Effects of Non-Thermal Plasma on Yeast Saccharomyces cerevisiae
Abstract
1. Introduction
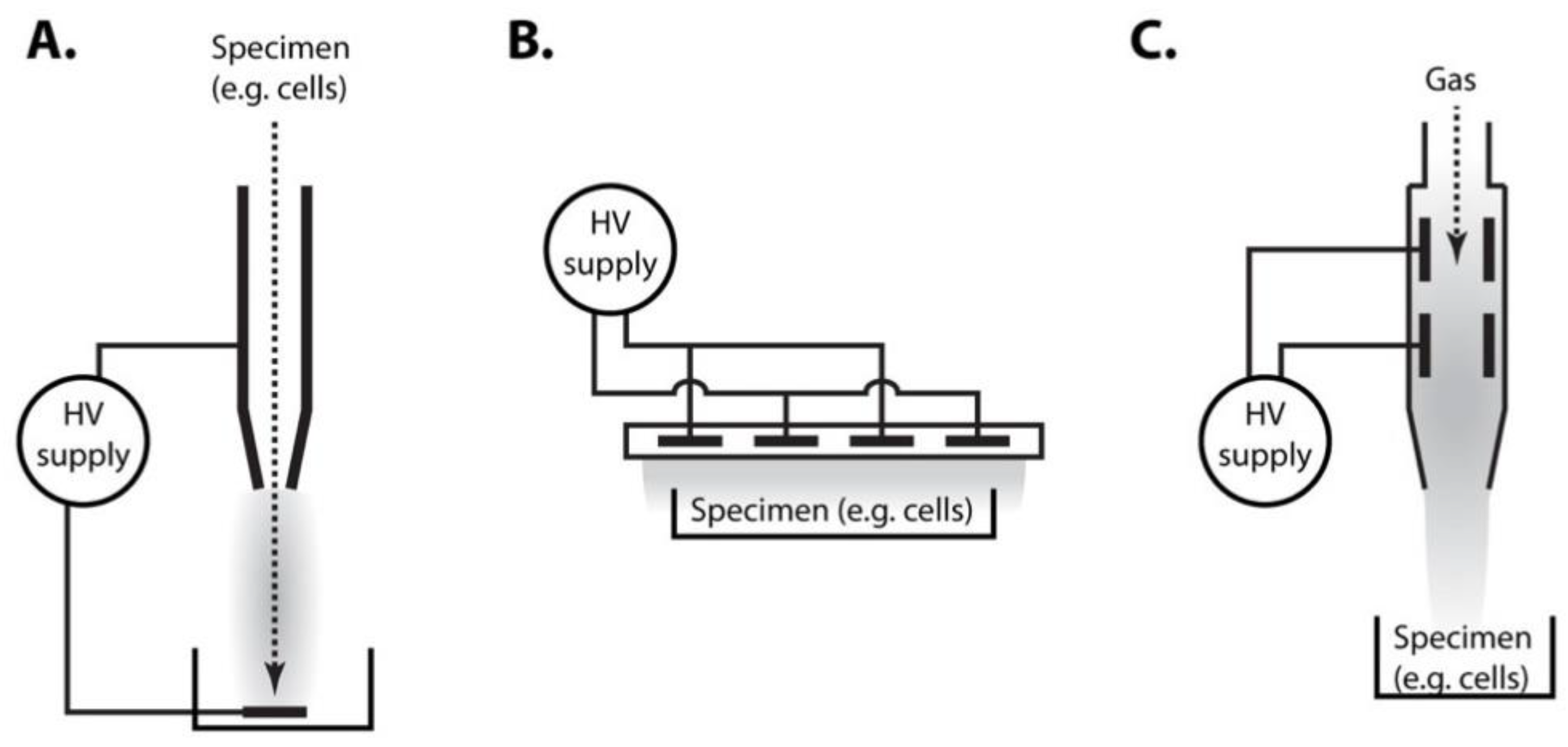
2. Plasma Treatment of Yeast Cells
3. Plasma-Induced Cell Death in Yeast
4. Effect of Plasma on Membranes and Energy Metabolism
5. Plasma Damage to DNA
6. Genetic Dissection of Plasma-Induced Changes in Yeast
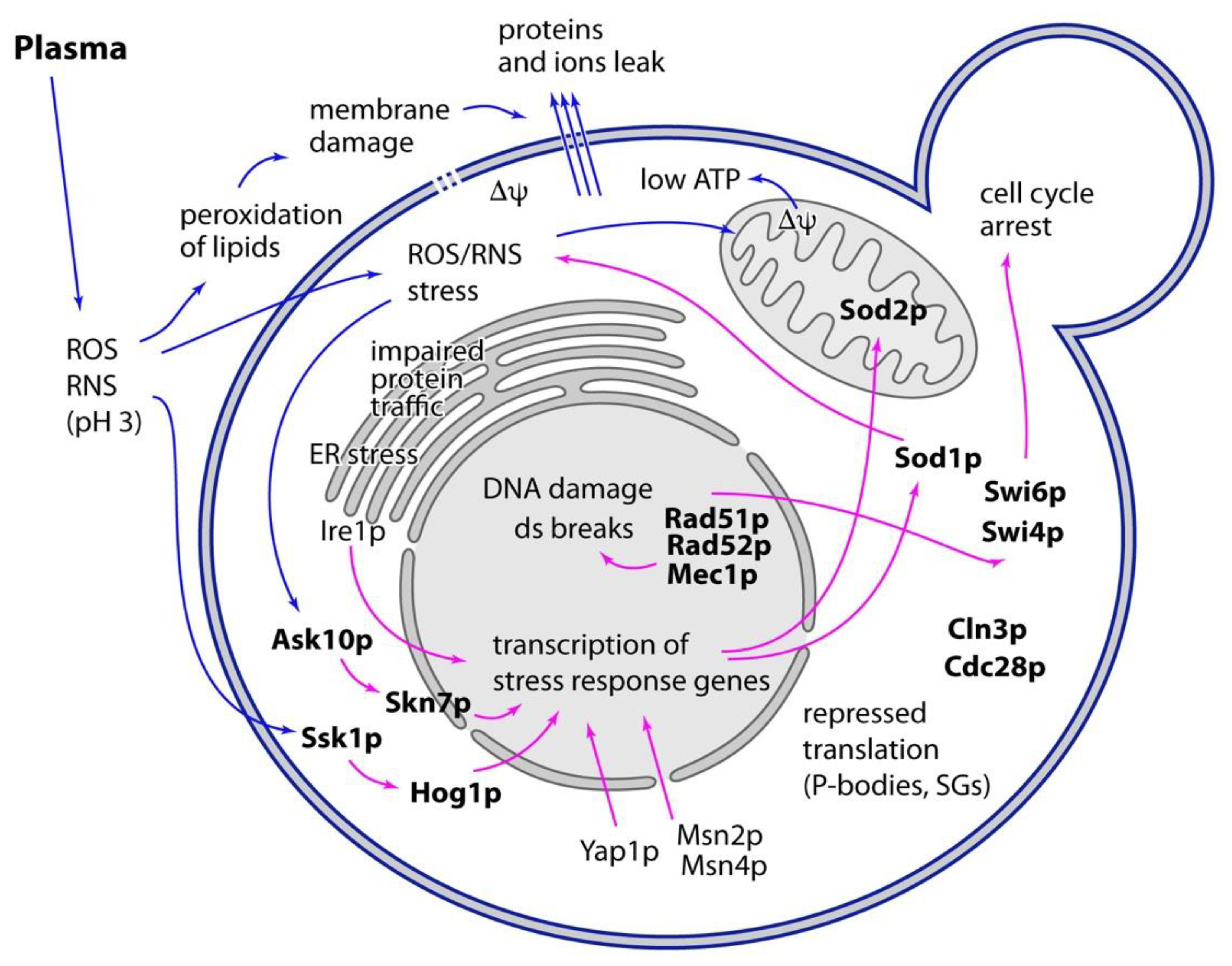
7. Plasma Treatment Induces Multiple Stress-Response Pathways
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Δψ | transmembrane potential |
| ARTP | atmospheric and room temperature plasmas |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DTT | dithiotreitol |
| ER | endoplasmic reticulum |
| GFP | green fluorescent protein |
| MAPK | mitogen-activated protein kinase |
| MDA | malonedialdehyde |
| PBS | phosphate buffered saline |
| PTP | permeability transition pore |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| SG | stress granule |
| STRE | stress response element |
| TMRM | tetramethylrhodamine methyl ester |
| YPD | yeast extract, peptone, dextrose (cultivation medium) |
References
- Laroussi, M.; Kong, M.; Morfill, G.; Stolz, W. Plasma Medicine: Applications of Low-Temperature Gas Plasmas in Medicine and Biology; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Machala, Z.; Hensel, K.; Akishev, Y. Plasma for bio-decontamination, medicine and food security. In NATO Science for Peace and Security Series A: Chemistry and Biology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Fridman, A.A.; Friedman, G.G. Plasma Medicine; John Wiley & Sons: Chichester, UK, 2013. [Google Scholar]
- Von Woedtke, T.; Reuter, S.; Masur, K.; Weltmann, K.-D. Plasmas for medicine. Phys. Rep. 2013, 530, 291–320. [Google Scholar] [CrossRef]
- Scholtz, V.; Pazlarová, J.; Soušková, H.; Khun, J.; Julák, J. Nonthermal plasma—A tool for decontamination and disinfection. Biotechnol. Adv. 2015, 33, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- Misra, N.; Segat, A.; Cullen, P. Atmospheric-Pressure Non-Thermal Plasma Decontamination of Foods. Adv. Food Biotechnol. 2015, 565–574. [Google Scholar]
- Puač, N.; Gherardi, M.; Shiratani, M. Plasma agriculture: A rapidly emerging field. Plasma Process. Polym. 2018, 15, 1700174. [Google Scholar] [CrossRef]
- Bernhardt, T.; Semmler, M.L.; Schäfer, M.; Bekeschus, S.; Emmert, S.; Boeckmann, L. Plasma medicine: Applications of cold atmospheric pressure plasma in dermatology. Oxid. Med. Cell. Longev. 2019, 2019, 3873928. [Google Scholar] [CrossRef]
- Bekeschus, S.; Schmidt, A.; Weltmann, K.-D.; von Woedtke, T. The plasma jet kINPen—A powerful tool for wound healing. Clin. Plasma Med. 2016, 4, 19–28. [Google Scholar] [CrossRef]
- Tanaka, H.; Ishikawa, K.; Mizuno, M.; Toyokuni, S.; Kajiyama, H.; Kikkawa, F.; Metelmann, H.-R.; Hori, M. State of the art in medical applications using non-thermal atmospheric pressure plasma. Rev. Mod. Plasma Phys. 2017, 1, 3. [Google Scholar] [CrossRef]
