Nitric Oxide (NO) Scaffolds the Peroxisomal Protein–Protein Interaction Network in Higher Plants
Abstract
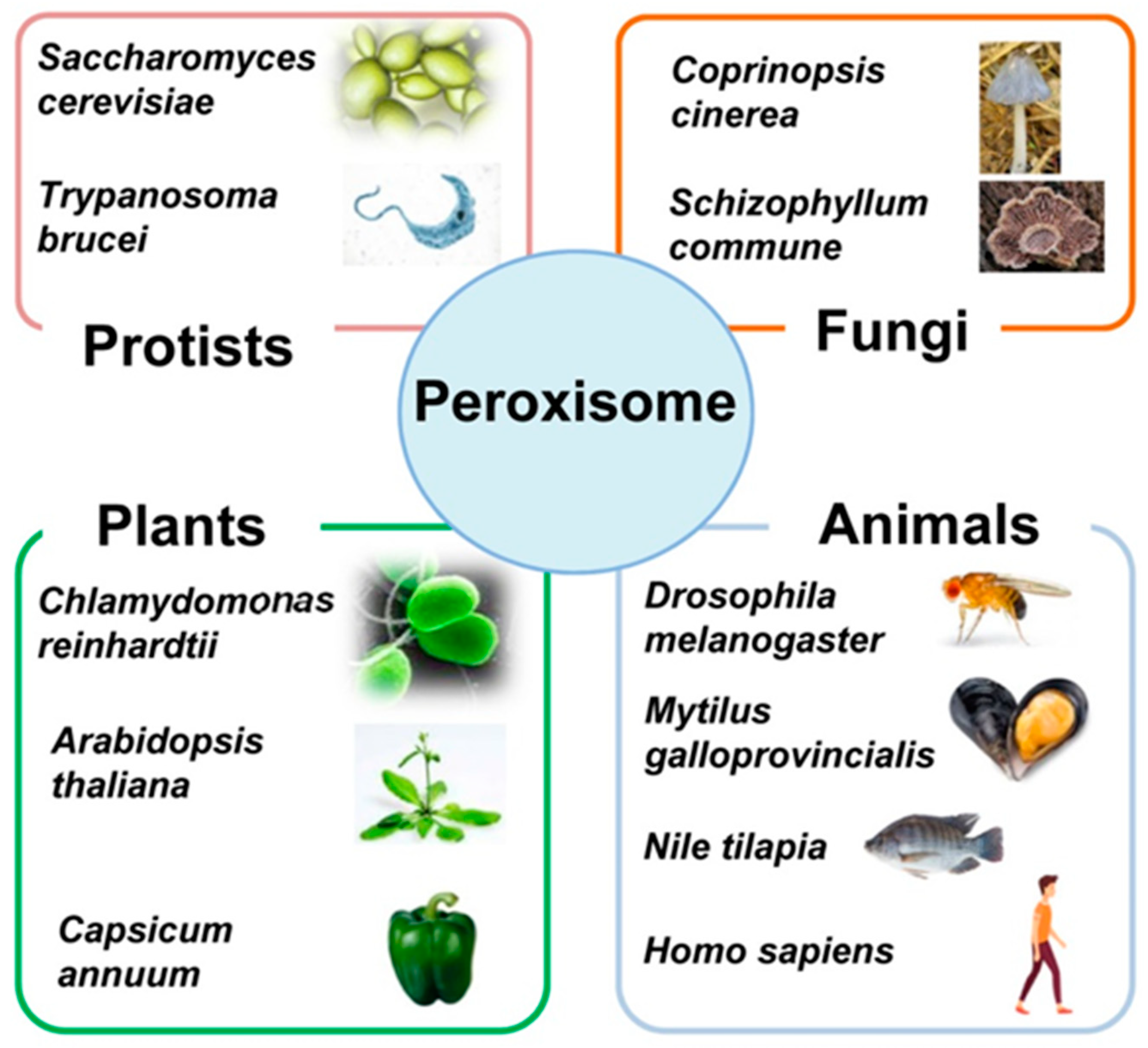
:1. Overview of the Diversity of Peroxisomes in Eukaryotic Cells
2. Hydrogen Peroxide (H2O2) and Nitric Oxide (NO) Metabolism in Plant Peroxisomes
3. NO and Protein–Protein Interactions (PPIs) in Plant Peroxisomes
4. Can NO Be a Signal Which Scaffolds Peroxisomal Function?
5. Conclusions and Future Trials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Islinger, M.; Cardoso, M.; Schrader, M. Be different—The diversity of peroxisomes in the animal kingdom. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1803, 881–897. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Baker, A.; Bartel, B.; Linka, N.; Mullen, R.T.; Reumann, S.; Zolman, B.K. Plant peroxisomes: Bio-genesis and function. Plant Cell 2012, 24. [Google Scholar] [CrossRef] [Green Version]
- Gabaldón, T.; Ginger, M.L.; Michels, P.A.M. Peroxisomes in parasitic protists. Mol. Biochem. Parasitol. 2016, 209, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Williams, C. Fungal Peroxisomes Proteomics. Cholest. Bind. Cholest. Transp. Proteins 2018, 89, 67–83. [Google Scholar] [CrossRef]
- Akşit, A.; van der Klei, I.J. Yeast peroxisomes: How are they formed and how do they grow? Int. J. Biochem. Cell Biol. 2018, 105, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Walter, T.; Erdmann, R. Current Advances in Protein Import into Peroxisomes. Protein J. 2019, 38, 351–362. [Google Scholar] [CrossRef]
- Rhodin, J.A.G. Correlation of Ultrastructural Organization and Function in Normal and Experimentally Changed Proximal Convoluted Tubule Cells of the Mouse Kidney. Ph.D. Thesis, Karolinska Institutet, Stockholm, Sweden, 1954. [Google Scholar]
- de Duve, C.; Beaufay, H.; Jacques, P.; Rahman-Li, Y.; Sellinger, O.Z.; Wattiaux, R.; de Coninck, S. Intracellular localization of catalase and of some oxidases in rat liver. Biochim. Biophys. Acta 1960, 40, 186–187. [Google Scholar] [CrossRef]
- De Duve, C.; Baudhuin, P. Peroxisomes (microbodies and related particles). Physiol Rev. 1966, 46, 323–357. [Google Scholar] [CrossRef]
- Tolbert, N.E. Metabolic Pathways in Peroxisomes and Glyoxysomes. Annu. Rev. Biochem. 1981, 50, 133–157. [Google Scholar] [CrossRef]
- del Río, L.A.; Sandalio, L.M.; Palma, J.M.; Bueno, P.; Corpas, F.J. Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radic. Biol. Med. 1992, 13, 557–580. [Google Scholar] [CrossRef]
- Hayashi, M.; Toriyama, K.; Kondo, M.; Kato, A.; Mano, S.; De Bellis, L.; Hayashi-Ishimaru, Y.; Yamaguchi, K.; Hayashi, H.; Nishimura, M. Functional transformation of plant peroxisomes. Cell Biophys. 2000, 32, 295–304. [Google Scholar] [CrossRef]
- Titorenko, V.I.; Nicaud, J.M.; Wang, H.; Chan, H.; Rachubinski, R.A. Acyl-CoA oxidase is imported as a heteropentameric, cofactor-containing complex into peroxisomes of Yarrowia lipolytica. J. Cell Biol. 2002, 156, 481–494. [Google Scholar] [CrossRef] [Green Version]
- van der Klei, I.; Veenhuis, M. Peroxisomes: Flexible and dynamic organelles. Curr. Opin. Cell Biol. 2002, 14, 500–505. [Google Scholar] [CrossRef] [Green Version]
- Cajaraville, M.P.; Cancio, I.; Ibabe, A.; Orbea, A. Peroxisome proliferation as a biomarker in environ-mental pollution assessment. Microsc. Res. Tech. 2003, 61, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Islinger, M.; Voelkl, A.; Fahimi, H.D.; Schrader, M. The peroxisome: An update on mysteries 2.0. Histochem. Cell Biol. 2018, 150, 443–471. [Google Scholar] [CrossRef] [Green Version]
- Dias, A.F.; Francisco, T.; Rodrigues, T.A.; Grou, C.P.; Azevedo, J.E. The first minutes in the life of a pe-roxisomal matrix protein. Biochim. Biophys. Acta 2016, 1863, 814–820. [Google Scholar] [CrossRef]
