Identification of Cyclophilin A as a Potential Anticancer Target of Novel Nargenicin A1 Analog in AGS Gastric Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. 2-DE Profiling of the Differentially Expressed Proteins in AGS Cells Following Treatment with Compound 9
2.2. Effect of Compound 9 on CypA/CD147 Pathway in AGS Cells
2.3. CypA Silencing Inhibits Proliferation, Migration, and Invasion of AGS Cells
2.4. CypA Silencing Downregulates CD147-Mediated MAPK Signaling Pathway in AGS Cells
2.5. CypA Silencing Induces Cell Cycle Arrest and Apoptosis of AGS Cells
2.6. CypA Silencing Inhibits AGS Cell-Induced Angiogenesis of HUVECs
2.7. Identification of Binding between Compound 9 and CypA using DARTS and CETSA
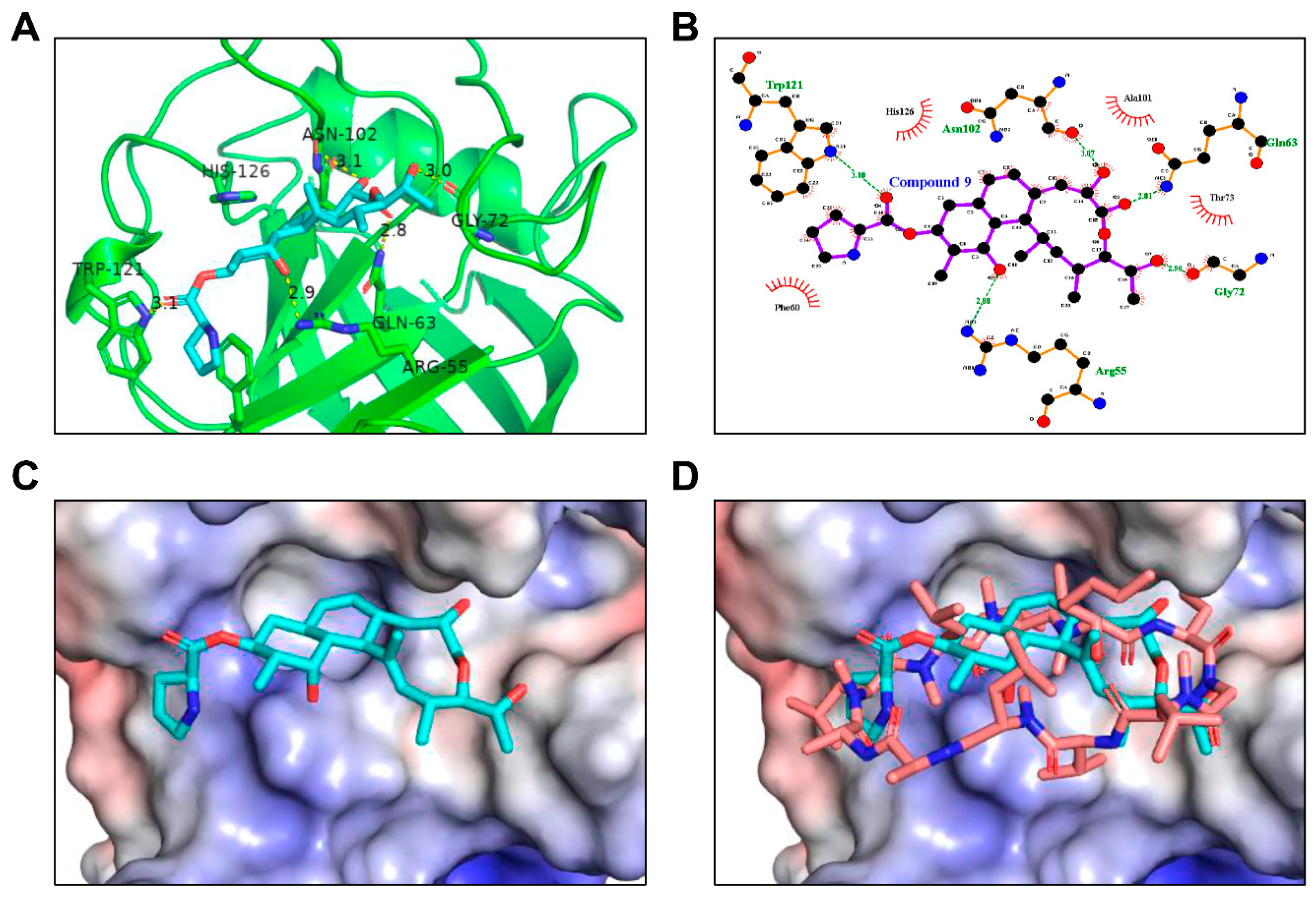
2.8. In Silico Docking Analysis of Interaction between Compound 9 and CypA
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Hypoxic Conditions
4.3. Preparation of Total Cellular Extract
4.4. Two-Dimensional Gel Electrophoresis (2-DE) and Image Analysis
4.5. Matrix Assisted Laser Desorption Ionization-Time of Flight/Mass Spectrometry (MALDI-TOF/MS) Analysis and Protein Identification
4.6. Western Blot Analysis
4.7. CypA-Directed RNA Interference
4.8. Cell Proliferation Assay
4.9. Wound Healing Assay
4.10. Invasion Assay
4.11. Cell Cycle Analysis
4.12. Apoptosis Analysis
4.13. Tumor Cell-Induced Proliferation Assay
4.14. Tumor Cell-Induced Invasion Assay
4.15. Measurement of VEGF by ELISA
4.16. DARTS Assay
4.17. CETSA
4.18. Ligand/Molecular Target Selection and Preparation
4.19. In Silico Docking Analysis
4.20. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Plummer, M.; Franceschi, S.; Vignat, J.; Forman, D.; de Martel, C. Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 2015, 136, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Karimi, P.; Islami, F.; Anandasabapathy, S.; Freedman, N.D.; Kamangar, F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 700–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Zhou, Y.; Yang, P.; Wu, X.T. Glutathione S-transferase M1 gene polymorphism and gastric cancer risk: An updated analysis. Arch. Med. Res. 2010, 41, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.X.; Wang, K.; Lv, F.F.; Chen, Z.Y.; Liu, X.; Zheng, C.L.; Li, W.H.; Zhu, X.D.; Guo, W.J.; Li, J. GSTM1 null allele is a risk factor for gastric cancer development in Asians. Cytokine 2011, 55, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Necula, L.; Matei, L.; Dragu, D.; Neagu, A.I.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044. [Google Scholar] [CrossRef]
- Izuishi, K.; Mori, H. Recent strategies for treating stage IV gastric cancer: Roles of palliative gastrectomy, chemotherapy, and radiotherapy. J. Gastrointestin. Liver Dis. 2016, 25, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, J.S.; Smalley, S.R.; Benedetti, J.; Hundahl, S.A.; Estes, N.C.; Stemmermann, G.N.; Haller, D.G.; Ajani, J.A.; Gunderson, L.L.; Jessup, J.M.; et al. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N. Engl. J. Med. 2001, 345, 725–730. [Google Scholar] [CrossRef]
- Arai, H.; Nakajima, T.E. Recent developments of systemic chemotherapy for gastric cancer. Cancers 2020, 12, 1100. [Google Scholar] [CrossRef]