- Metelmann, H.-R.; Von Woedtke, T.; Weltmann, K.-D. Comprehensive Clinical Plasma Medicine: Cold Physical Plasma for Medical Application; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Botstein, D.; Chervitz, S.A.; Cherry, J.M. Yeast as a model organism. Science 1997, 277, 1259–1260. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Engelhardt, S.; Herker, E.; Lehmann, N.; Maldener, C.; Proksch, A.; Wissing, S.; Frohlich, K.U. Apoptosis in yeast: A new model system with applications in cell biology and medicine. Curr. Genet. 2002, 41, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Sarto-Jackson, I.; Tomaska, L. How to bake a brain: Yeast as a model neuron. Curr. Genet. 2016, 62, 347–370. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.Y. Yeast for virus research. Microb. Cell 2017, 4, 311–330. [Google Scholar] [CrossRef]
- Mager, W.H.; Winderickx, J. Yeast as a model for medical and medicinal research. Trends Pharmacol. Sci. 2005, 26, 265–273. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.; Kim, T.; Lee, J.-S. The functional study of human proteins using humanized yeast. J. Microbiol. 2020, 58, 343–349. [Google Scholar] [CrossRef]
- Lee, K.; Paek, K.H.; Ju, W.T.; Lee, Y. Sterilization of bacteria, yeast, and bacterial endospores by atmospheric-pressure cold plasma using helium and oxygen. J. Microbiol. 2006, 44, 269–275. [Google Scholar] [PubMed]
- Machala, Z.; Jedlovský, I.; Chládeková, L.; Pongrác, B.; Giertl, D.; Janda, M.; Šikurová, L.; Polčic, P. DC discharges in atmospheric air for bio-decontamination–spectroscopic methods for mechanism identification. Eur. Phys. J. D 2009, 54, 195–204. [Google Scholar] [CrossRef]
- Barekzi, N.; Laroussi, M. Dose-dependent killing of leukemia cells by low-temperature plasma. J. Phys. D Appl. Phys. 2012, 45, 422002. [Google Scholar] [CrossRef]
- Bauer, G. Intercellular singlet oxygen-mediated bystander signaling triggered by long-lived species of cold atmospheric plasma and plasma-activated medium. Redox Biol. 2019, 26, 101301. [Google Scholar] [CrossRef] [PubMed]
- Sousa, J.S.; Bauville, G.; Lacour, B.; Puech, V.; Touzeau, M.; Ravanat, J.-L. DNA oxidation by singlet delta oxygen produced by atmospheric pressure microdischarges. Appl. Phys. Lett. 2010, 97, 141502. [Google Scholar] [CrossRef]
- Machala, Z.; Tarabova, B.; Hensel, K.; Spetlikova, E.; Sikurová, L.; Lukes, P. Formation of ROS and RNS in Water Electro-S prayed through Transient Spark Discharge in Air and their Bactericidal Effects. Plasma Process. Polym. 2013, 10, 649–659. [Google Scholar] [CrossRef]
- Lukes, P.; Dolezalova, E.; Sisrova, I.; Clupek, M. Aqueous-phase chemistry and bactericidal effects from an air discharge plasma in contact with water: Evidence for the formation of peroxynitrite through a pseudo-second-order post-discharge reaction of H2O2 and HNO2. Plasma Sources Sci. Technol. 2014, 23, 015019. [Google Scholar] [CrossRef]
- Polčic, P.; Pakosová, L.; Chovančíková, P.; Machala, Z. Reactive cold plasma particles generate oxidative stress in yeast but do not trigger apoptosis. Can. J. Microbiol. 2018, 64, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.H.; Kim, Y.H.; Lee, J.Y.; Shim, G.B.; Uhm, H.S.; Park, G.; Choi, E.H. Effects of background fluid on the efficiency of inactivating yeast with non-thermal atmospheric pressure plasma. PLoS ONE 2013, 8, e66231. [Google Scholar] [CrossRef]
- Carmelo, V.; Bogaerts, P.; Sá-Correia, I. Activity of plasma membrane H+-ATPase and expression of PMA1 and PMA2 genes in Saccharomyces cerevisiae cells grown at optimal and low pH. Arch. Microbiol. 1996, 166, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Chung, T.; Bae, S.; Leem, S. Induction of apoptosis in human breast cancer cells by a pulsed atmospheric pressure plasma jet. Appl. Phys. Lett. 2010, 97, 023702. [Google Scholar] [CrossRef]