- Navarro-Espíndola, R.; Suaste-Olmos, F.; Peraza-Reyes, L. Dynamic regulation of peroxisomes and mitochondria during fungal development. J. Fungi. 2020, 6, 302. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Hashimoto, M.; Williams, T.A.; Hirawake-Mogi, H.; Makiuchi, T.; Tsubouchi, A.; Kaga, N.; Taka, H.; Fujimura, T.; Koike, M.; et al. Differential remodelling of peroxisome function underpins the environmental and metabolic adaptability of diplonemids and kinetoplastids. Proc. R. Soc. B Boil. Sci. 2016, 283, 20160520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peraza-Reyes, L.; Berteaux-Lecellier, V. Peroxisomes and sexual development in fungi. Front. Physiol. 2013, 4, 244. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.-C.; Chen, C.-W.; Choo, C.Y.L.; Chen, Y.-K.; Yago, J.I.; Chung, K.-R. Proper Functions of Peroxisomes Are Vital for Pathogenesis of Citrus Brown Spot Disease Caused by Alternaria alternata. J. Fungi 2020, 6, 248. [Google Scholar] [CrossRef]
- Kong, F.; Liang, Y.; Légeret, B.; Beyly-Adriano, A.; Blangy, S.; Haslam, R.P.; Napier, J.A.; Beisson, F.; Peltier, G.; Li-Beisson, Y. Chlamydomonas carries out fatty acid beta-oxidation in ancestral peroxisomes using a bona fide acyl-CoA oxidase. Plant. J. 2017, 90, 358–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, F.; Burlacot, A.; Liang, Y.; Légeret, B.; Alseekh, S.; Brotman, Y.; Fernie, A.R.; Krieger-Liszkay, A.; Beisson, F.; Peltier, G.; et al. Interorganelle Communication: Peroxisomal malate DE-HYDROGENASE2 connects lipid catabolism to photosynthesis through redox coupling in Chla-mydomonas. Plant Cell 2018, 30, 1824–1847. [Google Scholar] [CrossRef] [Green Version]
- De Bellis, L.; Picciarelli, P.; Pistelli, L.; Alpi, A. Localization of glyoxylate-cycle marker enzymes in peroxisomes of senescent leaves and green cotyledons. Planta 1990, 180, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; Palma, J.M.; Lupiáñez, J.A.; Del Río, L.A. Peroxisomal NADP-Dependent Isocitrate Dehydrogenase. Characterization and Activity Regulation during Natural Senescence. Plant. Physiol. 1999, 121, 921–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzniak, E.; Skłodowska, M. Compartment-specific role of the ascorbate-glutathione cycle in the response of tomato leaf cells to Botrytis cinerea infection. J. Exp. Bot. 2005, 56, 921–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuźniak, E.; Skłodowska, M. Fungal pathogen-induced changes in the antioxidant systems of leaf peroxisomes from infected tomato plants. Planta 2005, 222, 192–200. [Google Scholar] [CrossRef]
- Leterrier, M.; Barroso, J.B.; Valderrama, R.; Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Luque, F.; Viñegla, B.; Palma, J.M.; Corpas, F.J. Peroxisomal NADP-isocitrate dehydrogenase is required for Ara-bidopsis stomatal movement. Protoplasma 2016, 253, 403–415. [Google Scholar] [CrossRef]
- Kataya, A.R.A.; Elshobaky, A.; Heidari, B.; Dugassa, N.F.; Thelen, J.J.; Lillo, C. Multi-targeted treha-lose-6-phosphate phosphatase I harbors a novel peroxisomal targeting signal 1 and is essential for flowering and development. Planta 2020, 251, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Chen, Z.; Han, J.; Zhao, H.; Liu, J.; Yu, Y. Suppression of chorismate synthase, which is localized in chloroplasts and peroxisomes, results in abnormal flower development and anthocyanin reduction in petunia. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Cajaraville, M.P.; Ortiz-Zarragoitia, M. Specificity of the peroxisome proliferation response in mus-sels exposed to environmental pollutants. Aquat Toxicol. 