Controlling Ratios of Plasmid-Based Double Cut Donor and CRISPR/Cas9 Components to Enhance Targeted Integration of Transgenes in Chinese Hamster Ovary Cells
Abstract
1. Introduction
2. Results and Discussion
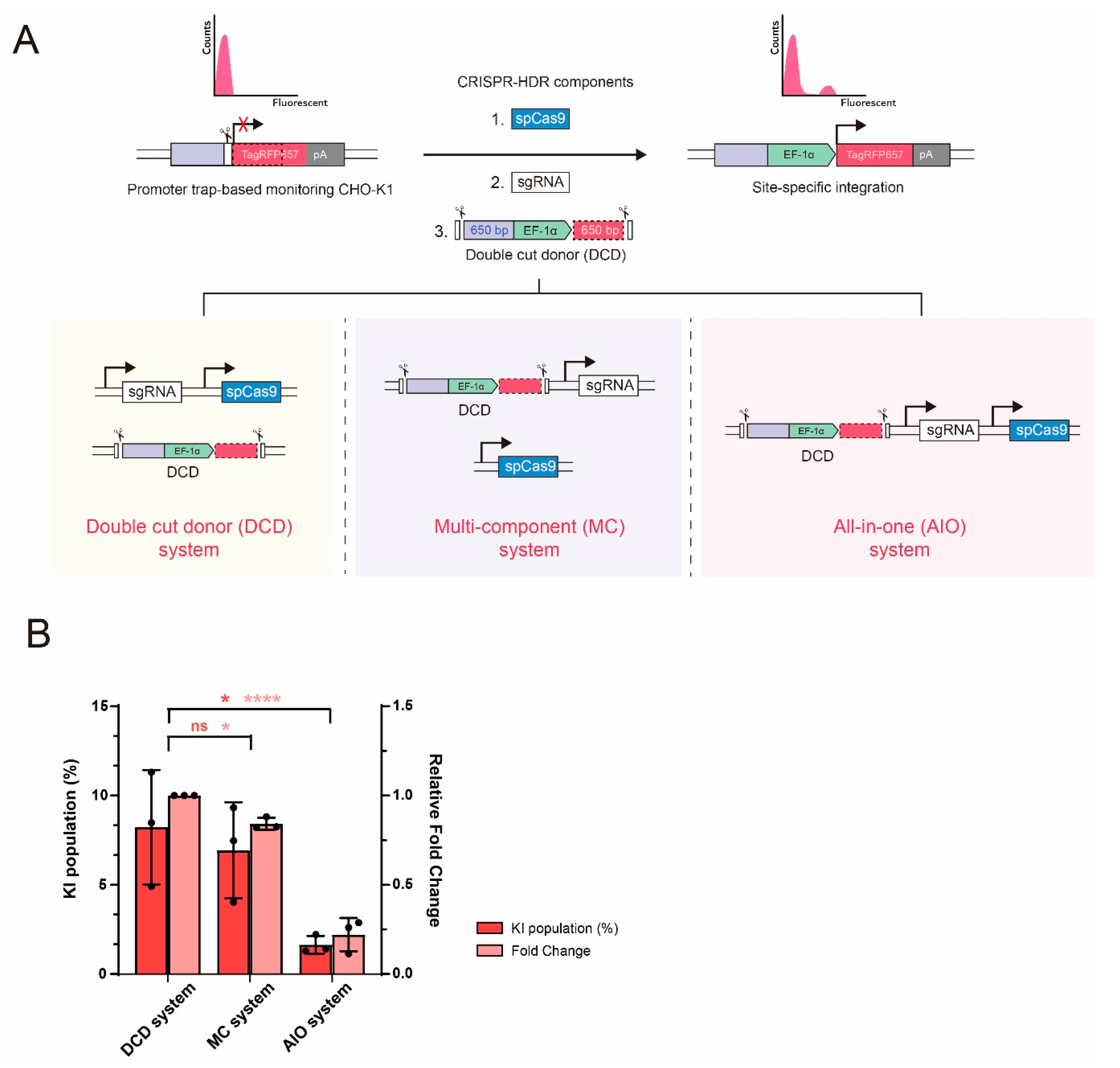
2.1. Effect of Different CRISPR-HDR Vector Configuration on KI Efficiency in CHO Cells
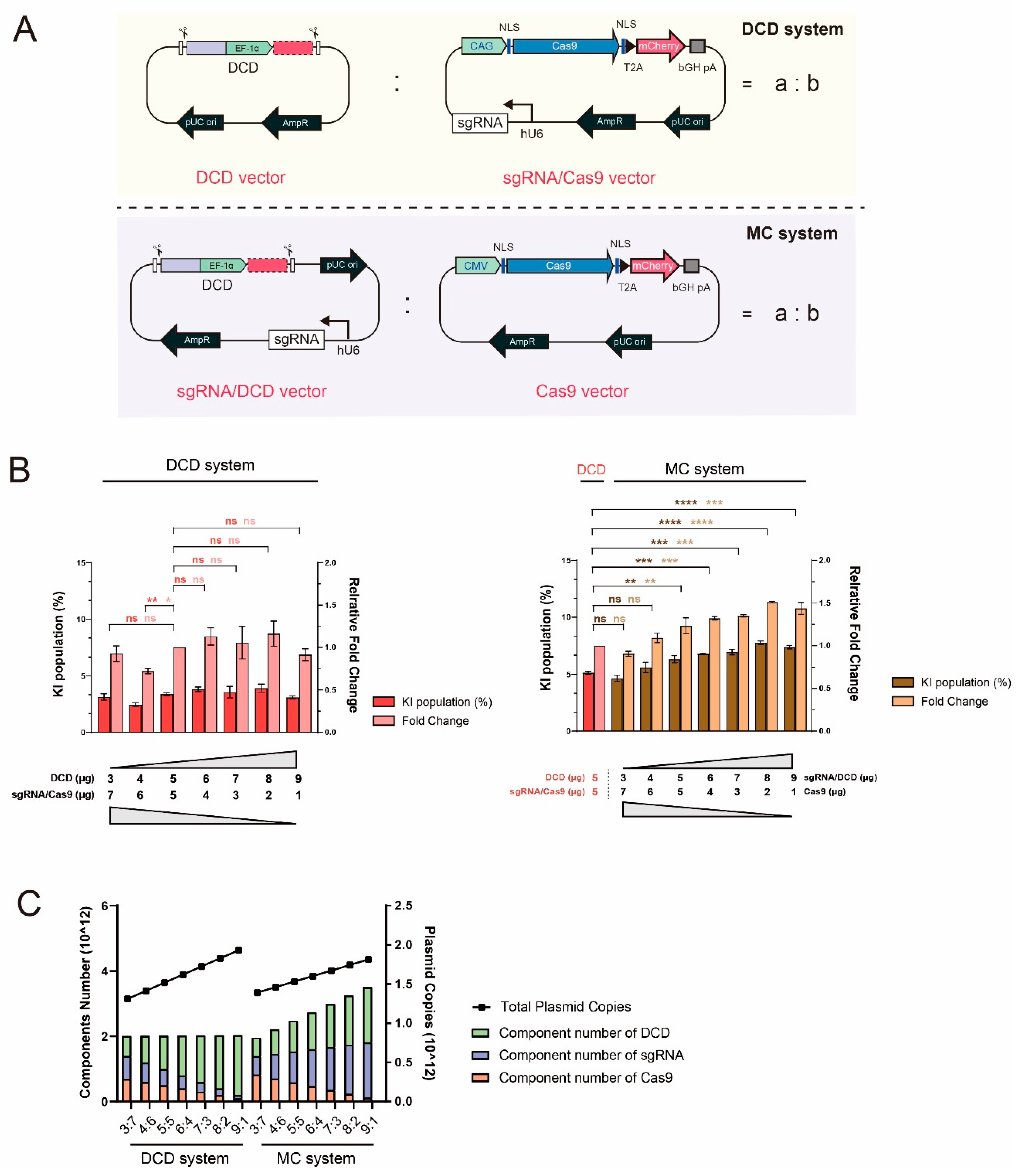
2.2. CRISPR-HDR Component Titration and Ratio Optimization in the DCD and MC Systems
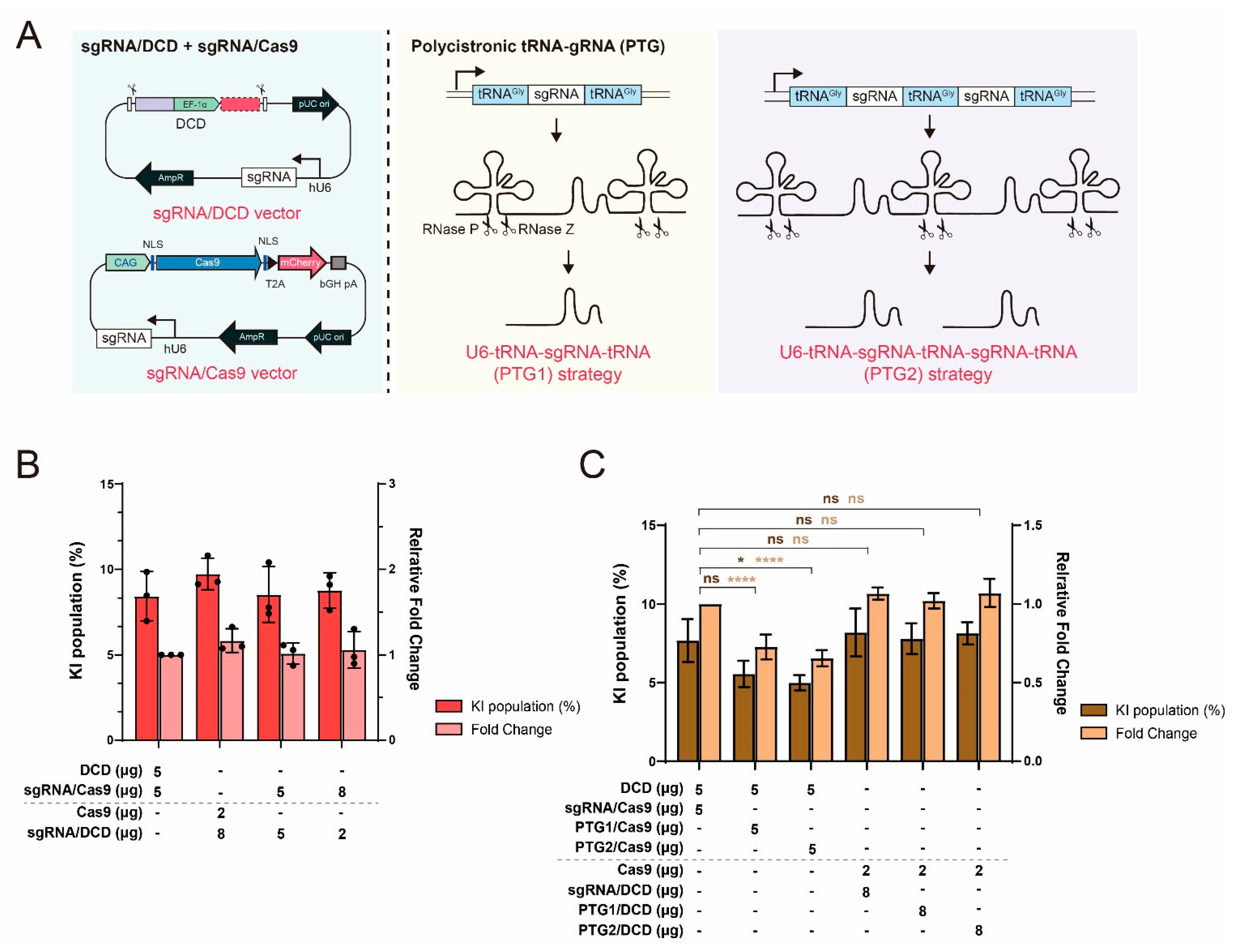
2.3. Limited Effect of sgRNA Supplementation in the MC System
2.4. Application of Simultaneous MC–TI and Split-Intein MC System on Double TI
3. Materials and Methods
3.1. Plasmid Design and Construction
3.2. Cell Lines and Cell Culture
3.3. KI Efficiency Measurement Using Flow Cytometry
3.4. Plasmid Copy Number Calculation
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, J.Y.; Kim, Y.G.; Lee, G.M. CHO cells in biotechnology for production of recombinant proteins: Current state and further potential. Appl. Microbiol. Biotechnol. 2012, 93, 917–930. [Google Scholar] [CrossRef]
- Shin, S.W.; Lee, J.S. CHO Cell Line Development and Engineering via Site-specific Integration: Challenges and Opportunities. Biotechnol. Bioproc. E. 2020, 25, 633–645. [Google Scholar] [CrossRef]
- Lee, J.S.; Kallehauge, T.B.; Pedersen, L.E.; Kildegaard, H.F. Site-specific integration in CHO cells mediated by CRISPR/Cas9 and homology-directed DNA repair pathway. Sci. Rep. 2015, 5, 8572. [Google Scholar] [CrossRef]
