F.; Bonomo, R.A. Exploring additional dimensions of complexity in inhibitor design for serine β-lactamases: Mechanistic and intra- and inter-molecular chemistry approaches. Front. Microbiol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Papp-Wallace, K.M.; Nguyen, N.Q.; Jacobs, M.R.; Bethel, C.R.; Barnes, M.D.; Kumar, V.; Bajaksouzian, S.; Rudin, S.D.; Rather, P.N.; Bhavsar, S.; et al. Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using β-Lactamase Inhibitors and β-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234. J. Med. Chem. 2018, 61, 4067–4086. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Tang, C.; Bethel, C.R.; Papp-Wallace, K.M.; Wyatt, J.; Desarbre, E.; Bonomo, R.A.; van den Akker, F. Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3. Antimicrob. Agents Chemother. 2020, 64, 1–12. [Google Scholar] [CrossRef]
- Goldberg, J.A.; Nguyen, H.; Kumar, V.; Spencer, E.J.; Hoyer, D.; Marshall, E.K.; Cmolik, A.; O’Shea, M.; Marshall, S.H.; Hujer, A.M.; et al. A γ-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli. J. Med. Chem. 2020, 63, 5990–6002. [Google Scholar] [CrossRef]
- Solioz, M. Copper Homeostasis in Gram-Negative Bacteria. In Copper and Bacteria. Springer Briefs in Molecular Science; Springer: Cham, Switzerland, 2018; pp. 49–80. [Google Scholar]
- Andrei, A.; Öztürk, Y.; Khalfaoui-Hassani, B.; Rauch, J.; Marckmann, D.; Trasnea, P.I.; Daldal, F.; Koch, H.G. Cu homeostasis in bacteria: The ins and outs. Membranes 2020, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, H.; Nakae, T. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 1996, 142, 2137–2144. [Google Scholar] [CrossRef] [Green Version]
- Porcheron, G.; Garénaux, A.; Proulx, J.; Sabri, M.; Dozois, C.M. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: Correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell Infect. Microbiol. 2013, 3, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Cha, J.S.; Cooksey, D.A. Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl. Environ. Microbiol. 1993, 59, 1671–1674. [Google Scholar] [CrossRef] [Green Version]
- Ekici, S.; Yang, H.; Koch, H.G.; Daldal, F. Novel transporter required for biogenesis of cbb3-type Cytochrome C oxidase in Rhodobacter capsulatus. MBio 2012, 3, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Parmar, J.H.; Quintana, J.; Ramírez, D.; Laubenbacher, R.; Argüello, J.M.; Mendes, P. An important role for periplasmic storage in Pseudomonas aeruginosa copper homeostasis revealed by a combined experimental and computational modeling study. Mol. Microbiol. 2018, 110, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Yan, N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef]
- Ekici, S.; Turkarslan, S.; Pawlik, G.; Dancis, A.; Baliga, N.S.; Koch, H.G.; Daldal, F. Intracytoplasmic copper homeostasis controls cytochrome c oxidase production. MBio 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandel, J.; Humbert, N.; Elhabiri, M.; Schalk, I.J.; Mislin, G.L.A.; Albrecht-Gary, A.M. Pyochelin, a siderophore of Pseudomonas aeruginosa: Physicochemical characterization of the iron(iii), copper(ii) and zinc(ii) complexes. Dalt. Trans. 2012, 41, 2820–2834. [Google Scholar] [CrossRef]
- Xiao, R.; Kisaalita, W.S. Purification of pyoverdines of Pseudomonas fluorescens 2-79 by copper- chelate chromatography. Appl. Environ. Microbiol. 1995, 61, 3769–3774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braud, A.; Geoffroy, V.; Hoegy, F.; Mislin, G.L.A.; Schalk, I.J. Presence of the siderophores pyoverdine and pyochelin in the extracellular medium reduces toxic metal accumulation in Pseudomonas aeruginosa and increases bacterial metal toleranceemi. Environ. Microbiol. Rep. 2010, 2, 419–425. [Google Scholar] [CrossRef]