- Ajani, J.A.; Lee, J.; Sano, T.; Janjigian, Y.Y.; Fan, D.; Song, S. Gastric adenocarcinoma. Nat. Rev. Dis. Primers 2017, 3, 17036. [Google Scholar] [CrossRef]
- Ruan, T.; Liu, W.; Tao, K.; Wu, C. A Review of research progress in multidrug-resistance mechanisms in gastric cancer. Onco Targets Ther. 2020, 13, 1797–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mushtaq, S.; Abbasi, B.H.; Uzair, B.; Abbasi, R. Natural products as reservoirs of novel therapeutic agents. EXCLI J. 2018, 17, 420–451. [Google Scholar] [PubMed]
- Painter, R.E.; Adam, G.C.; Arocho, M.; DiNunzio, E.; Donald, R.G.; Dorso, K.; Genilloud, O.; Gill, C.; Goetz, M.; Hairston, N.N.; et al. Elucidation of DnaE as the antibacterial target of the natural product, nargenicin. Chem. Biol. 2015, 22, 1362–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhakal, D.; Han, J.M.; Mishra, R.; Pandey, R.P.; Kim, T.S.; Rayamajhi, V.; Jung, H.J.; Yamaguchi, T.; Sohng, J.K. Characterization of tailoring steps of nargenicin A1 biosynthesis reveals a novel analogue with anticancer activities. ACS Chem. Biol. 2020, 15, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Han, J.M.; Choi, Y.S.; Dhakal, D.; Sohng, J.K.; Jung, H.J. Novel nargenicin A1 analog inhibits angiogenesis by downregulating the endothelial VEGF/VEGFR2 signaling and tumoral HIF-1α/VEGF pathway. Biomedicines 2020, 8, 252. [Google Scholar] [CrossRef]
- Bai, Z.; Ye, Y.; Liang, B.; Xu, F.; Zhang, H.; Zhang, Y.; Peng, J.; Shen, D.; Cui, Z.; Zhang, Z.; et al. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int. J. Oncol. 2011, 38, 375–383. [Google Scholar] [PubMed] [Green Version]
- Wang, P.; Heitman, J. The cyclophilins. Genome Biol. 2005, 6, 226. [Google Scholar] [CrossRef] [Green Version]
- Lee, J. Role of cyclophilin a during oncogenesis. Arch. Pharm. Res. 2010, 33, 181–187. [Google Scholar] [CrossRef]
- Yurchenko, V.; Zybarth, G.; O’Connor, M.; Dai, W.W.; Franchin, G.; Hao, T.; Guo, H.; Hung, H.C.; Toole, B.; Gallay, P.; et al. Active site residues of cyclophilin A are crucial for its signaling activity via CD147. J. Biol. Chem. 2002, 277, 22959–22965. [Google Scholar] [CrossRef] [Green Version]
- Muramatsu, T. Basigin: A multifunctional membrane protein with an emerging role in infections by malaria parasites. Expert Opin. Ther. Targets 2012, 16, 999–1001. [Google Scholar] [CrossRef]
- Yang, H.; Chen, J.; Yang, J.; Qiao, S.; Zhao, S.; Yu, L. Cyclophilin A is upregulated in small cell lung cancer and activates ERK1/2 signal. Biochem. Biophys. Res. Commun. 2007, 361, 763–767. [Google Scholar] [CrossRef]
- Bergers, G.; Benjamin, L.E. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer 2003, 3, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Lomenick, B.; Jung, G.; Wohlschlegel, J.A.; Huang, J. Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 2011, 3, 163–180. [Google Scholar] [CrossRef] [Green Version]