- Lee, J.S.; Grav, L.M.; Lewis, N.E.; Faustrup Kildegaard, H. CRISPR/Cas9-mediated genome engineering of CHO cell factories: Application and perspectives. Biotechnol. J. 2015, 10, 979–994. [Google Scholar] [CrossRef]
- Bosshard, S.; Duroy, P.O.; Mermod, N. A role for alternative end-joining factors in homologous recombination and genome editing in Chinese hamster ovary cells. DNA Repair 2019, 82, 102691. [Google Scholar] [CrossRef]
- Lee, J.S.; Grav, L.M.; Pedersen, L.E.; Lee, G.M.; Kildegaard, H.F. Accelerated homology-directed targeted integration of transgenes in Chinese hamster ovary cells via CRISPR/Cas9 and fluorescent enrichment. Biotechnol. Bioeng. 2016, 113, 2518–2523. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, J.; Luo, M.; Luo, H.; Zhao, M.; Han, L.; Zhang, M.; Yang, H.; Xie, Y.; Jiang, H.; et al. Rapid development of stable transgene CHO cell lines by CRISPR/Cas9-mediated site-specific integration into C12orf35. Appl. Microbiol. Biotechnol. 2018, 102, 6105–6117. [Google Scholar] [CrossRef]
- Grav, L.M.; Sergeeva, D.; Lee, J.S.; de Mas, I.M.; Lewis, N.E.; Andersen, M.R.; Nielsen, L.K.; Lee, G.M.; Kildegaard, H.F. Minimizing Clonal Variation during Mammalian Cell Line Engineering for Improved Systems Biology Data Generation. ACS Synth. Biol. 2018, 7, 2148–2159. [Google Scholar] [CrossRef]
- Kouranova, E.; Forbes, K.; Zhao, G.; Warren, J.; Bartels, A.; Wu, Y.; Cui, X. CRISPRs for Optimal Targeting: Delivery of CRISPR Components as DNA, RNA, and Protein into Cultured Cells and Single-Cell Embryos. Hum. Gene Ther. 2016, 27, 464–475. [Google Scholar] [CrossRef]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef]