2006, 78, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, S.L.; Luo, Y.; Li, L.Y.; Chen, L.Q.; Zhang, M.L.; Du, Z.Y. Impaired peroxisomal fat oxidation induces hepatic lipid accumulation and oxidative damage in Nile tilapia. Fish Physiol. Biochem. 2020, 46, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Yamada, J.; Ogawa, S.; Horie, S.; Watanabe, T.; Suga, T. Participation of peroxisomes in the metab-olism of xenobiotic acyl compounds: Comparison between peroxisomal and mitochondrial be-ta-oxidation of omega-phenyl fatty acids in rat liver. Biochim. Biophys. Acta 1987, 921, 292–301. [Google Scholar]
- Santos, M.J.; Imanaka, T.; Shio, H.; Small, G.M.; Lazarow, P.B. Peroxisomal membrane ghosts in Zellwe-ger syndrome--aberrant organelle assembly. Science 1988, 239, 1536–1538. [Google Scholar] [CrossRef] [PubMed]
- Waterham, H.R.; Ferdinandusse, S.; Wanders, R.J. Human disorders of peroxisome metabolism and biogenesis. Biochim. Ebiophys. Acta (BBA) Bioenerg. 2016, 1863, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Lismont, C.; Revenco, I.; Fransen, M. Peroxisomal Hydrogen Peroxide Metabolism and Signaling in Health and Disease. Int. J. Mol. Sci. 2019, 20, 3673. [Google Scholar] [CrossRef] [Green Version]
- Cook, K.C.; Moreno, J.A.; Jean Beltran, P.M.; Cristea, I.M. peroxisome plasticity at the virus-host inter-face. Trends Microbiol. 2019, 27, 906–914. [Google Scholar] [CrossRef]
- Di Cara, F. Peroxisomes in host defense. PLoS Pathog. 2020, 16, e1008636. [Google Scholar] [CrossRef]
- Kao, Y.-T.; Gonzalez, K.L.; Bartel, B. Peroxisome Function, Biogenesis, and Dynamics in Plants. Plant. Physiol. 2018, 176, 162–177. [Google Scholar] [CrossRef] [Green Version]
- Imoto, Y.; Itoh, K.; Fujiki, Y. Molecular Basis of Mitochondrial and Peroxisomal Division Machineries. Int. J. Mol. Sci. 2020, 21, 5452. [Google Scholar] [CrossRef]
- Fujiki, Y.; Abe, Y.; Imoto, Y.; Tanaka, A.J.; Okumoto, K.; Honsho, M.; Tamura, S.; Miyata, N.; Yamashita, T.; Chung, W.K.; et al. Recent insights into peroxisome biogenesis and associated diseases. J. Cell Sci. 2020, 133, jcs236943. [Google Scholar] [CrossRef]
- Dubreuil, M.M.; Morgens, D.W.; Okumoto, K.; Honsho, M.; Contrepois, K.; Lee-McMullen, B.; Traber, G.M.; Sood, R.S.; Dixon, S.J.; Snyder, M.P.; et al. Systematic Identification of Regulators of Oxidative Stress Reveals Non-canonical Roles for Peroxisomal Import and the Pentose Phosphate Pathway. Cell Rep. 2020, 30, 1417–1433.e7. [Google Scholar] [CrossRef] [PubMed]
- Wright, Z.J.; Bartel, B. Peroxisomes form intralumenal vesicles with roles in fatty acid catabolism and protein compartmentalization in Arabidopsis. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M.; Sandalio, L.M.; López-Huertas, E.; Romero-Puertas, M.C.; Barroso, J.B.; Del Río, L.A. Purification of catalase from pea leaf peroxisomes: Identification of five different isoforms. Free Radic. Res 1999, 31, S235–S241. [Google Scholar] [CrossRef]
- Schrader, M.; Fahimi, H. Peroxisomes and oxidative stress. Biochim. Biophys. Acta (BBA) Bioenerg. 2006, 1763, 1755–1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fransen, M.; Lismont, C. Redox Signaling from and to Peroxisomes: Progress, Challenges, and Prospects. Antioxid. Redox Signal. 2019, 30, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Witte, C.P. A kinase and a glycosylase catabolize pseudouridine in the peroxisome to pre-vent toxic pseudouridine monophosphate accumulation. Plant Cell 2020, 32, 722–739. [Google Scholar] [CrossRef]
- Lazarow, P.B. What is a peroxisome? Toxicol. Ind. Health. 1987, 3, 1–6. [Google Scholar] [CrossRef]
