Structure–Function Relationship Study of a Secretory Amoebic Phosphatase: A Computational-Experimental Approach
Abstract
:1. Introduction
2. Results and Discussion
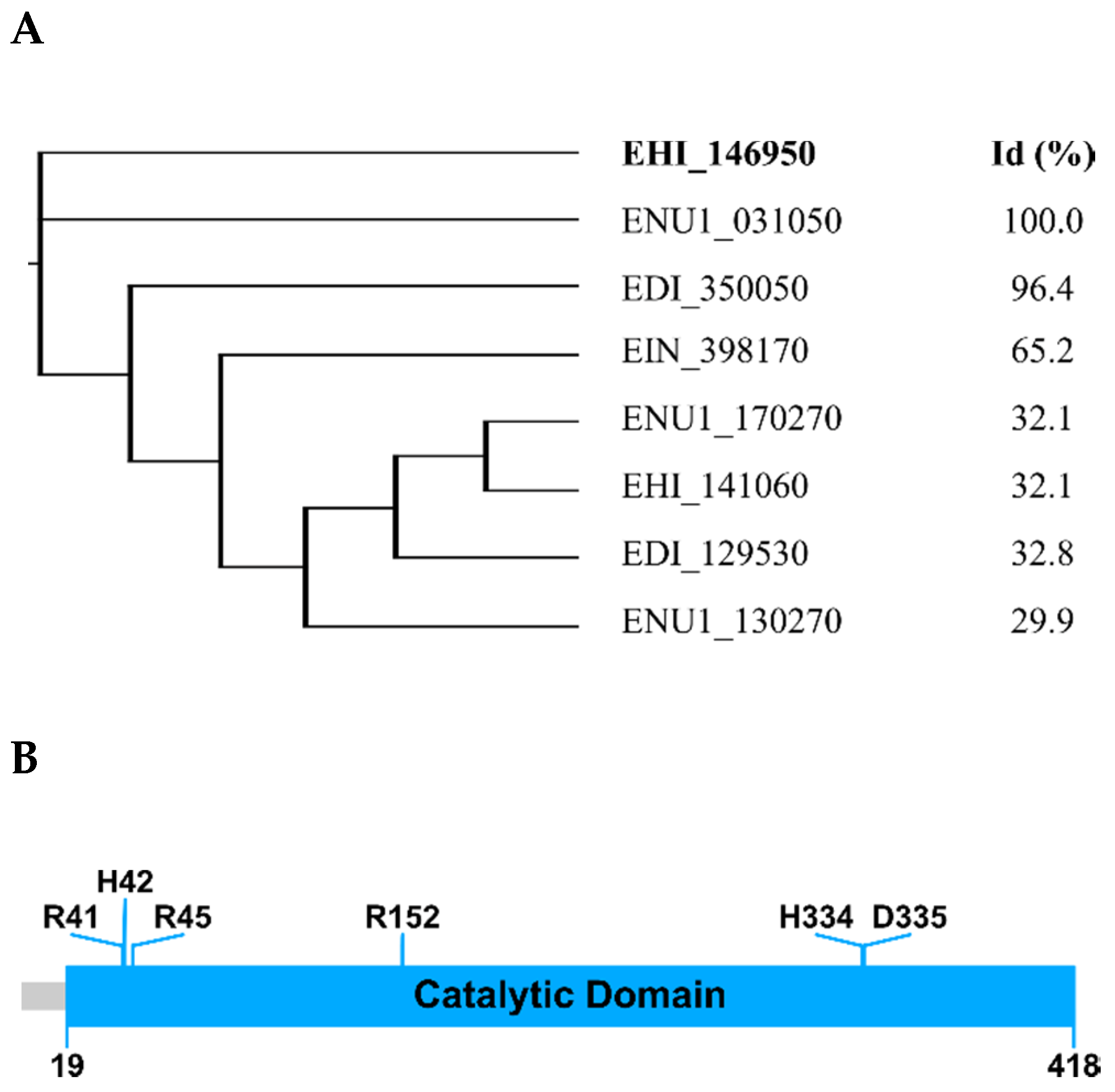
2.1. E. histolytica Encodes a HAP/Phytase-Like Protein: EhHAPp49
2.2. rEhHAPp49 Is an Active Enzyme
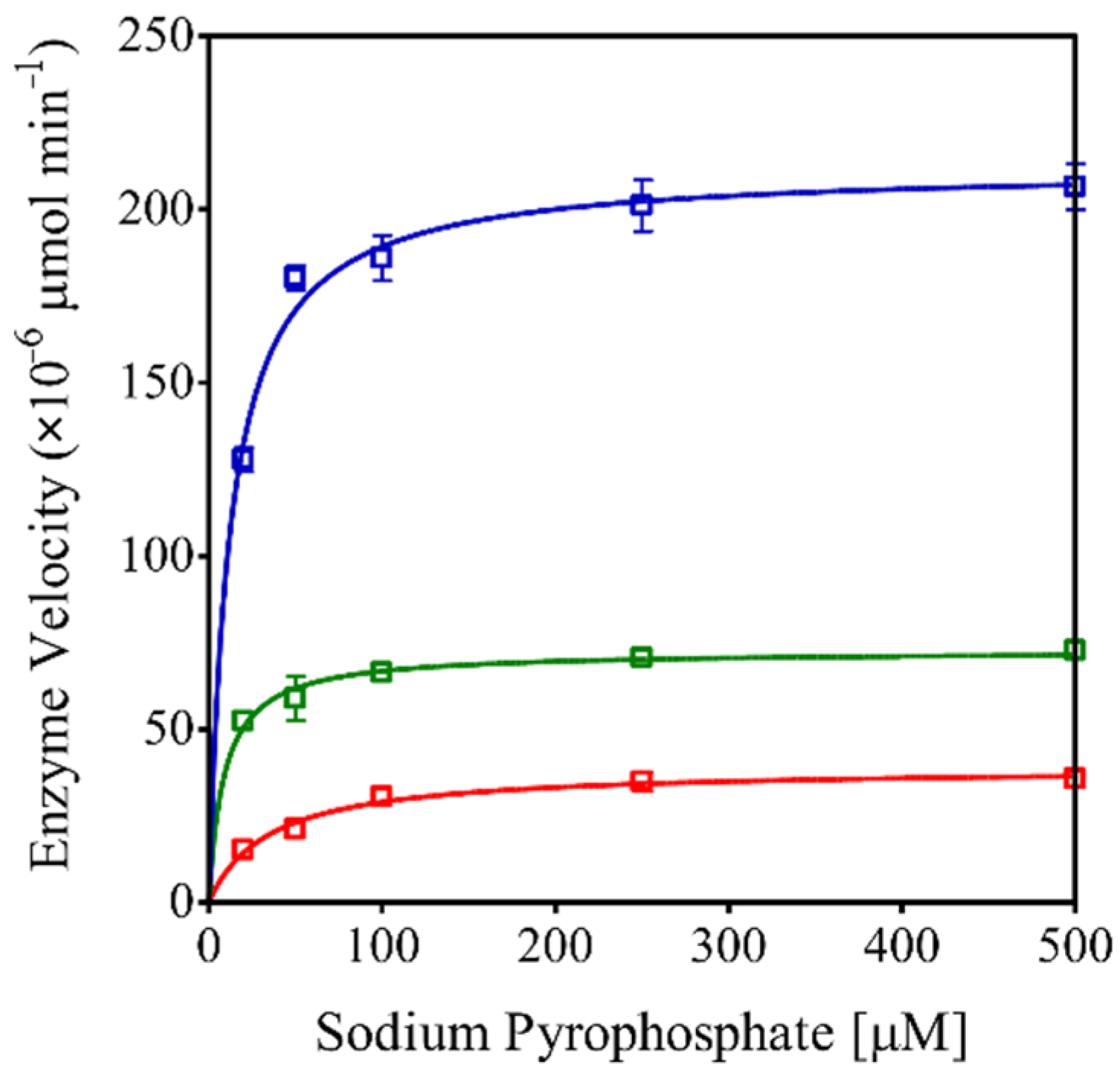
2.3. rEhHAPp49 Exhibits Pyrophosphatase Activity
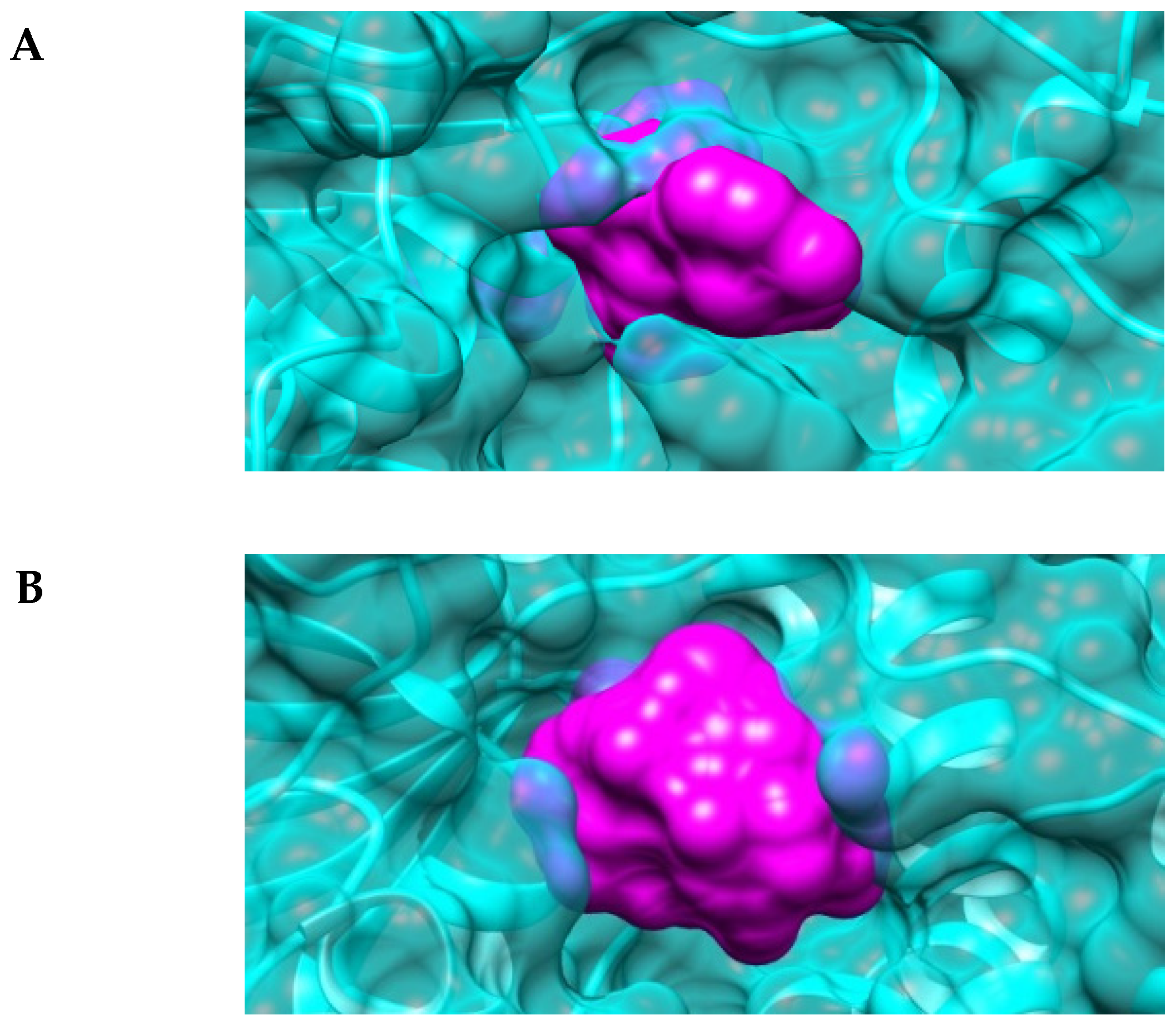
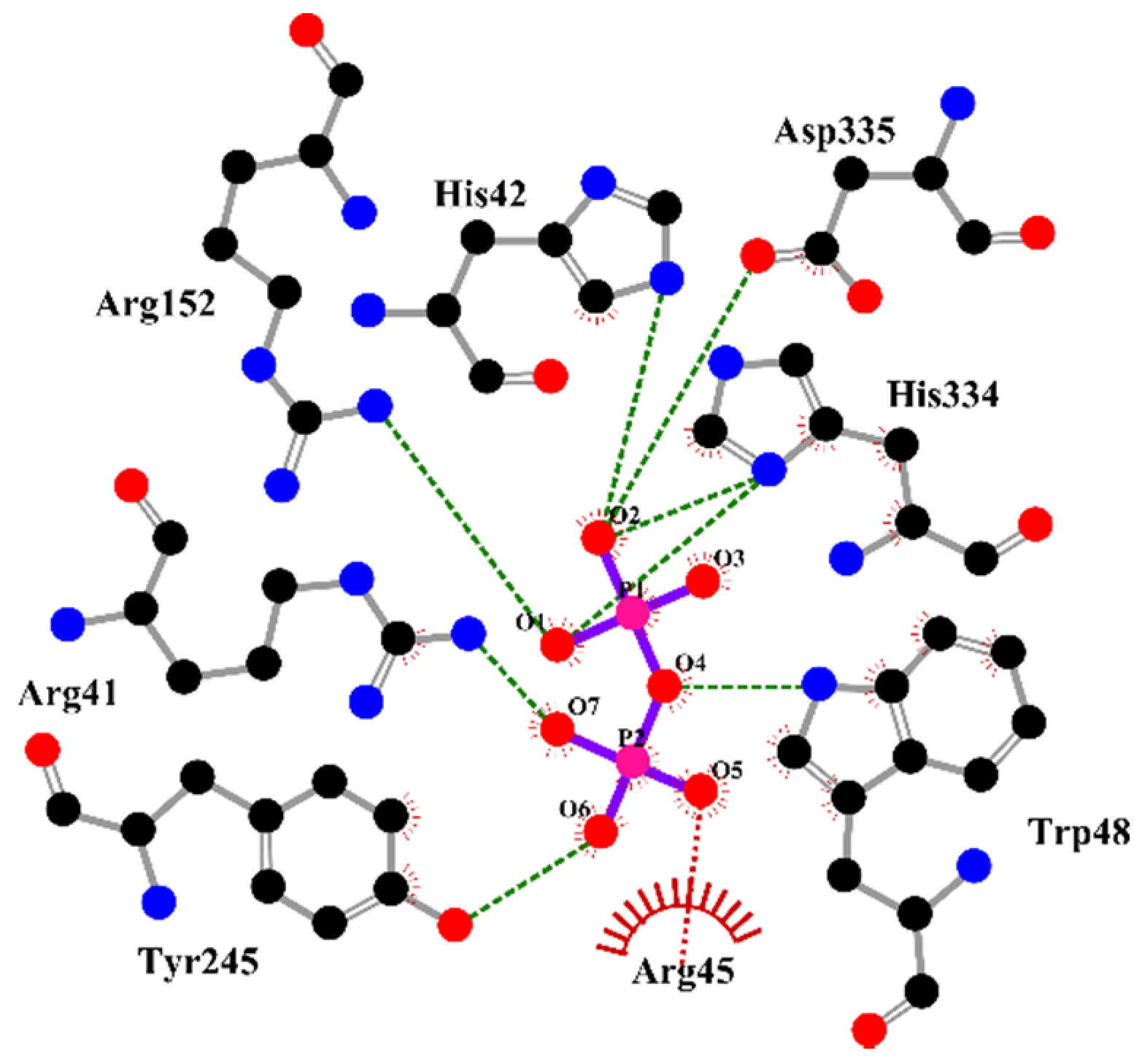
2.4. The Active-Site Entrance of EhHAPp49, an Apparent Molecular Sieve
2.5. EhHAPp49 Is a Non-Canonical Phosphatase
3. Materials and Methods
3.1. Database Searching and Protein Structure Analyses
3.2. Materials
3.3. Strains, Plasmids and Primers
3.4. EhHAPp49 PCR Amplification
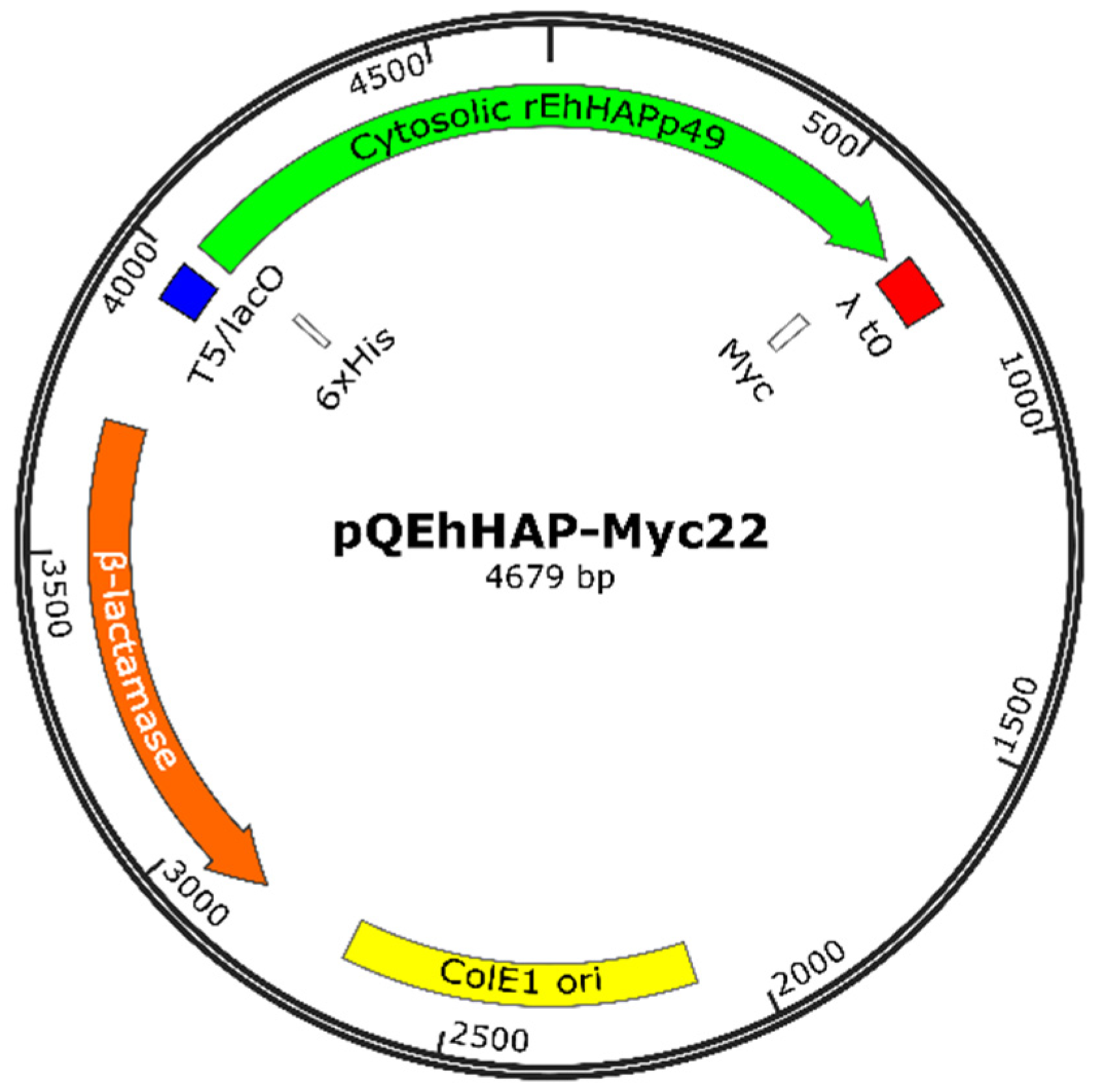
3.5. Construction of Recombinant Plasmids Harboring EhHAPp49
3.6. Construction of the Recombinant Plasmid Expressing EhHAPp49
3.7. Production of rEhHAPp49
3.8. EhHAPp49 Enzyme Activity Assays
3.8.1. Phosphatase Activity Assay
3.8.2. Phytase Activity Assay
3.8.3. Pyrophosphatase Activity Assay
3.8.4. MG Colorimetric Assay
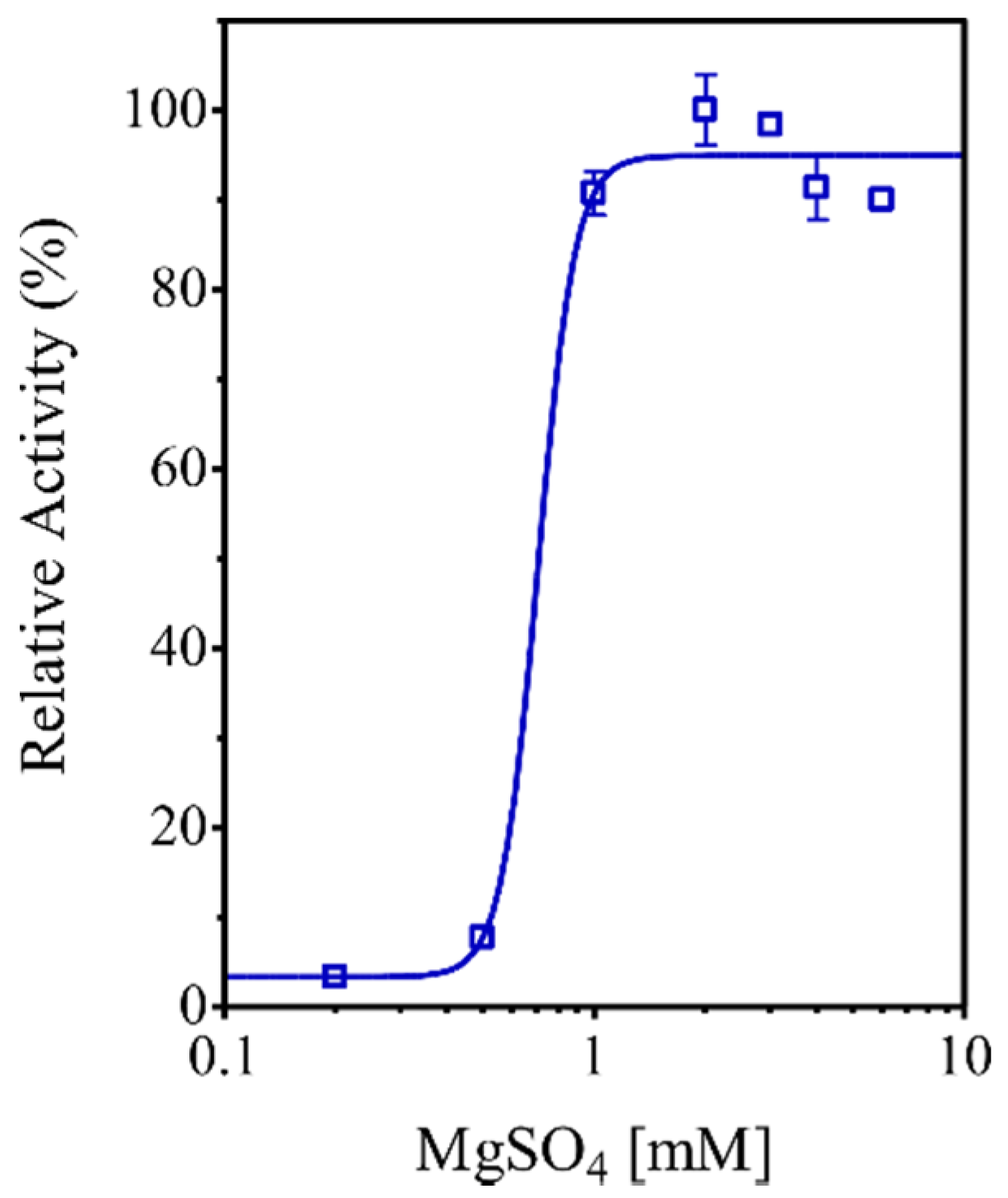
3.9. Characterization of the EhHAPp49 PPase Activity
3.10. Data Analysis
3.11. In Silico Analysis of the EhHAPp49 Ligand-Binding Site
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dick, C.F.; Dos-Santos, A.L.A.; Meyer-Fernandes, J.R. Inorganic Phosphate as an Important Regulator of Phosphatases. Enzyme Res. 2011, 2011, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Fontanillo, M.; Köhn, M. Phosphatases: Their Roles in Cancer and Their Chemical Modulators. Adv. Exp. Med. Biol. 2016, 917, 209–240. [Google Scholar] [CrossRef]
- Dotaniya, M.L.; Aparna, K.; Dotaniya, C.K.; Singh, M.; Regar, K.L. Role of Soil Enzymes in Sustainable Crop Production. In Enzymes in Food Biotechnology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 569–589. ISBN 978-0-12-813280-7. [Google Scholar]