- Hannauer, M.; Braud, A.; Hoegy, F.; Ronot, P.; Boos, A.; Schalk, I.J. The PvdRT-OpmQ efflux pump controls the metal selectivity of the iron uptake pathway mediated by the siderophore pyoverdine in Pseudomonas aeruginosa. Environ. Microbiol. 2012, 14, 1696–1708. [Google Scholar] [CrossRef]
- Braud, A.; Hoegy, F.; Jezequel, K.; Lebeau, T.; Schalk, I.J. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 2009, 11, 1079–1091. [Google Scholar] [CrossRef]
- Poole, R.K. Microbiology of Metal Ions; Academic Press: Cambridge, MA, USA, 2017; p. 379. [Google Scholar]
- Thaden, J.T.; Lory, S.; Gardner, T.S. Quorum-sensing regulation of a copper toxicity system in Pseudomonas aeruginosa. J. Bacteriol. 2010, 192, 2557–2568. [Google Scholar] [CrossRef] [Green Version]
- Habjanič, J.; Mathew, A.; Eberl, L.; Freisinger, E. Deciphering the Enigmatic Function of Pseudomonas Metallothioneins. Front. Microbiol. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Teitzel, G.M.; Geddie, A.; De Long, S.K.; Kirisits, M.J.; Whiteley, M.; Parsek, M.R. Survival and growth in the presence of elevated copper: Transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7242–7256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, S.D.; Jasalavich, C.A.; Cooksey, D.A. A Two-Component Regulatory System Required for Copper- Inducible Expression of the Copper Resistance Operon of Pseudomonas syringae. J. Bacteriol. 1993, 175, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Alquethamy, S.F.; Khorvash, M.; Pederick, V.G.; Whittall, J.J.; Paton, J.C.; Paulsen, I.T.; Hassan, K.A.; McDevitt, C.A.; Eijkelkamp, B.A. The role of the copA copper efflux system in acinetobacter baumannii virulence. Int. J. Mol. Sci. 2019, 20, 575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsen, S.; Ragno, M.; Attree, I. PtrA is a periplasmic protein involved in Cu tolerance in Pseudomonas aeruginosa. J. Bacteriology 2011, 193, 3376–3378. [Google Scholar] [CrossRef] [Green Version]
- Palm-Espling, M.E.; Niemiec, M.S.; Wittung-Stafshede, P. Role of metal in folding and stability of copper proteins in vitro. Biochim. Biophys. Acta 2012, 1823, 1594–1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, A.B.; Stoj, C.S.; Ziegler, L.; Kosman, D.J.; Hart, P.J. The copper-iron connection in biology: Structure of the metallo-oxidase Fet3p. Proc. Natl. Acad. Sci. USA 2005, 102, 15459–15464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Pannuri, A.A.; Ni, D.; Zhou, H.; Cao, X.; Lu, X.; Romeo, T.; Huang, Y. Structural basis for translocation of a biofilm-supporting exopolysaccharide across the bacterial outer membrane. J. Biol. Chem. 2016, 291, 10046–10057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Outten, F.W.; Outten, C.E.; Hale, J.; O’Halloran, T.V. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J. Biol. Chem. 2000, 275, 31024–31029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouch, D.A.; Brown, N.L. Copper-induci ble transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology 1997, 143, 1191–1202. [Google Scholar] [CrossRef] [Green Version]
- Munson, G.P.; Lam, D.L.; Outten, F.W. Identification of a Copper-Responsive Two-Component System on the Chromosome of Escherichia coli K-12. J. Bacteriol. 2000, 182, 5864–5871. [Google Scholar] [CrossRef] [Green Version]
- Franke, S.; Grass, G.; Rensing, C.; Nies, D.H. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003, 185, 3804–3812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, M.; Udagedara, S.R.; Sze, C.M.; Ryan, T.M.; Howlett, G.J.; Xiao, Z.; Wedd, A.G. PcoE—A metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J. Inorg. Biochem. 2012, 115, 186–197. [Google Scholar] [CrossRef]
- Wernimont, A.K.; Huffman, D.L.; Finney, L.A.; Demeler, B.; O’Halloran, T.V.; Rosenzweig, A.C. Crystal structure and dimerization equilibria of PcoC, a methionine-rich copper resistance protein from Escherichia coli. J. Biol. Inorg. Chem. 2003, 8, 185–194. [Google Scholar] [CrossRef]