- Trelease, R.N.; Becker, W.M.; Gruber, P.J. Newcomb EH. Microbodies (glyoxysomes and peroxisomes) in cucumber cotyledons: Correlative biochemical and ultrastructural study in light- and dark-grown seedlings. Plant. Physiol. 1971, 48, 461–475. [Google Scholar] [CrossRef] [Green Version]
- Beevers, H. Microbodies in Higher Plants. Annu. Rev. Plant. Physiol. 1979, 30, 159–193. [Google Scholar] [CrossRef]
- Vicentini, F.; Matile, P. Gerontosomes, a Multifunctional Type of Peroxisome in Senescent Leaves. J. Plant. Physiol. 1993, 142, 50–56. [Google Scholar] [CrossRef]
- Pracharoenwattana, I.; Smith, S.M. When is a peroxisome not a peroxisome? Trends Plant. Sci. 2008, 13, 522–525. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant Peroxisomes: A Factory of Reactive Species. Front. Plant. Sci. 2020, 11, 853. [Google Scholar] [CrossRef] [PubMed]
- del Río, L.A.; Lyon, D.S.; Olah, I.; Glick, B.; Salin, M.L. Immunocytochemical evidence for a pe-roxisomal localization of manganese superoxide dismutase in leaf protoplasts from a higher plant. Planta 1983, 158, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Sandalio, L.M.; del Río, L.A.; Trelease, R.N. Copper–zinc superoxide dismutase is a constit-uent enzyme of the matrix of peroxisomes in the cotyledons of oilseed plants. New Phytol. 1998, 138, 307–314. [Google Scholar] [CrossRef]
- Corpas, F.J.; Fernández-Ocaña, A.; Carreras, A.; Valderrama, R.; Luque, F.; Esteban, F.J. The expres-sion of different superoxide dismutase forms is cell-type dependent in olive (Olea europaea L.) leaves. Plant. Cell Physiol. 2006, 47, 984–994. [Google Scholar] [CrossRef] [Green Version]
- Bunkelmann, J.R.; Trelease, R.N. Ascorbate peroxidase. A prominent membrane protein in oilseed glyoxysomes. Plant. Physiol. 1996, 110, 589–598. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, K.; Mori, H.; Nishimura, M. A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant. Cell Physiol. 1995, 36, 1157–1162. [Google Scholar] [CrossRef]
- Corpas, F.J.; Trelease, R.N. Differential expression of ascorbate peroxidase and a putative molecular chaperone in the boundary membrane of differentiating cucumber seedling peroxisomes. J. Plant. Physiol. 1998, 153, 332–338. [Google Scholar] [CrossRef]
- Narendra, S.; Venkataramani, S.; Shen, G.; Wang, J.; Pasapula, V.; Lin, Y.; Kornyeyev, D.; Holaday, A.S.; Zhang, H. The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 2006, 57, 3033–3042. [Google Scholar] [CrossRef] [Green Version]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; del Río, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant. Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef] [Green Version]
- Lisenbee, C.S.; Lingard, M.J.; Trelease, R.N. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant. J. 2005, 43, 900–914. [Google Scholar] [CrossRef]
- Jiménez, A.; Hernandez, J.A.; del Rio, L.A.; Sevilla, F. Evidence for the presence of the ascor-bate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant. Physiol. 1997, 114, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Palma, J.M.; Jiménez, A.; Sandalio, L.M.; Corpas, F.J.; Lundqvist, M.; Gómez, M.; Sevilla, F.; Del Río, L.A. Antioxidative enzymes from chloroplasts, mitochondria, and peroxisomes during leaf senescence of nodulated pea plants. J. Exp. Bot. 2006, 57, 1747–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reumann, S.; Quan, S.; Aung, K.; Yang, P.; Manandhar-Shrestha, K.; Holbrook, D.; Linka, N.; Switzenberg, R.; Wilkerson, C.G.; Weber, A.P.; et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant. Physiol. 2009, 150, 125–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zechmann, B.; Zellnig, G. Ultrastructural localization of glutathione in Cucurbita pepo plants. Protoplasma 2004, 223, 213–219. [Google Scholar] [CrossRef]