- Vincent, J.B.; Crowder, M.W.; Averill, B.A. Hydrolysis of Phosphate Monoesters: A Biological Problem with Multiple Chemical Solutions. Trends Biochem. Sci. 1992, 17, 105–110. [Google Scholar] [CrossRef]
- Rigden, D.J. The Histidine Phosphatase Superfamily: Structure and Function. Biochem. J. 2008, 409, 333–348. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Wang, F.; Li, H.; Ji, J.; Cao, Z.; Lyu, J.; Shi, X.; Zhu, Y.; Zhang, C.; Guo, F.; et al. Prostatic Acid Phosphatase (PAP) Predicts Prostate Cancer Progress in a Population-Based Study: The Renewal of PAP? Dis. Markers 2019, 2019, 1–10. [Google Scholar] [CrossRef]
- Bhavsar, K.; Khire, J.M. Current Research and Future Perspectives of Phytase Bioprocessing. RSC Adv. 2014, 4, 26677–26691. [Google Scholar] [CrossRef]
- Reddy, C.S.; Kim, S.-C.; Kaul, T. Genetically Modified Phytase Crops Role in Sustainable Plant and Animal Nutrition and Ecological Development: A Review. 3 Biotech 2017, 7, 195. [Google Scholar] [CrossRef]
- Jatuwong, K.; Suwannarach, N.; Kumla, J.; Penkhrue, W.; Kakumyan, P.; Lumyong, S. Bioprocess for Production, Characteristics, and Biotechnological Applications of Fungal Phytases. Front. Microbiol. 2020, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Kumanan, T.; Sujanitha, V.; Sreeharan, N. Amoebic Liver Abscess: A Neglected Tropical Disease. Lancet Infect. Dis. 2020, 20, 160–162. [Google Scholar] [CrossRef]
- Loftus, B.; Anderson, I.; Davies, R.; Alsmark, U.C.M.; Samuelson, J.; Amedeo, P.; Roncaglia, P.; Berriman, M.; Hirt, R.P.; Mann, B.J.; et al. The Genome of the Protist Parasite Entamoeba histolytica. Nature 2005, 433, 865–868. [Google Scholar] [CrossRef]
- Anwar, T.; Gourinath, S. Analysis of the Protein Phosphotome of Entamoeba histolytica Reveals an Intricate Phosphorylation Network. PLoS ONE 2013, 8, e78714. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Huston, C.D.; Oue, M.; Mann, B.J.; Petri, W.A.; Kita, K.; Nozaki, T. Kinetics and Strain Variation of Phagosome Proteins of Entamoeba histolytica by Proteomic Analysis. Mol. Biochem. Parasitol. 2006, 145, 171–183. [Google Scholar] [CrossRef] [PubMed]