- Ahn, H.J.; Kim, K.I.; Kim, G.; Moon, E.; Yang, S.S.; Lee, J.S. Atmospheric-pressure plasma jet induces apoptosis involving mitochondria via generation of free radicals. PLoS ONE 2011, 6, e28154. [Google Scholar] [CrossRef]
- Tan, X.; Zhao, S.; Lei, Q.; Lu, X.; He, G.; Ostrikov, K. Single-cell-precision microplasma-induced cancer cell apoptosis. PLoS ONE 2014, 9, e101299. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Li, W.; Zhou, B. Caspase-independent apoptosis in yeast. Biochim. Biophys. Acta 2008, 1783, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, C.; Falcone, C. Caspase-dependent apoptosis in yeast. Biochim. Biophys. Acta 2008, 1783, 1320–1327. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Eisenberg, T.; Buttner, S.; Meisinger, C.; Kroemer, G.; Madeo, F. Apoptosis in yeast: Triggers, pathways, subroutines. Cell Death Differ. 2010, 17, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Strich, R. Programmed Cell Death Initiation and Execution in Budding Yeast. Genetics 2015, 200, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Aguilera, A.; Austriaco, N.; Ayscough, K.; Balzan, R.; Bar-Nun, S.; Barrientos, A.; Belenky, P.; et al. Guidelines and recommendations on yeast cell death nomenclature. Microb. Cell 2018, 5, 4–31. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Feng, H.; Liang, Y.; Zhang, Q.; Tian, Y.; Su, B.; Zhang, J.; Fang, J. An atmospheric-pressure cold plasma leads to apoptosis in Saccharomyces cerevisiae by accumulating intracellular reactive oxygen species and calcium. J. Phys. D Appl. Phys. 2013, 46, 285401. [Google Scholar] [CrossRef]
- Čtvrtečková, L.; Pichová, A.; Scholtz, V.; Khun, J.; Julák, J. Non-thermal plasma-induced apoptosis in yeast Saccharomyces cerevisiae. Contrib. Plasma Phys. 2019, 59, e201800064. [Google Scholar] [CrossRef]
- Madeo, F.; Herker, E.; Maldener, C.; Wissing, S.; Lachelt, S.; Herlan, M.; Fehr, M.; Lauber, K.; Sigrist, S.J.; Wesselborg, S.; et al. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 2002, 9, 911–917. [Google Scholar] [CrossRef]
- Wissing, S.; Ludovico, P.; Herker, E.; Buttner, S.; Engelhardt, S.M.; Decker, T.; Link, A.; Proksch, A.; Rodrigues, F.; Corte-Real, M.; et al. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 2004, 166, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Buttner, S.; Eisenberg, T.; Carmona-Gutierrez, D.; Ruli, D.; Knauer, H.; Ruckenstuhl, C.; Sigrist, C.; Wissing, S.; Kollroser, M.; Frohlich, K.U.; et al. Endonuclease G regulates budding yeast life and death. Mol. Cell 2007, 25, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Munoz, M.F.; Arguelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed]
- Reilly, C.A.; Aust, S.D. Measurement of lipid peroxidation. Curr. Protoc. Toxicol. 2001. [Google Scholar] [CrossRef]
- Joshi, S.G.; Cooper, M.; Yost, A.; Paff, M.; Ercan, U.K.; Fridman, G.; Friedman, G.; Fridman, A.; Brooks, A.D. Nonthermal dielectric-barrier discharge plasma-induced inactivation involves oxidative DNA damage and membrane lipid peroxidation in Escherichia coli. Antimicrob. Agents Chemother. 2011, 55, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Doležalová, E.; Lukeš, P. Membrane damage and active but nonculturable state in liquid cultures of Escherichia coli treated with an atmospheric pressure plasma jet. Bioelectrochemistry 2015, 103, 7–14. [Google Scholar] [CrossRef]
