Structure of the Signal Transduction Domain in Second-Generation CAR Regulates the Input Efficiency of CAR Signals
Abstract
:1. Introduction
2. Results
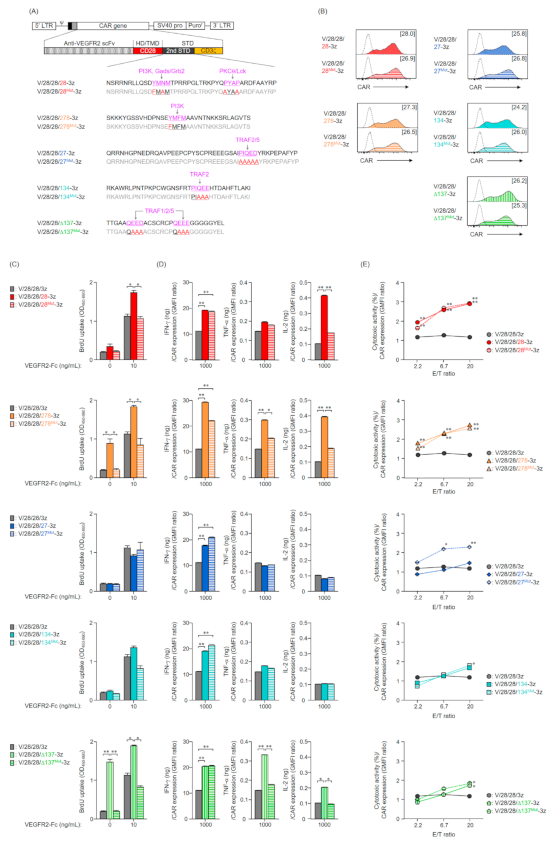
2.1. Expression of Various Second-Generation CARs on Mouse T Cells
2.2. Function of Mouse T Cells Expressing Various Second-Generation CARs
2.3. Expression and Function in Mouse T Cells of CAR Mutants Deficient in the Co-Stimulatory Signal Input Motif
2.4. Expression and Function in Mouse T Cells of Second-Generation CARs with an Altered Order of STDs
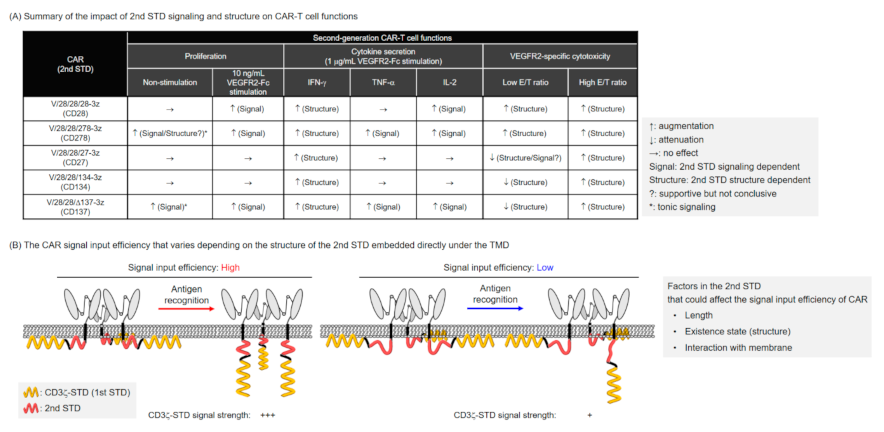
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Mice
4.3. Construction of CAR Structural Variants
4.4. Production of CAR-T Cells
4.5. RT-qPCR Analysis for CAR mRNA Expression
4.6. Flow Cytometry Analysis for CAR Surface Expression
4.7. BrdU Proliferation Assay and Cytokine ELISA
4.8. Cytotoxicity Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onea, A.S.; Jazirehi, A.R. CD19 chimeric antigen receptor (CD19 CAR)-redirected adoptive T-cell immunotherapy for the treatment of relapsed or refractory B-cell Non-Hodgkin’s Lymphomas. Am. J. Cancer Res. 2016, 6, 403–424. [Google Scholar] [PubMed]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef]
- Lee, D.W.; Kochenderfer, J.N.; Stetler-Stevenson, M.; Cui, Y.K.; Delbrook, C.; Feldman, S.A.; Fry, T.J.; Orentas, R.; Sabatino, M.; Shah, N.N.; et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: A phase 1 dose-escalation trial. Lancet 2015, 385, 517–528. [Google Scholar] [CrossRef]
- Morgan, R.A.; Yang, J.C.; Kitano, M.; Dudley, M.E.; Laurencot, C.M.; Rosenberg, S.A. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 2010, 18, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Maus, M.V.; Alexander, S.; Bishop, M.R.; Brudno, J.N.; Callahan, C.; Davila, M.L.; Diamonte, C.; Dietrich, J.; Fitzgerald, J.C.; Frigault, M.J.; et al. Society for Immunotherapy of Cancer (SITC) clinical practice guideline on immune effector cell-related adverse events. J. Immunother. Cancer 2020, 8, e001511. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Gardner, R.; Porter, D.L.; Louis, C.U.; Ahmed, N.; Jensen, M.; Grupp, S.A.; Mackall, C.L. Current concepts in the diagnosis and management of cytokine release syndrome. Blood 2014, 124, 188–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Hao, H.; Yang, G.; Zhang, Y.; Fu, Y. Immunotherapy with CAR-modified T cells: Toxicities and overcoming strategies. J. Immunol. Res. 2018, 2018, 2386187. [Google Scholar] [CrossRef]
- Watanabe, K.; Kurammitsu, S.; Posey, A.D., Jr.; June, C.H. Expanding the therapeutic window for CAR T cell therapy in solid tumors: The knowns and unknowns of CAR T cell biology. Front. Immunol. 2018, 9, 2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, K.; Tsunei, A.; Kusabuka, H.; Ogaki, E.; Tachibana, M.; Okada, N. Hinge and transmembrane domains of chimeric antigen receptor regulate receptor expression and signaling threshold. Cells 2020, 9, 1182. [Google Scholar] [CrossRef]