- LaCount, M.W.; Handy, G.; Lebioda, L. Structural Origins of L(+)-Tartrate Inhibition of Human Prostatic Acid Phosphatase. J. Biol. Chem. 1998, 273, 30406–30409. [Google Scholar] [CrossRef] [Green Version]
- Jakob, C.G.; Lewinski, K.; Kuciel, R.; Ostrowski, W.; Lebioda, L. Crystal Structure of Human Prostatic Acid Phosphatase. Prostate 2000, 42, 211–218. [Google Scholar] [CrossRef]
- Ortlund, E.; LaCount, M.W.; Lebioda, L. Crystal Structures of Human Prostatic Acid Phosphatase in Complex with a Phosphate Ion and Alpha-Benzylaminobenzylphosphonic Acid Update the Mechanistic Picture and Offer New Insights into Inhibitor Design. Biochemistry 2003, 42, 383–389. [Google Scholar] [CrossRef]
- Li, J.; Dong, Y.; Lü, X.; Wang, L.; Peng, W.; Zhang, X.C.; Rao, Z. Crystal Structures and Biochemical Studies of Human Lysophosphatidic Acid Phosphatase Type 6. Protein Cell 2013, 4, 548–561. [Google Scholar] [CrossRef] [Green Version]
- Dhatwalia, R.; Singh, H.; Reilly, T.J.; Tanner, J.J. Crystal Structure and Tartrate Inhibition of Legionella pneumophila Histidine Acid Phosphatase. Arch. Biochem. Biophys. 2015, 585, 32–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechtel, T.J.; Weerapana, E. From Structure to Redox: The Diverse Functional Roles of Disulfides and Implications in Disease. Proteomics 2017, 17, 1600391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sowdhamini, R.; Srinivasan, N.; Shoichet, B.; Santi, D.V.; Ramakrishnan, C.; Balaram, P. Stereochemical Modeling of Disulfide Bridges. Criteria for Introduction into Proteins by Site-Directed Mutagenesis. Protein Eng. Des. Sel. 1989, 3, 95–103. [Google Scholar] [CrossRef]