- Martin, C.E.; Oh, C.S.; Jiang, Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 2007, 1771, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Stukey, J.E.; McDonough, V.M.; Martin, C.E. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 1989, 264, 16537–16544. [Google Scholar] [CrossRef]
- Xu, H.; Zhu, Y.; Du, M.; Wang, Y.; Ju, S.; Ma, R.; Jiao, Z. Subcellular mechanism of microbial inactivation during water disinfection by cold atmospheric-pressure plasma. Water Res. 2021, 188, 116513. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, P.; Rasola, A.; Forte, M.; Lippe, G. The Mitochondrial Permeability Transition Pore: Channel Formation by F-ATP Synthase, Integration in Signal Transduction, and Role in Pathophysiology. Physiol. Rev. 2015, 95, 1111–1155. [Google Scholar] [CrossRef]
- Yamada, A.; Yamamoto, T.; Yoshimura, Y.; Gouda, S.; Kawashima, S.; Yamazaki, N.; Yamashita, K.; Kataoka, M.; Nagata, T.; Terada, H.; et al. Ca2+-induced permeability transition can be observed even in yeast mitochondria under optimized experimental conditions. Biochim. Biophys. Acta 2009, 1787, 1486–1491. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carraro, M.; Bernardi, P. Calcium and reactive oxygen species in regulation of the mitochondrial permeability transition and of programmed cell death in yeast. Cell Calcium 2016, 60, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Arjunan, K.P.; Sharma, V.K.; Ptasinska, S. Effects of atmospheric pressure plasmas on isolated and cellular DNA—A review. Int. J. Mol. Sci. 2015, 16, 2971–3016. [Google Scholar] [CrossRef]
- Wende, K.; Straßenburg, S.; Haertel, B.; Harms, M.; Holtz, S.; Barton, A.; Masur, K.; von Woedtke, T.; Lindequist, U. Atmospheric pressure plasma jet treatment evokes transient oxidative stress in HaCaT keratinocytes and influences cell physiology. Cell Biol. Int. 2014, 38, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Brun, P.; Brun, P.; Vono, M.; Venier, P.; Tarricone, E.; Deligianni, V.; Martines, E.; Zuin, M.; Spagnolo, S.; Cavazzana, R.; et al. Disinfection of ocular cells and tissues by atmospheric-pressure cold plasma. PLoS ONE 2012, 7, e33245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.F.; Li, H.P.; Wang, L.Y.; Zhang, C.; Xing, X.H.; Bao, C.Y. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl. Microbiol. Biotechnol. 2014, 98, 5387–5396. [Google Scholar] [CrossRef] [PubMed]
- Ottenheim, C.; Nawrath, M.; Wu, J.C. Microbial mutagenesis by atmospheric and room-temperature plasma (ARTP): The latest development. Bioresour. Bioprocess. 2018, 5, 12. [Google Scholar] [CrossRef]
- Tian, T.; Wu, D.; Ng, C.T.; Yang, H.; Sun, J.; Liu, J.; Lu, J. A multiple-step strategy for screening Saccharomyces cerevisiae strains with improved acid tolerance and aroma profiles. Appl. Microbiol. Biotechnol. 2020, 104, 3097–3107. [Google Scholar] [CrossRef]