- Walker, A.J.; Majzner, R.G.; Zhang, L.; Wanhainen, K.; Long, A.H.; Nguyen, S.M.; Lopomo, P.; Vigny, M.; Fry, T.J.; Orentas, R.J.; et al. Tumor antigen and receptor densities regulate efficacy of a chimeric antigen receptor targeting anaplastic lymphoma kinase. Mol. Ther. 2017, 25, 2189–2201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoiber, S.; Cadilha, B.L.; Benmebarek, M.R.; Lesch, S.; Endres, S.; Kobold, S. Limitations in the design of chimeric antigen receptors for cancer therapy. Cells 2019, 8, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eshhar, Z.; Waks, T.; Gross, G.; Schindler, D.G. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc. Natl. Acad. Sci. USA 1993, 90, 720–724. [Google Scholar] [CrossRef] [Green Version]
- Rafiq, S.; Hackett, C.S.; Brentjens, R.J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin Oncol. 2020, 17, 147–167. [Google Scholar] [CrossRef]
- Lim, W.A.; June, C.H. The principles of engineering immune cells to treat cancer. Cell 2017, 168, 724–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinthus, J.H.; Waks, T.; Kaufman-Francis, K.; Schindler, D.G.; Harmelin, A.; Kanety, H.; Ramon, J.; Eshhar, Z. Immuno-gene therapy of established prostate tumors using chimeric receptor-redirected human lymphocytes. Cancer Res. 2003, 63, 2470–2476. [Google Scholar]
- Carpenito, C.; Milone, M.C.; Hassan, R.; Simonet, J.C.; Lakhal, M.; Suhoski, M.M.; Varela-Rohena, A.; Haines, K.M.; Heitjan, D.F.; Albelda, S.M.; et al. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc. Natl. Acad. Sci. USA 2009, 106, 3360–3365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkie, S.; Picco, G.; Foster, J.; Davies, D.M.; Julien, S.; Cooper, L.; Arif, S.; Mather, S.J.; Taylor-Papapdimitriou, J.; Burchell, M.; et al. Retargeting of human T cells to tumor-associated MUC1: The evolution of a chimeric antigen receptor. J. Immunol. 2008, 180, 4901–4909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milone, M.C.; Fish, J.D.; Carpenito, C.; Carroll, R.G.; Binder, G.K.; Teachey, D.; Samanta, M.; Lakhal, M.; Gloss, B. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 2009, 17, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Finney, H.M.; Akbar, A.N.; Lawson, A.D.G. Activation of resting human primary T cells with chimeric receptors: Costimulation from CD28, inducible costimulator, CD134, and CD137 in series with signals from the TCR zeta chain. J. Immunol. 2004, 172, 104–113. [Google Scholar] [CrossRef]
- Weinkove, R.; George, P.; Dasyam, N.; McLellan, A.D. Selecting costimulatory domains for chimeric antigen receptors: Functional and clinical considerations. Clin. Transl. Immunol. 2019, 8, e1049. [Google Scholar] [CrossRef] [Green Version]
- Savoldo, B.; Ramos, C.A.; Liu, E.; Mims, M.P.; Keating, M.J.; Carrum, G.; Kamble, R.T.; Bollard, C.M.; Gee, A.P.; Mei, Z.; et al. CD28 costimulation improves expansion and persistence of chimeric antigen receptor-modified T cells in lymphoma patients. J. Clin. Investig. 2011, 121, 1822–1826. [Google Scholar] [CrossRef] [Green Version]
- Salter, A.I.; Ivey, R.G.; Kennedy, J.J.; Voillet, V.V.; Rajan, A.; Alderman, E.J.; Voytovich, U.J.; Lin, C.; Sommermeyer, D.; Liu, L.; et al. Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci. Signal. 2018, 11, eaat6753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, A.H.; Haso, W.M.; Shern, J.F.; Wanhainen, K.M.; Murgai, M.; Ingaramo, M.; Smith, J.P.; Walker, A.J.; Kohler, M.E.; Venkateshwara, V.R.; et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 2015, 21, 581–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guedan, S.; Posey, A.D.; Jr Shaw, C.; Wing, A.; Da, T.; Patel, P.R.; McGettigan, S.E.; Casado-Medrano, V.; Kawalekar, O.U.; Uribe-Herranz, M.; et al. Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight 2018, 3, 96976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, Z.; He, T.; Wang, X.; Zheng, W.; Lin, N.; Tu, M.; Xie, Y.; Ping, L.; Zhang, C.; Liu, W.; et al. Parallel comparison of 4-1BB or CD28 co-stimulated CD19-targeted CAR-T cells for B cell Non-Hodgkin’s lymphoma. Mol. Ther. Oncolytics 2019, 15, 60–68. [Google Scholar] [CrossRef] [Green Version]