- Hudson, D.A.; Gannon, S.A.; Thorpe, C. Oxidative Protein Folding: From Thiol-Disulfide Exchange Reactions to the Redox Poise of the Endoplasmic Reticulum. Free Radic. Biol. Med. 2015, 80, 171–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, P.K. Enzymes: Principles and Biotechnological Applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]
- Colussi, F.; Garcia, W.; Rosseto, F.R.; de Mello, B.L.S.; de Oliveira Neto, M.; Polikarpov, I. Effect of pH and Temperature on the Global Compactness, Structure, and Activity of Cellobiohydrolase Cel7A from Trichoderma harzianum. Eur. Biophys. J. EBJ 2012, 41, 89–98. [Google Scholar] [CrossRef]
- Lim, D.; Golovan, S.; Forsberg, C.W.; Jia, Z. Crystal Structures of Escherichia coli Phytase and Its Complex with Phytate. Nat. Struct. Biol. 2000, 7, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Golovan, S.; Wang, G.; Zhang, J.; Forsberg, C.W. Characterization and Overproduction of the Escherichia coli AppA Encoded Bifunctional Enzyme That Exhibits Both Phytase and Acid Phosphatase Activities. Can. J. Microbiol. 2000, 46, 59–71. [Google Scholar] [CrossRef]
- Edelheit, O.; Hanukoglu, A.; Hanukoglu, I. Simple and Efficient Site-Directed Mutagenesis Using Two Single-Primer Reactions in Parallel to Generate Mutants for Protein Structure–function Studies. BMC Biotechnol. 2009, 9, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, K.M.; Lee, W.; Yu, J.-R.; Ahnn, J. PYP-1, Inorganic Pyrophosphatase, Is Required for Larval Development and Intestinal Function in C. elegans. FEBS Lett. 2007, 581, 5445–5453. [Google Scholar] [CrossRef] [Green Version]
- Kajander, T.; Kellosalo, J.; Goldman, A. Inorganic Pyrophosphatases: One Substrate, Three Mechanisms. FEBS Lett. 2013, 587, 1863–1869. [Google Scholar] [CrossRef] [Green Version]