- Djoko, K.Y.; Xiao, Z.; Wedd, A.G. Copper resistance in E. coli: The multicopper oxidase PcoA catalyzes oxidation of copper (I) in CuICuII-PcoC. ChemBioChem 2008, 9, 1579–1582. [Google Scholar] [CrossRef]
- Grass, G.; Rensing, C. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys. Res. Commun. 2001, 286, 902–908. [Google Scholar] [CrossRef]
- Koh, E.-I.; Robinson, A.E.; Bandara, N.; Rogers, B.E.; Henderson, J.P. Copper import in Escherichia coli by the yersiniabactin metallophore system. Nat. Chem. Biol. 2017, 13, 1016–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, N.L.; Barrett, S.R.; Camakaris, J.; Lee, B.T.O.O.; Rouch, D.A. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 1995, 17, 1153–1166. [Google Scholar] [CrossRef]
- Novoa-Aponte, L.; Ramírez, D.; Argüello, J.M. The interplay of the metallosensor CueR with two distinct CopZ chaperones defines copper homeostasis in Pseudomonas aeruginosa. J. Biol. Chem. 2019, 294, 4934–4945. [Google Scholar] [CrossRef] [Green Version]
- Raimunda, D.; Padilla-Benavides, T.; Vogt, S.; Boutigny, S.; Tomkinson, K.N.; Finney, L.A.; Argüello, J.M. Periplasmic response upon disruption of transmembrane Cu transport in Pseudomonas aeruginosa. Metallomics 2013, 5, 144–151. [Google Scholar] [CrossRef]
- Quintana, J.; Novoa-Aponte, L.; Argüello, J.M. Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J. Biol. Chem. 2017, 292, 15691–15704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, N.L.; Stoyanov, J.V.; Kidd, S.P.; Hobman, J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. [Google Scholar] [CrossRef] [Green Version]
- Sameach, H.; Narunsky, A.; Azoulay-Ginsburg, S.; Gevorkyan-Aiapetov, L.; Zehavi, Y.; Moskovitz, Y.; Juven-Gershon, T.; Ben-Tal, N.; Ruthstein, S. Structural and Dynamics Characterization of the MerR Family Metalloregulator CueR in its Repression and Activation States. Structure 2017, 25, 988–996.e3. [Google Scholar] [CrossRef] [Green Version]
- Sameach, H.; Ghosh, S.; Gevorkyan-Airapetov, L.; Saxena, S.; Ruthstein, S. EPR Spectroscopy Detects Various Active State Conformations of the Transcriptional Regulator CueR. Angew. Chem. 2019, 58, 3053–3056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, C.; Philips, S.J.; Wu, X.; Chen, K.; Shi, J.; Shen, L.; Xu, J.; Feng, Y.; O’Halloran, T.V.; Zhang, Y. CueR activates transcription through a DNA distortion mechanism. Nat. Chem. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Changela, A.; Chen, K.; Xue, Y.; Holschen, J.; Outten, C.E.; O’Halloran, T.V.; Mondragón, A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 2003, 301, 1383–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, D.; Piergentili, C.; Chen, J.; Chakrabarti, B.; Foster, A.W.; Lurie-Luke, E.; Huggins, T.G.; Robinson, N.J. Generating a metal-responsive transcriptional regulator to test what confers metal sensing in cells. J. Biol. Chem. 2015, 290, 19806–19822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Wang, T.; Chen, G.; Pu, Q.; Liu, Q.; Zhang, Y.; Xu, L.; Wu, M.; Liang, H. A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 2019, 15, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Wright, B.W.; Kamath, K.S.; Krisp, C.; Molloy, M.P. Proteome profiling of Pseudomonas aeruginosa PAO1 identifies novel responders to copper stress. BMC Microbiol. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haider, S.; Pal, R. Integrated Analysis of Transcriptomic and Proteomic Data. Curr. Genomics 2013, 14, 91–110. [Google Scholar] [CrossRef]