- Symington, L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002, 66, 630–670. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, K.; Kang, K.T.; Lee, J.S.; Yang, S.S.; Chung, W.H. Atmospheric-pressure plasma jet induces DNA double-strand breaks that require a Rad51-mediated homologous recombination for repair in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2014, 560, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gasior, S.L.; Olivares, H.; Ear, U.; Hari, D.M.; Weichselbaum, R.; Bishop, D.K. Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 2001, 98, 8411–8418. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Kowalczykowski, S.C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002, 277, 31663–31672. [Google Scholar] [CrossRef] [PubMed]
- Weinert, T.A.; Kiser, G.L.; Hartwell, L.H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994, 8, 652–665. [Google Scholar] [CrossRef]
- Longhese, M.P.; Foiani, M.; Muzi-Falconi, M.; Lucchini, G.; Plevani, P. DNA damage checkpoint in budding yeast. EMBO J. 1998, 17, 5525–5528. [Google Scholar] [CrossRef]
- Guo, L.; Zhao, Y.; Liu, D.; Liu, Z.; Chen, C.; Xu, R.; Tian, M.; Wang, X.; Chen, H.; Kong, M.G. Cold atmospheric-pressure plasma induces DNA-protein crosslinks through protein oxidation. Free Radic. Res. 2018, 52, 783–798. [Google Scholar] [CrossRef]
- Fielden, J.; Ruggiano, A.; Popovic, M.; Ramadan, K. DNA protein crosslink proteolysis repair: From yeast to premature ageing and cancer in humans. DNA Repair 2018, 71, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Chvalova, K.; Brabec, V.; Kasparkova, J. Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic Acids Res. 2007, 35, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Groehler, A.T.; Michaelson-Richie, E.D.; Villalta, P.W.; Campbell, C.; Tretyakova, N.Y. Mass Spectrometry Based Proteomics Study of Cisplatin-Induced DNA-Protein Cross-Linking in Human Fibrosarcoma (HT1080) Cells. Chem. Res. Toxicol. 2017, 30, 980–995. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef]
- Rothstein, R. [19] Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991, 194, 281–301. [Google Scholar] [PubMed]
- Gardner, J.M.; Jaspersen, S.L. Manipulating the yeast genome: Deletion, mutation, and tagging by PCR. In Yeast Genetics; Springer: Berlin/Heidelberg, Germany, 2014; pp. 45–78. [Google Scholar]
- Chang, E.C.; Crawford, B.F.; Hong, Z.; Bilinski, T.; Kosman, D.J. Genetic and biochemical characterization of Cu,Zn superoxide dismutase mutants in Saccharomyces cerevisiae. J. Biol. Chem. 1991, 266, 4417–4424. [Google Scholar] [CrossRef]
- Costa, V.; Amorim, M.A.; Reis, E.; Quintanilha, A.; Moradas-Ferreira, P. Mitochondrial superoxide dismutase is essential for ethanol tolerance of Saccharomyces cerevisiae in the post-diauxic phase. Microbiology 1997, 143 Pt 5, 1649–1656. [Google Scholar] [CrossRef]
- Galiazzo, F.; Labbe-Bois, R. Regulation of Cu,Zn- and Mn-superoxide dismutase transcription in Saccharomyces cerevisiae. FEBS Lett. 1993, 315, 197–200. [Google Scholar] [CrossRef]
- Spevak, W.; Fessl, F.; Rytka, J.; Traczyk, A.; Skoneczny, M.; Ruis, H. Isolation of the catalase T structural gene of Saccharomyces cerevisiae by functional complementation. Mol. Cell. Biol. 1983, 3, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.; Fessl, F.; Traczyk, A.; Rytka, J.; Ruis, H. Isolation of the catalase A gene of Saccharomyces cerevisiae by complementation of the cta1 mutation. Mol. Gen. Genet. 1985, 200, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.Y.; Drescher, D.; Kujumdzieva, A.V.; Schmitt, M.J. Dual targeting of yeast catalase A to peroxisomes and mitochondria. Biochem. J. 2004, 380, 393–400. [Google Scholar] [CrossRef]