- Farquharson, K.L. Life of PPi: Soluble PPases and H+-PPase Act Cooperatively to Keep Pyrophosphate Levels in Check. Plant Cell 2018, 30, 951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baykov, A.A.; Anashkin, V.A.; Salminen, A.; Lahti, R. Inorganic Pyrophosphatases of Family II-Two Decades after Their Discovery. FEBS Lett. 2017, 591, 3225–3234. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.S.; Cho, Y.; Kim, Y.-J.; Lho, T.-O.; Cha, S.-S.; Lee, J.-H.; Kang, S.G. A Novel Inorganic Pyrophosphatase in Thermococcus Onnurineus NA1. FEMS Microbiol. Lett. 2009, 300, 68–74. [Google Scholar] [CrossRef] [Green Version]
- May, A.; Berger, S.; Hertel, T.; Köck, M. The Arabidopsis thaliana Phosphate Starvation Responsive Gene AtPPsPase1 Encodes a Novel Type of Inorganic Pyrophosphatase. Biochim. Biophys. Acta BBA Gen. Subj. 2011, 1810, 178–185. [Google Scholar] [CrossRef]
- Huang, H.; Patskovsky, Y.; Toro, R.; Farelli, J.D.; Pandya, C.; Almo, S.C.; Allen, K.N.; Dunaway-Mariano, D. Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides Thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase. Biochemistry 2011, 50, 8937–8949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahs, S.; Lujan, P.; Köhn, M. Approaches to Study Phosphatases. ACS Chem. Biol. 2016, 11, 2944–2961. [Google Scholar] [CrossRef]
- Medvedev, K.E.; Kinch, L.N.; Schaeffer, R.D.; Grishin, N.V. Functional Analysis of Rossmann-like Domains Reveals Convergent Evolution of Topology and Reaction Pathways. PLOS Comput. Biol. 2019, 15, e1007569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooperman, B.S.; Baykov, A.A.; Lahti, R. Evolutionary Conservation of the Active Site of Soluble Inorganic Pyrophosphatase. Trends Biochem. Sci. 1992, 17, 262–266. [Google Scholar] [CrossRef]
- Heikinheimo, P.; Lehtonen, J.; Baykov, A.; Lahti, R.; Cooperman, B.S.; Goldman, A. The Structural Basis for Pyrophosphatase Catalysis. Structure 1996, 4, 1491–1508. [Google Scholar] [CrossRef] [Green Version]