- Mima, T.; Sekiya, H.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. Gene cloning and properties of the RND-type multidrug efflux pumps MexPQ-OpmE and MexMN-OprM from Pseudomonas aeruginosa. Microbiol. Immunol. 2005, 49, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Conry, R.R. Copper: Inorganic & Coordination Chemistry Based in part on the article Copper: Inorganic & Coordination Chemistry by Rebecca, R. Conry & Kenneth, D. Karlin which appeared in the Encyclopedia of Inorganic Chemistry. In Encyclopedia of Inorganic and Bioinorganic Chemistry, 1st ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2011. [Google Scholar]
- Novoa-Aponte, L.; Soncini, F.; Argüello, J. The two-component system CopRS maintains femtomolar levels of free copper in the periplasm of Pseudomonas aeruginosa using a phosphatase-based mechanism. mSphere 2020, 5, 1–15. [Google Scholar] [CrossRef]
- Grünberger, F.; Reichelt, R.; Waege, I.; Ned, V.; Bronner, K.; Kaljanac, M.; Weber, N.; El Ahmad, Z.; Knauss, L.; Madej, M.G.; et al. CopR, a Global Regulator of Transcription to Maintain Copper Homeostasis in Pyrococcus furiosus. Front. Microbiol. 2021, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Oc, S.; Eraslan, S.; Kirdar, B. Dynamic transcriptional response of Saccharomyces cerevisiae cells to copper. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Caille, O.; Rossier, C.; Perron, K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 4561–4568. [Google Scholar] [CrossRef] [Green Version]
- González-Guerrero, M.; Raimunda, D.; Cheng, X.; Argüello, J.M. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 2010, 78, 1246–1258. [Google Scholar] [CrossRef]
- Ducret, V.; Gonzalez, M.R.; Leoni, S.; Valentini, M.; Perron, K. The CzcCBA Efflux System Requires the CadA P-Type ATPase for Timely Expression Upon Zinc Excess in Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Nies, D.H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 1995, 177, 2707–2712. [Google Scholar] [CrossRef] [Green Version]
- Perron, K.; Caille, O.; Rossier, C.; Van Delden, C.; Dumas, J.L.; Köhler, T. CzcR-CzcS, a Two-component System Involved in Heavy Metal and Carbapenem Resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef] [Green Version]
- Ducret, V.; Gonzalez, M.; Scrignari, T.; Perron, K. OprD Repression upon Metal Treatment Requires the RNA Chaperone Hfq in Pseudomonas aeruginosa. Genes 2016, 7, 82. [Google Scholar] [CrossRef]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef] [Green Version]
- Bondarczuk, K.; Piotrowska-Seget, Z. Molecular basis of active copper resistance mechanisms in Gram-negative bacteria. Cell Biol. Toxicol. 2013, 29, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.; Grass, G.; Rensing, C.; Barrett, S.R.; Yates, C.J.D.; Stoyanov, J.V.; Brown, N.L. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 2002, 295, 616–620. [Google Scholar] [CrossRef]
- Cha, J.S.; Cooksey, D.A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 1991, 88, 8915–8919. [Google Scholar] [CrossRef] [Green Version]
- Huston, W.M.; Jennings, M.P.; McEwan, A.G. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 2002, 45, 1741–1750. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [Green Version]
- Ha, U.H.; Kim, J.; Badrane, H.; Jia, J.; Baker, H.V.; Wu, D.; Jin, S. An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system. Mol. Microbiol. 2004, 54, 307–320. [Google Scholar] [CrossRef]
- Steinmetzer, K. CopR binds and bends its target DNA: A footprinting and fluorescence resonance energy transfer study. Nucleic Acids Res. 2002, 30, 2052–2060. [Google Scholar] [CrossRef]