- Shin, S.W.; Lee, J.S. Optimized CRISPR/Cas9 strategy for homology-directed multiple targeted integration of transgenes in CHO cells. Biotechnol. Bioeng. 2020, 117, 1895–1903. [Google Scholar] [CrossRef]
- Zhang, J.P.; Li, X.L.; Li, G.H.; Chen, W.; Arakaki, C.; Botimer, G.D.; Baylink, D.; Zhang, L.; Wen, W.; Fu, Y.W.; et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017, 18, 35. [Google Scholar] [CrossRef]
- Pinder, J.; Salsman, J.; Dellaire, G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015, 43, 9379–9392. [Google Scholar] [CrossRef]
- Li, K.; Wang, G.; Andersen, T.; Zhou, P.; Pu, W.T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS ONE 2014, 9, e105779. [Google Scholar] [CrossRef]
- Song, F.; Stieger, K. Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks. Mol. Ther. Nucleic Acids 2017, 7, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Potter, J.; Kumar, S.; Ravinder, N.; Chesnut, J.D. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J. Biotechnol. 2017, 241, 136–146. [Google Scholar] [CrossRef]
- Sakuma, T.; Nishikawa, A.; Kume, S.; Chayama, K.; Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014, 4, 5400. [Google Scholar] [CrossRef]
- Moore, R.; Spinhirne, A.; Lai, M.J.; Preisser, S.; Li, Y.; Kang, T.; Bleris, L. CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res. 2015, 43, 1297–1303. [Google Scholar] [CrossRef]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar] [CrossRef] [PubMed]