- Brock, D.A.; van Egmond, W.N.; Shamoo, Y.; Hatton, R.D.; Gomer, R.H. A 60-Kilodalton Protein Component of the Counting Factor Complex Regulates Group Size in Dictyostelium discoideum. Eukaryot. Cell 2006, 5, 1532–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, W.; Gomer, R.H. Combining Experiments and Modelling to Understand Size Regulation in Dictyostelium Discoideum. J. R. Soc. Interface 2008, 5, S49–S58. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. Clifton NJ 2017, 1654, 39–54. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feig, M. Local Protein Structure Refinement via Molecular Dynamics Simulations with LocPREFMD. J. Chem. Inf. Model. 2016, 56, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation. Protein Sci. Publ. Protein Soc. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; ISBN 978-0-87969-577-4. [Google Scholar]
- Diamond, L.S.; Harlow, D.R.; Cunnick, C.C. A New Medium for the Axenic Cultivation of Entamoeba histolytica and Other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 431–432. [Google Scholar] [CrossRef]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight Regulation, Modulation, and High-Level Expression by Vectors Containing the Arabinose PBAD Promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Zor, T.; Selinger, Z. Linearization of the Bradford Protein Assay Increases Its Sensitivity: Theoretical and Experimental Studies. Anal. Biochem. 1996, 236, 302–308. [Google Scholar] [CrossRef] [Green Version]
- Baykov, A.A.; Evtushenko, O.A.; Avaeva, S.M. A Malachite Green Procedure for Orthophosphate Determination and Its Use in Alkaline Phosphatase-Based Enzyme Immunoassay. Anal. Biochem. 1988, 171, 266–270. [Google Scholar] [CrossRef]
- Roche, D.B.; McGuffin, L.J. In Silico Identification and Characterization of Protein-Ligand Binding Sites. Methods Mol. Biol. Clifton NJ 2016, 1414, 1–21. [Google Scholar] [CrossRef] [Green Version]
- McGuffin, L.J.; Adiyaman, R.; Maghrabi, A.H.A.; Shuid, A.N.; Brackenridge, D.A.; Nealon, J.O.; Philomina, L.S. IntFOLD: An Integrated Web Resource for High Performance Protein Structure and Function Prediction. Nucleic Acids Res. 2019, 47, W408–W413. [Google Scholar] [CrossRef]