- Fang, C.; Li, L.; Zhao, Y.; Wu, X.; Philips, S.J.; You, L.; Zhong, M.; Shi, X.; O’Halloran, T.V.; Li, Q.; et al. The bacterial multidrug resistance regulator BmrR distorts promoter DNA to activate transcription. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Philips, S.J.; Canalizo-Hernandez, M.; Yildirim, I.; Schatz, G.C.; Mondragón, A.; O’Halloran, T.V. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science 2015, 349, 877–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puzari, M.; Chetia, P. RND efflux pump mediated antibiotic resistance in Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: A major issue worldwide. World J. Microbiol. Biotechnol. 2017, 33, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Colclough, A.L.; Alav, I.; Whittle, E.E.; Pugh, H.L.; Darby, E.M.; Legood, S.W.; McNeil, H.E.; Blair, J.M.A. RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol. 2020, 15, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Checa, S.K.; Giri, G.F.; Espariz, M.; Argüello, J.M.; Soncini, F.C. Copper Handling in the Salmonella Cell Envelope and Its Impact on Virulence. Trends Microbiol. 2021. [Google Scholar] [CrossRef]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinforma. 2020, 72, 1–30. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V.A. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Hawass, Z. Hidden Treasures of the Egyptian Museum; Amercain University in Cairo Press: Cairo, Egypt, 2002. [Google Scholar]
- Borkow, G.; Gabbay, J. Copper, An Ancient Remedy Returning to Fight Microbial, Fungal and Viral Infections. Curr. Chem. Biol. 2009, 3, 272–278. [Google Scholar]
- Borkow, G.; Gabbay, J. Copper as a Biocidal Tool. Curr. Med. Chem. 2005, 12, 2163–2175. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, N.; Hiatt, C.W.; Haller, W. Mechanism of inactivation of bacteriophages by metals. BBA Spec. Sect. Nucleic Acids Relat. Subj. 1964, 91, 257–261. [Google Scholar] [CrossRef]
- Soliman, M.Y.M.; Medema, G.; Bonilla, B.E.; Brouns, S.J.J.; van Halem, D. Inactivation of RNA and DNA viruses in water by copper and silver ions and their synergistic effect. Water Res. X 2020, 9, 100077. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.G.; Attaway, H.H.; Fairey, S.E.; Steed, L.L.; Michels, H.T.; Salgado, C.D. Copper Continuously Limits the Concentration of Bacteria Resident on Bed Rails within the Intensive Care Unit. Infect. Control. Hosp. Epidemiol. 2013, 34, 530–533. [Google Scholar] [CrossRef] [Green Version]
- Salgado, C.D.; Sepkowitz, K.A.; John, J.F.; Cantey, J.R.; Attaway, H.H.; Freeman, K.D.; Sharpe, P.A.; Michels, H.T.; Schmidt, M.G. Copper Surfaces Reduce the Rate of Healthcare-Acquired Infections in the Intensive Care Unit. Infect. Control. Hosp. Epidemiol. 2013, 34, 479–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcus, E.L.; Yosef, H.; Borkow, G.; Caine, Y.; Sasson, A.; Moses, A.E. Reduction of health care–associated infection indicators by copper oxide–impregnated textiles: Crossover, double-blind controlled study in chronic ventilator-dependent patients. Am. J. Infect. Control. 2017, 45, 401–403. [Google Scholar] [CrossRef] [Green Version]
- Lazary, A.; Weinberg, I.; Vatine, J.J.; Jefidoff, A.; Bardenstein, R.; Borkow, G.; Ohana, N. Reduction of healthcare-associated infections in a long-term care brain injury ward by replacing regular linens with biocidal copper oxide impregnated linens. Int. J. Infect. Dis. 2014, 24, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Vincent, M.; Duval, R.E.; Hartemann, P.; Engels-Deutsch, M. Contact killing and antimicrobial properties of copper. J. Appl. Microbiol. 2018, 124, 1032–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, H.L.; Cronholm, P.; Hedberg, Y.; Tornberg, M.; De Battice, L.; Svedhem, S.; Wallinder, I. Cell membrane damage and protein interaction induced by copper containing nanoparticles-Importance of the metal release process. Toxicology 2013, 313, 59–69. [Google Scholar] [CrossRef]
- Pramanik, A.; Laha, D.; Bhattacharya, D.; Pramanik, P.; Karmakar, P. A novel study of antibacterial activity of copper iodide nanoparticle mediated by DNA and membrane damage. Colloids Surf. B Biointerfaces 2012, 96, 50–55. [Google Scholar] [CrossRef]
- Sagripanti, J.L.; Kraemer, K.H. Site-specific oxidative DNA damage at polyguanosines produced by copper plus hydrogen peroxide. J. Biol. Chem. 1989, 264, 1729–1734. [Google Scholar] [CrossRef]