- Zechmann, B. Compartment-Specific Importance of Ascorbate During Environmental Stress in Plants. Antioxid. Redox Signal. 2018, 29, 1488–1501. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B. Subcellular Roles of Glutathione in Mediating Plant Defense during Biotic Stress. Plants 2020, 9, 1067. [Google Scholar] [CrossRef]
- Mittova, V.; Tal, M.; Volokita, M.; Guy, M. Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii. Plant. Cell Environ. 2003, 26, 845–856. [Google Scholar] [CrossRef]
- Piacentini, D.; Corpas, F.J.; D’Angeli, S.; Altamura, M.M.; Falasca, G. Cadmium and arse-nic-induced-stress differentially modulates Arabidopsis root architecture, peroxisome distribution, enzymatic activities and their nitric oxide content. Plant. Physiol. Biochem. 2020, 148, 312–323. [Google Scholar] [CrossRef]
- Reumann, S.; Chowdhary, G.; Lingner, T. Characterization, prediction and evolution of plant peroxi-somal targeting signals type 1 (PTS1s). Biochim. Biophys. Acta 2016, 1863, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Mullen, R.T.; Lee, M.S.; Trelease, R.N. Identification of the peroxisomal targeting signal for cottonseed catalase. Plant. J. 1997, 12, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Kamigaki, A.; Mano, S.; Terauchi, K.; Nishi, Y.; Tachibe-Kinoshita, Y.; Nito, K.; Kondo, M.; Hayashi, M.; Nishimura, M.; Esaka, M. Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant. J. 2003, 33, 161–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, Y.; Kamigaki, A.; Nakamori, C.; Mano, S.; Hayashi, M.; Nishimura, M.; Esaka, M. Plant Catalase is Imported into Peroxisomes by Pex5p but is Distinct from Typical PTS1 Import. Plant. Cell Physiol. 2008, 49, 671–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujikawa, Y.; Suekawa, M.; Endo, S.; Fukami, Y.; Mano, S.; Nishimura, M.; Esaka, M. Effect of mutation of C-terminal and heme binding region of Arabidopsis catalase on the import to peroxisomes. Biosci. Biotechnol. Biochem. 2019, 83, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Walton, P.A.; Brees, C.; Lismont, C.; Apanasets, O.; Fransen, M. The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1864, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Nitric oxide: A radical molecule with potential biotech-nological applications in fruit ripening. J. Biotechnol. 2020, 324, 211–219. [Google Scholar] [CrossRef]
- Corpas, F.J.; Chaki, M.; Leterrier, M.; Barroso, J.B. Protein tyrosine nitration: A new challenge in plants. Plant. Signal. Behav. 2009, 4, 920–923. [Google Scholar] [CrossRef] [Green Version]
- Bartesaghi, S.; Radi, R. Fundamentals on the biochemistry of peroxynitrite and protein tyrosine nitration. Redox Biol. 2018, 14, 618–625. [Google Scholar] [CrossRef]
- Mohn, M.A.; Thaqi, B.; Fischer-Schrader, K. Isoform-Specific NO Synthesis by Arabidopsis thaliana Nitrate Reductase. Plants 2019, 8, 67. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; Barroso, J.B. Nitric oxide synthase-like activity in higher plants. Nitric Oxide 2017, 68, 5–6. [Google Scholar] [CrossRef]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Sandalio, L.M.; Valderrama, R.; Palma, J.; Lupiáñez, J.A.; Del Río, L.A. Localization of Nitric-oxide Synthase in Plant Peroxisomes. J. Biol. Chem. 1999, 274, 36729–36733. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; Barroso, J.B.; Carreras, A.; Quirós, M.; León, A.M.; Romero-Puertas, M.C.; Esteban, F.J.; Valderrama, R.; Palma, J.M.; Sandalio, L.M.; et al. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant. Physiol. 2004, 136, 2722–2733. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; Barroso, J.B. Calmodulin antagonist affects peroxisomal functionality by disrupting both peroxisomal Ca2+and protein import. J. Cell Sci. 2017, 131, jcs201467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpas, F.J.; Palma, J.M.; Sandalio, L.M.; Valderrama, R.; Barroso, J.B.; Del Río, L.A. Peroxisomal xanthine oxidoreductase: Characterization of the enzyme from pea (Pisum sativum L.) leaves. J. Plant. Physiol. 2008, 165, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Peroxynitrite (ONOO-) is endogenously produced in Arabidopsis peroxi-somes and is overproduced under cadmium stress. Ann. Bot. 2014, 113, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Kissner, R.; Nauser, T.; Bugnon, P.; Lye, P.G.; Koppenol, W.H. Formation and properties of peroxynitrite as studied by laser flash photolysis, high-pressure stopped-flow technique, and pulse radiolysis. Chem. Res. Toxicol. 1997, 10, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Sueta, G.; Campolo, N.; Trujillo, M.; Bartesaghi, S.; Carballal, S.; Romero, N.; Alvarez, B.; Radi, R. Biochemistry of Peroxynitrite and Protein Tyrosine Nitration. Chem. Rev. 2018, 118, 1338–1408. [Google Scholar] [CrossRef]
- Broniowska, K.A.; Diers, A.R.; Hogg, N. S-nitrosoglutathione. Biochim. Biophys. Acta 2013, 1830, 3173–3181. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; Alché, J.D.D.; Barroso, J.B. Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant. Sci. 2013, 4, 126. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; Distefano, S.; Palma, J.M.; Lupiáñez, J.A.; del Río, L.A.; Baune, M.C.; Lansing, H.; Fischer, K.; et al. The Ara-bidopsis plastidial glucose-6-phosphate transporter GPT1 is dually targeted to peroxisomes via the endoplasmic reticulum. Plant Cell 2020, 32, 1703–1726. [Google Scholar]
- A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem J. 1998, 330, 777–784. [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Nitric oxide (NO) and hydrogen sulfide (H2S) modulate the NADPH-generating enzymatic system in higher plants. J. Exp. Bot. 2021, 72, 830–847. [Google Scholar] [CrossRef]
- Alamillo, J.M.; García-Olmedo, F. Effects of urate, a natural inhibitor of peroxyni-trite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudo-monas syringae. Plant. J. 2001, 25, 529–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Signorelli, S.; Imparatta, C.; Rodríguez-Ruiz, M.; Borsani, O.; Corpas, F.J.; Jorge Monza, J. In vivo and in vitro approaches demonstrate proline is not directly involved in the protection against superox-ide, nitric oxide, nitrogen dioxide and peroxynitrite. Funct. Plant Biol. 2016, 43, 870–879. [Google Scholar] [CrossRef]
- Corpas, F.J.; de la Colina, C.; Sánchez-Rasero, F.; del Río, L.A. A role for leaf peroxisomes in the catabolism of purines. J. Plant. Physiol. 1997, 151, 246–250. [Google Scholar] [CrossRef]
- Hauck, O.K.; Scharnberg, J.; Escobar, N.M.; Wanner, G.; Giavalisco, P.; Witte, C.P. Uric acid ac-cumulation in an Arabidopsis urate oxidase mutant impairs seedling establishment by blocking pe-roxisome maintenance. Plant. Cell 2014, 26, 3090–3100. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palma, J.M.; Mateos, R.M.; López-Jaramillo, J.; Rodríguez-Ruiz, M.; González-Gordo, S.; Lechuga-Sancho, A.M.; Corpas, F.J. Plant catalases as NO and H2S targets. Redox Biol. 2020, 34, 101525. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, R.; Feng, J.; Feng, T.; Wang, C.; Hu, J.; Zhan, N.; Li, Y.; Ma, X.; Ren, B.; et al. Transnitrosylation Mediated by the Non-canonical Catalase ROG1 Regulates Nitric Oxide Signaling in Plants. Dev. Cell 1319, 53, 444–457.e5. [Google Scholar] [CrossRef]