- Roche, D.B.; Tetchner, S.J.; McGuffin, L.J. FunFOLD: An Improved Automated Method for the Prediction of Ligand Binding Residues Using 3D Models of Proteins. BMC Bioinform. 2011, 12, 160. [Google Scholar] [CrossRef] [Green Version]
- Roche, D.B.; Buenavista, M.T.; McGuffin, L.J. The FunFOLD2 Server for the Prediction of Protein-Ligand Interactions. Nucleic Acids Res. 2013, 41, W303–W307. [Google Scholar] [CrossRef] [PubMed]
- Roche, D.B.; Buenavista, M.T.; McGuffin, L.J. FunFOLDQA: A Quality Assessment Tool for Protein-Ligand Binding Site Residue Predictions. PLoS ONE 2012, 7, e38219. [Google Scholar] [CrossRef] [Green Version]
- Bitencourt-Ferreira, G.; de Azevedo, W.F. Molecular Docking Simulations with ArgusLab. Methods Mol. Biol. Clifton NJ 2019, 2053, 203–220. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminformatics 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully Automated Protein–Ligand Interaction Profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]


| pH (Buffer §) | Substrate | ||
|---|---|---|---|
| p-Nitrophenyl Phosphate 1 | Phytic Acid 2 | Sodium Pyrophosphate 3 | |
| 5.0 (Na-acetate) | 0.05 ± 0.005 | 0.02 ± 0.002 | 0.2 ± 0.02 |
| 7.0 (Tris-HCl) | Not significant | Not significant | 2.8 ± 0.04 |
| 9.0 (Tris-HCl) | Not significant | Not significant | 9.3 ± 0.05 |
| pH | KM (µM) | kcat (min−1) | kcat/KM (µM−1 min−1) § |
|---|---|---|---|
| 5.0 | 34 ± 6 | 0.2 ± 0.0 | 0.006 |
| 7.0 | 9 ± 2 | 3.6 ± 0.1 | 0.400 |
| 9.0 | 12 ± 2 | 10.6 ± 0.2 | 0.883 |
| Strain | Genotype | Source |
|---|---|---|
| ER2738 | F´ proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10(TetR) / fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5 | NEB 1 |
| SHuffle | fhuA2 [lon] ompT ahpC gal λatt::pNEB3-r1-cDsbC (SpecR, lacIq) ΔtrxB sulA11 R(mcr-73::miniTn10-TetS)2 [dcm] R(zgb-210::Tn10 -TetS) endA1 Δgor ∆(mcrC-mrr)114::IS10 | NEB 1 |