- Handschumacher, R.E.; Harding, M.W.; Rice, J.; Drugge, R.J.; Speicher, D.W. Cyclophilin: A specific cytosolic binding protein for cyclosporin A. Science 1984, 226, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, K.; Das, A.; Walsh, C.T. Hsp90 chaperonins possess ATPase activity and bind heat shock transcription factors and peptidyl prolyl isomerases. J. Biol. Chem. 1993, 268, 1479–1487. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Siegelin, M.D.; Dohi, T.; Altieri, D.C. Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 2010, 70, 8988–8993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uittenbogaard, A.; Ying, Y.; Smart, E.J. Characterization of a cytosolic heat-shock protein-caveolin chaperone complex. Involvement in cholesterol trafficking. J. Biol. Chem. 1998, 273, 6525–6532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundback, T.; Nordlund, P.; Martinez Molina, D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef]
- Mikol, V.; Kallen, J.; Pflügl, G.; Walkinshaw, M.D. X-ray structure of a monomeric cyclophilin A-cyclosporin A crystal complex at 2.1 A resolution. J. Mol. Biol. 1993, 234, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.; Schmid, F.X.; Fischer, G. Catalysis of protein folding by prolyl isomerase. Nature 1987, 329, 268–270. [Google Scholar] [CrossRef]
- Muramatsu, T.; Miyauchi, T. Basigin (CD147): A multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol. Histopathol. 2003, 18, 981–987. [Google Scholar]
- Feng, W.; Xin, Y.; Xiao, Y.; Li, W.; Sun, D. Cyclophilin A enhances cell proliferation and xenografted tumor growth of early gastric cancer. Dig. Dis. Sci. 2015, 60, 2700–2711. [Google Scholar] [CrossRef]
- Campa, M.J.; Wang, M.Z.; Howard, B.; Fitzgerald, M.C.; Patz, E.F. Protein expression profiling identifies macrophage migration inhibitory factor and cyclophilin a as potential molecular targets in non-small cell lung cancer. Cancer Res. 2003, 63, 1652–1656. [Google Scholar]
- Li, M.; Zhai, Q.; Bharadwaj, U.; Wang, H.; Li, F.; Fisher, W.E.; Chen, C.; Yao, Q. Cyclophilin A is overexpressed in human pancreatic cancer cells and stimulates cell proliferation through CD147. Cancer 2006, 106, 2284–2294. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Su, J.; Zhou, Y.; Xie, X.; Peng, C.; Yuan, Z.; Chen, X. Repressing CD147 is a novel therapeutic strategy for malignant melanoma. Oncotarget 2017, 8, 25806–25813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Koblinski, J.E.; Dutson, L.V.; Feeney, Y.B.; Clevenger, C.V. Prolyl isomerase cyclophilin A regulation of Janus-activated kinase 2 and the progression of human breast cancer. Cancer Res. 2008, 68, 7769–7778. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Kim, H.; Suk, K.; Less, W.H. Activation of CD147 with cyclophilin a induces the expression of IFITM1 through ERK and PI3K in THP-1 cells. Med. Inflamm. 2010, 2010, 821940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigro, P.; Pompilio, G.; Capogrossi, M.C. Cyclophilin A: A key player for human disease. Cell Death Dis. 2013, 4, e888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.C.; Takahashi, H.; Murai, Y.; Cui, Z.G.; Nomoto, K.; Miwa, S.; Tsuneyama, K.; Takano, Y. Upregulated EMMPRIN/CD147 might contribute to growth and angiogenesis of gastric carcinoma: A good marker for local invasion and prognosis. Br. J. Cancer 2006, 95, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Zhu, S.; Li, J.; Ji, G.; Wang, W.; Wu, G.; Zheng, J. CD147 expression in human gastric cancer is associated with tumor recurrence and prognosis. PLoS ONE 2014, 9, e101027. [Google Scholar] [CrossRef]