- Wang, Y.; Chu, C. S-nitrosylation control of ROS and RNS homeostasis in plants: The switch-ing function of catalase. Mol. Plant. 2020, 13, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, J.; Wang, G.; Cha, J.Y.; Li, G.; Chen, S.; Li, Z.; Guo, J.; Zhang, C.; Yang, Y.; et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 2015, 27, 908–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.J.; Li, X.D.; Ratnasekera, D.; Wang, C.; Liu, W.X.; Song, L.F.; Zhang, W.Z.; Wu, W.H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, N.; Wang, C.; Chen, L.; Yang, H.; Feng, J.; Gong, X.; Ren, B.; Wu, R.; Mu, J.; Li, Y.; et al. S-Nitrosylation targets GSNO reductase for selective autophagy during hypoxia responses in plants. Mol. Cell 2018, 71, 142–154.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, M.; Oikawa, K.; Yoshimoto, K.; Goto-Yamada, S.; Mano, S.; Yamada, K.; Kondo, M.; Hayashi, M.; Sakamoto, W.; Ohsumi, Y.; et al. Plant autophagy is responsible for peroxisomal transition and plays an important role in the maintenance of peroxisomal quality. Autophagy 2014, 10, 936–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, M.; Oikawa, K.; Yoshimoto, K.; Kondo, M.; Mano, S.; Yamada, K.; Hayashi, M.; Sakamoto, W.; Ohsumi, Y.; Nishimura, M. Highly Oxidized Peroxisomes Are Selectively Degraded via Autophagy in Arabidopsis. Plant. Cell 2013, 25, 4967–4983. [Google Scholar] [CrossRef] [Green Version]
- Young, P.G.; Bartel, B. Pexophagy and peroxisomal protein turnover in plants. Biochim. Biophys. Acta (BBA) Bioenerg. 2016, 1863, 999–1005. [Google Scholar] [CrossRef]
- Luo, M.; Zhuang, X. Review: Selective degradation of peroxisome by autophagy in plants: Mecha-nisms, functions, and perspectives. Plant. Sci. 2018, 274, 485–491. [Google Scholar] [CrossRef]
- Borek, S.; Stefaniak, S.; Śliwiński, J.; Garnczarska, M.; Pietrowska-Borek, M. Autophagic Machinery of Plant Peroxisomes. Int. J. Mol. Sci. 2019, 20, 4754. [Google Scholar] [CrossRef] [Green Version]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant. Sci. 2020, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Anand, P.; Kwak, Y.; Simha, R.; Donaldson, R.P. Hydrogen peroxide induced oxidation of peroxisomal malate synthase and catalase. Arch. Biochem. Biophys. 2009, 491, 25–31. [Google Scholar] [CrossRef]
- Pena, L.B.; Azpilicueta, C.E.; Gallego, S.M. Sunflower cotyledons cope with copper stress by inducing catalase subunits less sensitive to oxidation. J. Trace Elem. Med. Biol. 2011, 25, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, M.; González-Gordo, S.; Cañas, A.; Campos, M.J.; Paradela, A.; Corpas, F.J.; Palma, J.M. Sweet Pepper (Capsicum annuum L.) fruits contain an atypical peroxisomal catalase that is modu-lated by reactive oxygen and nitrogen species. Antioxidants 2019, 8, 374. [Google Scholar]
- Yoshimoto, K.; Shibata, M.; Kondo, M.; Oikawa, K.; Sato, M.; Toyooka, K.; Shirasu, K.; Nishimura, M.; Ohsumi, Y. Organ-specific quality control of plant peroxisomes is mediated by autophagy. J. Cell Sci. 2014, 127 Pt 6, 1161–1168. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, S.; Mano, S.; Oikawa, K.; Hikino, K.; Teshima, K.M.; Kimori, Y.; Nishimura, M.; Shimazaki, K.I.; Takemiya, A. Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc. Natl. Acad. Sci. USA 2019, 116, 19187–19192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Wang, P.; Wei, Y.; Bai, Y.; Lu, Y.; Zeng, H.; Liu, G.; Reiter, R.J.; He, C.; Shi, H. The dual interplay of RAV5 in activating nitrate reductases and repressing catalase activity to improve disease resistance in cassava. Plant. Biotechnol. J. 2020. [Google Scholar] [CrossRef] [PubMed]




| Organism/Organ (Species) | Peroxisomal Function | Ref. |
|---|---|---|
| Free-living marine diplonemid (Diplonema papillatum) | Peroxisome undergoes remodeling metabolism involving the housing gluconeogenesis | [19] |