- Ihry, R.J.; Worringer, K.A.; Salick, M.R.; Frias, E.; Ho, D.; Theriault, K.; Kommineni, S.; Chen, J.; Sondey, M.; Ye, C.; et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018, 24, 939–946. [Google Scholar] [CrossRef]
- Geisinger, J.M.; Stearns, T. CRISPR/Cas9 treatment causes extended TP53-dependent cell cycle arrest in human cells. Nucleic Acids Res. 2020, 48, 9067–9081. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Dong, F.; Xie, K.; Chen, Y.; Yang, Y.; Mao, Y. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem. Biophys. Res. Commun. 2017, 482, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Knapp, D.J.H.F.; Michaels, Y.S.; Jamilly, M.; Ferry, Q.R.V.; Barbosa, H.; Milne, T.A.; Fulga, T.A. Decoupling tRNA promoter and processing activities enables specific Pol-II Cas9 guide RNA expression. Nat. Commun. 2019, 10, 1490. [Google Scholar] [CrossRef]
- Yan, Q.; Xu, K.; Xing, J.; Zhang, T.; Wang, X.; Wei, Z.; Ren, C.; Liu, Z.; Shao, S.; Zhang, Z. Multiplex CRISPR/Cas9-based genome engineering enhanced by Drosha-mediated sgRNA-shRNA structure. Sci. Rep. 2016, 6, 38970. [Google Scholar] [CrossRef]
- Dean, D.A.; Dean, B.S.; Muller, S.; Smith, L.C. Sequence requirements for plasmid nuclear import. Exp. Cell Res. 1999, 253, 713–722. [Google Scholar] [CrossRef]
- Mesika, A.; Grigoreva, I.; Zohar, M.; Reich, Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol. Ther. 2001, 3, 653–657. [Google Scholar] [CrossRef]