- Shiraishi, S.; Yokoo, H.; Kobayashi, H.; Yanagita, T.; Uezono, Y.; Minami, S.; Takasaki, M.; Wada, A. Post-translational reduction of cell surface expression of insulin receptors by cyclosporin A, FK506 and rapamycin in bovine adrenal chromaffin cells. Neurosci. Lett. 2000, 293, 211–215. [Google Scholar] [CrossRef]
- Chklovskaia, E.; Nissen, C.; Landmann, L.; Rahner, C.; Pfister, O.; Wodnar-Filipowicz, A. Cell-surface trafficking and release of flt3 ligand from T lymphocytes is induced by common cytokine receptor gamma-chain signaling and inhibited by cyclosporin A. Blood 2001, 97, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, V.; Pushkarsky, T.; Li, J.H.; Dai, W.W.; Sherry, B.; Bukrinsky, M. Regulation of CD147 cell surface expression: Involvement of the proline residue in the CD147 transmembrane domain. J. Biol. Chem. 2005, 280, 17013–17019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihatsch, M.J.; Kyo, M.; Morozumi, K.; Yamaguchi, Y.; Nickeleit, V.; Ryffel, B. The side-effects of ciclosporine-A and tacrolimus. Clin. Nephrol. 1998, 49, 356–363. [Google Scholar] [PubMed]










| Spot no. | Uniprot ID | Protein Name | Accession no. | Sequence Coverage (%)/ Peptides Matched | Mascot Score | Protein MW (kDa) | pI Value | Downregulation by Compound 9 (Fold) |
|---|---|---|---|---|---|---|---|---|
| 271 | MIC60_HUMAN | motor protein | BAA04656 | 27/12 | 211 | 83.678 | 6.08 | 3.5 |
| 516 | CH60A_ARATH | heat shock protein 60 (HSP60) | NP_189041 | 19/17 | 249 | 61.281 | 5.7 | 5.3 |
| 736 | ANXA7_HUMAN | synexin (Annexin VII) | AAA36616 | 17/11 | 525 | 50.316 | 6.25 | 3.9 |
| 1165 | MLEC_HUMAN | Malectin | Q14165 | 22/10 | 128 | 32.234 | 5.27 | 5.2 |
| 1216 | CAPZB_HUMAN | F-actin capping protein beta subunit | AAA87395 | 15/14 | 226 | 30.629 | 5.69 | 5.3 |
| 1257 | LEG3_HUMAN | IgE-binding protein | AAA35607 | 26/14 | 200 | 26.188 | 8.58 | 3.0 |
| 1541 | CBX3_HUMAN | HP1-gamma (CBX3) | AAB48101 | 26/20 | 344 | 19.72 | 5.03 | 3.1 |
| 1687 | MYL6_HUMAN | non-muscle myosin light chain | AAA59893 | 21/16 | 105 | 16.961 | 4.46 | 3.7 |
| 1699 | PPIA_HUMAN | peptidylprolyl isomerase A (Cyclophilin A) | CAA37039 | 27/23 | 402 | 18.013 | 7.68 | 3.2 |
| 1703 | ATPD_HUMAN | H(+)-transporting ATP synthase | CAA45016 | 11/9 | 117 | 17.49 | 5.38 | 3.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.M.; Sohng, J.K.; Lee, W.-H.; Oh, T.-J.; Jung, H.J. Identification of Cyclophilin A as a Potential Anticancer Target of Novel Nargenicin A1 Analog in AGS Gastric Cancer Cells. Int. J. Mol. Sci. 2021, 22, 2473. https://doi.org/10.3390/ijms22052473
Han JM, Sohng JK, Lee W-H, Oh T-J, Jung HJ. Identification of Cyclophilin A as a Potential Anticancer Target of Novel Nargenicin A1 Analog in AGS Gastric Cancer Cells. International Journal of Molecular Sciences. 2021; 22(5):2473. https://doi.org/10.3390/ijms22052473
Chicago/Turabian StyleHan, Jang Mi, Jae Kyung Sohng, Woo-Haeng Lee, Tae-Jin Oh, and Hye Jin Jung. 2021. "Identification of Cyclophilin A as a Potential Anticancer Target of Novel Nargenicin A1 Analog in AGS Gastric Cancer Cells" International Journal of Molecular Sciences 22, no. 5: 2473. https://doi.org/10.3390/ijms22052473