- Izawa, S.; Inoue, Y.; Kimura, A. Importance of catalase in the adaptive response to hydrogen peroxide: Analysis of acatalasaemic Saccharomyces cerevisiae. Biochem. J. 1996, 320 Pt 1, 61–67. [Google Scholar] [CrossRef]
- Feng, H.; Wang, R.; Sun, P.; Wu, H.; Liu, Q.; Fang, J.; Zhu, W.; Li, F.; Zhang, J. A study of eukaryotic response mechanisms to atmospheric pressure cold plasma by using Saccharomyces cerevisiae single gene mutants. Appl. Phys. Lett. 2010, 97, 131501. [Google Scholar] [CrossRef]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 2005, 569, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Proft, M.; Remize, F.; Tamas, M.; Serrano, R.; Thevelein, J.M.; Hohmann, S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 2001, 40, 1067–1083. [Google Scholar] [CrossRef] [PubMed]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- Maeda, T.; Wurgler-Murphy, S.M.; Saito, H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 1994, 369, 242–245. [Google Scholar] [CrossRef]
- Li, S.; Ault, A.; Malone, C.L.; Raitt, D.; Dean, S.; Johnston, L.H.; Deschenes, R.J.; Fassler, J.S. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998, 17, 6952–6962. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; North, S.; Bussey, H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 1993, 175, 6908–6915. [Google Scholar] [CrossRef] [PubMed]
- Krems, B.; Charizanis, C.; Entian, K.D. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 1996, 29, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar] [CrossRef]
- Page, N.; Sheraton, J.; Brown, J.L.; Stewart, R.C.; Bussey, H. Identification of ASK10 as a multicopy activator of Skn7p-dependent transcription of a HIS3 reporter gene. Yeast 1996, 12, 267–272. [Google Scholar] [CrossRef]
- Cohen, T.J.; Lee, K.; Rutkowski, L.H.; Strich, R. Ask10p mediates the oxidative stress-induced destruction of the Saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p. Eukaryot. Cell 2003, 2, 962–970. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mendenhall, M.D.; Hodge, A.E. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998, 62, 1191–1243. [Google Scholar] [CrossRef]
- de Bruin, R.A.; McDonald, W.H.; Kalashnikova, T.I.; Yates, J., 3rd; Wittenberg, C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 2004, 117, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Nishikawa, J.L.; Tang, X.; Millman, J.S.; Schub, O.; Breitkreuz, K.; Dewar, D.; Rupes, I.; Andrews, B.; Tyers, M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 2004, 117, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Wittenberg, C.; Reed, S.I. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene 2005, 24, 2746–2755. [Google Scholar] [CrossRef]
- Sidorova, J.M.; Breeden, L.L. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997, 11, 3032–3045. [Google Scholar] [CrossRef] [PubMed]
- Sidorova, J.M.; Breeden, L.L. Rad53 checkpoint kinase phosphorylation site preference identified in the Swi6 protein of Saccharomyces cerevisiae. Mol. Cell. Biol. 2003, 23, 3405–3416. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.; Tactacan, C.M.; Tan, S.X.; Lin, R.C.; Wouters, M.A.; Dawes, I.W. Cell cycle sensing of oxidative stress in Saccharomyces cerevisiae by oxidation of a specific cysteine residue in the transcription factor Swi6p. J. Biol. Chem. 2011, 286, 5204–5214. [Google Scholar] [CrossRef] [PubMed]