- Daer, R.M.; Cutts, J.P.; Brafman, D.A.; Haynes, K.A. The Impact of Chromatin Dynamics on Cas9-Mediated Genome Editing in Human Cells. ACS Synth. Biol. 2017, 6, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, J.; Janssen, J.M.; Gonçalves, M.A.F.V. The Chromatin Structure Differentially Impacts High-Specificity CRISPR-Cas9 Nuclease Strategies. Mol. Ther. Nucleic Acids 2017, 8, 558–563. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.W.; Kim, D.; Lee, J.S. Controlling Ratios of Plasmid-Based Double Cut Donor and CRISPR/Cas9 Components to Enhance Targeted Integration of Transgenes in Chinese Hamster Ovary Cells. Int. J. Mol. Sci. 2021, 22, 2407. https://doi.org/10.3390/ijms22052407
Shin SW, Kim D, Lee JS. Controlling Ratios of Plasmid-Based Double Cut Donor and CRISPR/Cas9 Components to Enhance Targeted Integration of Transgenes in Chinese Hamster Ovary Cells. International Journal of Molecular Sciences. 2021; 22(5):2407. https://doi.org/10.3390/ijms22052407
Chicago/Turabian StyleShin, Sung Wook, Dongwoo Kim, and Jae Seong Lee. 2021. "Controlling Ratios of Plasmid-Based Double Cut Donor and CRISPR/Cas9 Components to Enhance Targeted Integration of Transgenes in Chinese Hamster Ovary Cells" International Journal of Molecular Sciences 22, no. 5: 2407. https://doi.org/10.3390/ijms22052407
APA StyleShin, S. W., Kim, D., & Lee, J. S. (2021). Controlling Ratios of Plasmid-Based Double Cut Donor and CRISPR/Cas9 Components to Enhance Targeted Integration of Transgenes in Chinese Hamster Ovary Cells. International Journal of Molecular Sciences, 22(5), 2407. https://doi.org/10.3390/ijms22052407