| Ascomycete (Sclerotinia sclerotiorum) | Peroxisome is involved in the fungi sexual development | [20] |
| Fungus (Alternaria alternate) | Peroxisome of the fungus is necessary for its pathogenesis in citrus | [21] |
| Chlamydomonas | Peroxisomal malate dehydrogenase 2 connects lipid catabolism to photosynthesis | [22,23] |
| Pea leaves (Pisum sativum) | Peroxisomes involve in leaf senescence | [24,25] |
| Leaf tomato (Solanum lycopersicum) | Peroxisomes involve in pathogen defence | [26,27] |
| Leaves (Arabidopsis thaliana) | Peroxisomal NADP-isocitrate dehydrogenase is required for stomatal movementPeroxisomal trehalose-6-phosphate phosphatase I is essential for flowering and development | [28,29] |
| Petunia (Petunia hybrida) | Peroxisomal and chloroplastic chorismate synthase, involved in shikimate pathway, are need for flower development | [30] |
| Mussels (Mytilus edulis) | Peroxisome proliferation in response to environmental pollutants | [31] |
| Nile tilapia (Oreochromis niloticus) | Impaired of peroxisomal fat oxidation induces hepatic lipid accumulation and oxidative damage | [32] |
| Rat liver (Rattus norvegicus) | Peroxisomes participate in the metabolism of xenobiotic acyl compounds | [33] |
| Human (Homo sapiens) | Defects in genes encoding peroxisomal proteins lead to a variety of human diseases. For example, X-linked adrenoleukodystrophy, acatalasemia, cerebro-hepato-renal syndrome, etc.Host defense | [34,35,36,37,38] |
| Peroxisomal Enzyme | NO-Derived PTM | Effect on Activity |
|---|---|---|
| Antioxidants | ||
| Catalase (CAT) | Tyr-nitration S-nitrosation | Inhibition Inhibition |
| Monodehydroascorbate reductase (MDAR) | Tyr-nitration S-nitrosation | Inhibition Inhibition |
| Ascorbate peroxidase (APX) | Tyr-nitration S-nitrosation | Inhibition Activation |
| CuZn-superoxide dismutase (CSD3) | Tyr-nitration | Inhibition |
| Photorespiration | ||
| Hydroxypyruvate reductase (HPR) | Tyr-nitration S-nitrosation | Inhibition Inhibition |
| Glycolate oxidase (GOX) | S-nitrosation | Inhibition |
| Fatty acid β-oxidation | ||
| Malate dehydrogenase (MDH) | Tyr-nitration S-nitrosation | Inhibition Inhibition |
| Isocitrate lyase (ICL) | S-nitrosation | Not reported |
| Multifunctional protein AIM1 isoform | S-nitrosation | Not reported |
| Glyoxylate cycle | ||
| Isocitrate lyase (ICL) | S-nitrosation | Not reported |
| Peroxisomal protein import | ||
| Lon protease homolog 2 | S-nitrosation | Not reported |
| NADPH supply | ||
| NADP-isocitrate dehydrogenase (NADP-ICDH) | Tyr-nitration S-nitrosation | Inhibition Inhibition |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corpas, F.J.; González-Gordo, S.; Palma, J.M. Nitric Oxide (NO) Scaffolds the Peroxisomal Protein–Protein Interaction Network in Higher Plants. Int. J. Mol. Sci. 2021, 22, 2444. https://doi.org/10.3390/ijms22052444
Corpas FJ, González-Gordo S, Palma JM. Nitric Oxide (NO) Scaffolds the Peroxisomal Protein–Protein Interaction Network in Higher Plants. International Journal of Molecular Sciences. 2021; 22(5):2444. https://doi.org/10.3390/ijms22052444
Chicago/Turabian StyleCorpas, Francisco J., Salvador González-Gordo, and José M. Palma. 2021. "Nitric Oxide (NO) Scaffolds the Peroxisomal Protein–Protein Interaction Network in Higher Plants" International Journal of Molecular Sciences 22, no. 5: 2444. https://doi.org/10.3390/ijms22052444
APA StyleCorpas, F. J., González-Gordo, S., & Palma, J. M. (2021). Nitric Oxide (NO) Scaffolds the Peroxisomal Protein–Protein Interaction Network in Higher Plants. International Journal of Molecular Sciences, 22(5), 2444. https://doi.org/10.3390/ijms22052444