| Plasmid | Features | Source |
| pBluescript SK(-) | Lactose regulation, ColE1 origin, AmpR | Stratagene |
| pBPelB-BHX-Myc | pBluescript-based, PelB signal, Myc tag | This study |
| pBPelB-EhHAP-Myc | pBluescript-based, periplasmic EhHAPp49 (Myc-tagged) | This study |
| pBAD33 | Arabinose regulation, p15A origin, CmR | ATCC 2 |
| pBAD-PelB-EhHAP-Myc | pBAD33-based, periplasmic EhHAPp49 (Myc-tagged) | This study |
| pQE30 | Lactose regulation, ColE1 origin, AmpR | Qiagen |
| pQEhHAP-Myc22 | pQE30-based, cytosolic EhHAPp49 (6xHis- and Myc-tagged) | This study |
| Primer | Sequence (5´ to 3´) | Endonuclease |
| EHHAPF | catcatggatccgatttaacatactgtgaagtacctgaa tt | BamHI |
| EHHAPR | catcatctcgagctgatttttggcattacagtctga | XbaI |
| M13_RV | caggaaacagctatgac | None |
| H3MYCR | catcataagcttttacagatcctcttcagagatgagt | HindIII |
| BAD_FW | cggcgtcacactttgctatgc | None |
| BAD_RV | tgggaccaccgcgctactgcc | None |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terán-Ramírez, C.; Mares-Alejandre, R.E.; Estrada-González, A.L.; Muñoz-Muñoz, P.L.A.; Ramos-Ibarra, M.A. Structure–Function Relationship Study of a Secretory Amoebic Phosphatase: A Computational-Experimental Approach. Int. J. Mol. Sci. 2021, 22, 2164. https://doi.org/10.3390/ijms22042164
Terán-Ramírez C, Mares-Alejandre RE, Estrada-González AL, Muñoz-Muñoz PLA, Ramos-Ibarra MA. Structure–Function Relationship Study of a Secretory Amoebic Phosphatase: A Computational-Experimental Approach. International Journal of Molecular Sciences. 2021; 22(4):2164. https://doi.org/10.3390/ijms22042164
Chicago/Turabian StyleTerán-Ramírez, Celina, Rosa E. Mares-Alejandre, Ana L. Estrada-González, Patricia L. A. Muñoz-Muñoz, and Marco A. Ramos-Ibarra. 2021. "Structure–Function Relationship Study of a Secretory Amoebic Phosphatase: A Computational-Experimental Approach" International Journal of Molecular Sciences 22, no. 4: 2164. https://doi.org/10.3390/ijms22042164