- Enserink, J.M.; Hombauer, H.; Huang, M.E.; Kolodner, R.D. Cdc28/Cdk1 positively and negatively affects genome stability in S. cerevisiae. J. Cell Biol. 2009, 185, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Temple, M.D.; Perrone, G.G.; Dawes, I.W. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 2005, 15, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Kuge, S.; Arita, M.; Murayama, A.; Maeta, K.; Izawa, S.; Inoue, Y.; Nomoto, A. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 2001, 21, 6139–6150. [Google Scholar] [CrossRef]
- Okazaki, S.; Tachibana, T.; Naganuma, A.; Mano, N.; Kuge, S. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol. Cell 2007, 27, 675–688. [Google Scholar] [CrossRef]
- Itooka, K.; Takahashi, K.; Izawa, S. Fluorescence microscopic analysis of antifungal effects of cold atmospheric pressure plasma in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2016, 100, 9295–9304. [Google Scholar] [CrossRef]
- Martinez-Pastor, M.T.; Marchler, G.; Schuller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996, 15, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Görner, W.; Durchschlag, E.; Martinez-Pastor, M.T.; Estruch, F.; Ammerer, G.; Hamilton, B.; Ruis, H.; Schüller, C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998, 12, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.P.; McEntee, K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5777–5782. [Google Scholar] [CrossRef] [PubMed]
- Kelley, R.; Ideker, T. Genome-wide fitness and expression profiling implicate Mga2 in adaptation to hydrogen peroxide. PLoS Genet. 2009, 5, e1000488. [Google Scholar] [CrossRef] [PubMed]
- MacIsaac, K.D.; Wang, T.; Gordon, D.B.; Gifford, D.K.; Stormo, G.D.; Fraenkel, E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinf. 2006, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Itooka, K.; Takahashi, K.; Kimata, Y.; Izawa, S. Cold atmospheric pressure plasma causes protein denaturation and endoplasmic reticulum stress in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2018, 102, 2279–2288. [Google Scholar] [CrossRef]
- Bosl, B.; Grimminger, V.; Walter, S. The molecular chaperone Hsp104--a molecular machine for protein disaggregation. J. Struct. Biol. 2006, 156, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Zhou, Y.; Ng, R.W.; Kung Hf, H.F.; Jin, D.Y. Cooperation of yeast peroxiredoxins Tsa1p and Tsa2p in the cellular defense against oxidative and nitrosative stress. J. Biol. Chem. 2002, 277, 5385–5394. [Google Scholar] [CrossRef]
- Rand, J.D.; Grant, C.M. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell 2006, 17, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.A.; Jacobsen, K. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell 1984, 38, 841–849. [Google Scholar] [CrossRef]
- Werner-Washburne, M.; Stone, D.E.; Craig, E.A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 2568–2577. [Google Scholar] [CrossRef]