- White, C.; Lee, J.; Kambe, T.; Fritsche, K.; Petris, M.J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 2009, 284, 33949–33956. [Google Scholar] [CrossRef] [Green Version]
- Achard, M.E.S.; Stafford, S.L.; Bokil, N.J.; Chartres, J.; Bernhardt, P.V.; Schembri, M.A.; Sweet, M.J.; McEwan, A.G. Copper redistribution in murine macrophages in response to Salmonella infection. Biochem. J. 2012, 444, 51–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaturvedi, K.S.; Henderson, J.P. Pathogenic adaptations to host-derived antibacterial copper. Front. Cell Infect. Microbiol. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Hodgkinson, V.; Zhu, S.; Weisman, G.A.; Petris, M.J. Advances in the Understanding of Mammalian Copper Transporters. Adv. Nutr. 2011, 2, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalecki, A.G.; Haeili, M.; Shah, S.; Speer, A.; Niederweis, M.; Kutsch, O.; Wolschendorf, F. Disulfiram and copper ions kill Mycobacterium tuberculosis in a synergistic manner. Antimicrob. Agents Chemother. 2015, 59, 4835–4844. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Dalecki, A.G.; Malalasekera, A.P.; Crawford, C.L.; Michalek, S.M.; Kutsch, O.; Sun, J.; Bossmann, S.H.; Wolschendorf, F. 8-Hydroxyquinolines are boosting agents of copper-related toxicity in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2016, 60, 5765–5776. [Google Scholar] [CrossRef] [Green Version]
- Djoko, K.Y.; Paterson, B.M.; Donnelly, P.S.; McEwan, A.G. Antimicrobial effects of copper(ii) bis(thiosemicarbazonato) complexes provide new insight into their biochemical mode of action. Metallomics 2014, 6, 854–863. [Google Scholar] [CrossRef] [Green Version]
- Skalka, M.; Čejková, M. DNA cleavage in chromatin spacer segments by a non-enzymic probe, 1,10-phenanthroline-copper complex. Mol. Biol. Rep. 1985, 10, 163–168. [Google Scholar] [CrossRef]
- Smit, H.; van Der Goot, H.; Nauta, W.T.; Timmerman, H.; de Bolster, M.W.; Jochemsen, A.G.; Stouthamer, A.H.; Vis, R.D. Mode of action of the copper(I) complex of 2,9-dimethyl-1,10-phenanthroline on Mycoplasma gallisepticum. Antimicrob. Agents Chemother. 1981, 20, 455–462. [Google Scholar] [CrossRef] [Green Version]
- Perrin, D.M.; Hoang, V.M.; Xu, Y.; Mazumder, A.; Sigman, D.S. Inhibitors of Escherichia coli RNA polymerase specific for the single-stranded DNA of transcription intermediates. Tetrahedral cuprous chelates of 1,10-phenanthrolines. Biochemistry 1996, 35, 5318–5326. [Google Scholar] [CrossRef]
- Reeder, N.L.; Kaplan, J.; Xu, J.; Youngquist, R.S.; Wallace, J.; Hu, P.; Juhlin, K.D.; Schwartz, J.R.; Grant, R.A.; Fieno, A.; et al. Zinc pyrithione inhibits yeast growth through copper influx and inactivation of iron-sulfur proteins. Antimicrob. Agents Chemother. 2011, 55, 5753–5760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, F.; Zhao, Y.; Gu, X.; Gu, C.; Lee, C.C.C. Joint toxicity of tetracycline with copper(II) and cadmium(II) to Vibrio fischeri: Effect of complexation reaction. Ecotoxicology 2015, 24, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.F.; Ferreira, M.; Abreu, B.; Medforth, C.J.; Gameiro, P. Interactions of a non-fluorescent fluoroquinolone with biological membrane models: A multi-technique approach. Int. J. Pharm. 2015, 495, 761–770. [Google Scholar] [CrossRef]
- Rafiee, F.; Haghi, F.; Bikas, R.; Heidari, A.; Gholami, M.; Kozakiewicz, A.; Zeighami, H. Synthesis, characterization and assessment of anti-quorum sensing activity of copper(II)-ciprofloxacin complex against Pseudomonas aeruginosa PAO1. AMB Express 2020, 10, 82. [Google Scholar] [CrossRef] [Green Version]
- Szczepanik, W.; Kaczmarek, P.; Sobczak, J.; Bal, W.; Gatner, K.; Jezowska-Bojczuk, M. Copper(II) binding by kanamycin A and hydrogen peroxide activation by resulting complexes. New J. Chem. 2002, 26, 1507–1514. [Google Scholar] [CrossRef]