- Hernández-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Domínguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The unfolded protein response pathway in the yeast Kluyveromyces lactis. a comparative view among yeast species. Cells 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ng, B.S.; Thibault, G. Endoplasmic reticulum stress response in yeast and humans. Biosci. Rep. 2014, 34, e00118. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.S.; Shamu, C.E.; Walter, P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 1993, 73, 1197–1206. [Google Scholar] [CrossRef]
- Cox, J.S.; Walter, P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 1996, 87, 391–404. [Google Scholar] [CrossRef]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450. [Google Scholar] [CrossRef]
- Parker, R.; Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 2007, 25, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Franks, T.M.; Lykke-Andersen, J. The control of mRNA decapping and P-body formation. Mol. Cell 2008, 32, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Parker, R. Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Balagopal, V.; Parker, R. Polysomes, P bodies and stress granules: States and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 2009, 21, 403–408. [Google Scholar] [CrossRef]
- Buchan, J.R.; Muhlrad, D.; Parker, R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008, 183, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Yamamoto, Y.; Izawa, S. Severe ethanol stress induces assembly of stress granules in Saccharomyces cerevisiae. Yeast 2011, 28, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Groušl, T.; Ivanov, P.; Frydlová, I.; Vašicová, P.; Janda, F.; Vojtová, J.; Malínská, K.; Malcová, I.; Nováková, L.; Janošková, D.; et al. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 2009, 122, 2078–2088. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Izawa, S. Adaptive response in stress granule formation and bulk translational repression upon a combined stress of mild heat shock and mild ethanol stress in yeast. Genes Cells 2013, 18, 974–984. [Google Scholar] [CrossRef]
- Stulić, V.; Vukušić, T.; Butorac, A.; Popović, D.; Herceg, Z. Proteomic analysis of Saccharomyces cerevisiae response to plasma treatment. Int. J. Food Microbiol. 2019, 292, 171–183. [Google Scholar] [CrossRef]
- Guaragnella, N.; Stirpe, M.; Marzulli, D.; Mazzoni, C.; Giannattasio, S. Acid Stress Triggers Resistance to Acetic Acid-Induced Regulated Cell Death through Hog1 Activation Which Requires RTG2 in Yeast. Oxid. Med. Cell. Longev. 2019, 2019, 4651062. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polčic, P.; Machala, Z. Effects of Non-Thermal Plasma on Yeast Saccharomyces cerevisiae. Int. J. Mol. Sci. 2021, 22, 2247. https://doi.org/10.3390/ijms22052247
Polčic P, Machala Z. Effects of Non-Thermal Plasma on Yeast Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2021; 22(5):2247. https://doi.org/10.3390/ijms22052247
Chicago/Turabian StylePolčic, Peter, and Zdenko Machala. 2021. "Effects of Non-Thermal Plasma on Yeast Saccharomyces cerevisiae" International Journal of Molecular Sciences 22, no. 5: 2247. https://doi.org/10.3390/ijms22052247
APA StylePolčic, P., & Machala, Z. (2021). Effects of Non-Thermal Plasma on Yeast Saccharomyces cerevisiae. International Journal of Molecular Sciences, 22(5), 2247. https://doi.org/10.3390/ijms22052247