- Albert, A. Avidity of terramycin and aureomycin for metallic cations. Nature 1953. [Google Scholar] [CrossRef]
- Rogolino, D.; Gatti, A.; Carcelli, M.; Pelosi, G.; Bisceglie, F.; Restivo, F.M.; Degola, F.; Buschini, A.; Montalbano, S.; Feretti, D.; et al. Thiosemicarbazone scaffold for the design of antifungal and antiaflatoxigenic agents: Evaluation of ligands and related copper complexes. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Hofmann, L.; Tsybovsky, Y.; Alexander, N.S.; Babino, D.; Leung, N.Y.; Montell, C.; Banerjee, S.; Von Lintig, J.; Palczewski, K. Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family. Biochemistry 2016, 55, 6545–6557. [Google Scholar] [CrossRef] [Green Version]
- Albert, A.; Rees, C.W.; Tomlinson, A.J. The influence of chemical constitution on antibacterial activity. V.I.I.I. 2-Mercaptopyridine-N-oxide, and some general observations on metalbinding agents. Br. J. Exp. Pathol. 1956, 37, 500–511. [Google Scholar]
- Yori, J.L.; Lozada, K.L.; Seachrist, D.D.; Mosley, J.D.; Abdul-Karim, F.W.; Booth, C.N.; Flask, C.A.; Keri, R.A. Combined SFK/mTOR Inhibition Prevents Rapamycin-Induced Feedback Activation of AKT and Elicits Efficient Tumor Regression. Cancer Res. 2014, 74, 4762–4771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meir, A.; Lepechkin-Zilbermintz, V.; Kahremany, S.; Schwerdtfeger, F.; Gevorkyan-Airapetov, L.; Munder, A.; Viskind, O.; Gruzman, A.; Ruthstein, S. Inhibiting the copper efflux system in microbes as a novel approach for developing antibiotics. PLoS ONE 2019, 14, e0227070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonasgenomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Antibiotic | Mode of Copperaction | Reference |
|---|---|---|
| Ditiocarb (Disulfiram) | Copper complex, bypasses the copper homeostatic machinery in Mycobacterium tuberculosis | [161] |
| 8-Hydroxyquinoline | Copper complex and ionophore, thus facilitates the transfer of copper across hydrophobic membranes. | [162] |
| Thiosemicarbazones | Copper complex and ionophore, target NADPH dehydrogenases | [163] |
| Phenanthroline | Copper complex with nuclease activity of mainly double-stranded DNA, some interference with respiration and inhibition of RNA polymerase | [164,165,166] |
| Pyrithione | Copper complex and ionophore, facilitates copper influx | [167] |
| Tetracycline | Copper complex formation shows an antagonistic effect while complex formation with Cd(II) exhibits a synergistic effect | [168] |
| Fluoroquinolone | Copper complex formation but with only minor antimicrobial effect, facilitates the transfer of copper across hydrophobic membranes | [169,170] |
| Aminoglycoside | A weak copper complex formation that is not physiologically relevant, maybe a potential role in DNA damage and formation of reactive oxygen species | [171] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofmann, L.; Hirsch, M.; Ruthstein, S. Advances in Understanding of the Copper Homeostasis in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 2050. https://doi.org/10.3390/ijms22042050
Hofmann L, Hirsch M, Ruthstein S. Advances in Understanding of the Copper Homeostasis in Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2021; 22(4):2050. https://doi.org/10.3390/ijms22042050
Chicago/Turabian StyleHofmann, Lukas, Melanie Hirsch, and Sharon Ruthstein. 2021. "Advances in Understanding of the Copper Homeostasis in Pseudomonas aeruginosa" International Journal of Molecular Sciences 22, no. 4: 2050. https://doi.org/10.3390/ijms22042050
APA StyleHofmann, L., Hirsch, M., & Ruthstein, S. (2021). Advances in Understanding of the Copper Homeostasis in Pseudomonas aeruginosa. International Journal of Molecular Sciences, 22(4), 2050. https://doi.org/10.3390/ijms22042050