- McKay, M.J.; Afrose, F.; Koeppe, R.E.; Greathouse, D.V. Helix formation and stability in membranes. Biochim. Biophys. Acta. Biomembr. 2018, 1860, 2108–2117. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Ajina, A.; Maher, J. Strategies to address chimeric antigen receptor tonic signaling. Mol. Cancer Ther. 2018, 17, 1795–1815. [Google Scholar] [CrossRef] [Green Version]
- Calderon, H.; Mamonkin, M.; Guedan, S. Analysis of CAR-mediated tonic signaling. Methods Mol. Biol. 2020, 2086, 223–236. [Google Scholar] [PubMed]
- Alcover, A.; Alarcón, B.; Bartolo, V.D. Cell biology of T cell receptor expression and regulation. Annu. Rev. Immunol. 2018, 36, 103–125. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Gagnon, E.; Call, M.E.; Schnell, J.R.; Schwieters, C.D.; Carman, C.V.; Chou, J.J.; Wucherpfennig, K.W. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-Based motif. Cell 2008, 135, 702–713. [Google Scholar] [CrossRef] [Green Version]
- Maher, J.; Brentjens, R.J.; Gunset, G.; Rivière, I.; Sadelain, M. Human T-lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRzeta /CD28 receptor. Nat. Biotechnol. 2002, 20, 70–75. [Google Scholar] [CrossRef]
- Finney, H.M.; Lawson, A.D.; Bebbington, C.R.; Weir, A.N. Chimeric receptors providing both primary and costimulatory signaling in T cells from a single gene product. J. Immunol. 1998, 161, 2791–2797. [Google Scholar]
- Dobbins, J.; Gagnon, E.; Godec, J.; Pyrdol, J.; Vignali, D.A.A.; Sharpe, A.H.; Wucherpfennig, K.W. Binding of the cytoplasmic domain of CD28 to the plasma membrane inhibits Lck recruitment and signaling. Sci. Signal. 2016, 9, ra75. [Google Scholar] [CrossRef] [Green Version]
- Arch, R.H.; Thompson, C.B. 4-1BB and Ox40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor kappaB. Mol. Cell Biol. 1998, 18, 558–565. [Google Scholar] [CrossRef] [Green Version]
- Feucht, J.; Sun, J.; Eyquem, J.; Ho, Y.; Zhao, Z.; Leibold, J.; Dobrin, A.; Cabriolu, A.; Hamieh, M.; Sadelain, M.; et al. Calibration of CAR activation potential directs alternative T cell fates and therapeutic potency. Nat. Med. 2019, 25, 82–88. [Google Scholar] [CrossRef]
- Sun, C.; Shou, P.; Du, H.; Hirabayashi, K.; Chen, Y.; Herring, L.E.; Ahn, S.; Xu, Y.; Suzuki, K.; Li, G.; et al. THEMIS-SHP1 Recruitment by 4-1BB tunes Lck-mediated priming of chimeric antigen receptor-redirected T cells. Cancer Cell. 2020, 37, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.C.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017, 543, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Kanagawa, N.; Yanagawa, T.; Mukai, Y.; Yoshioka, Y.; Okada, N.; Nakagawa, S. Tumor-targeting CTL expressing a single-chain Fv specific for VEGFR2. Biochem. Biophys. Res. Commun. 2010, 394, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Kusabuka, H.; Fujiwara, K.; Tokunaga, Y.; Hirobe, S.; Nakagawa, S.; Okada, N. Highly efficient gene transfer using a retroviral vector into murine T cells for preclinical chimeric antigen receptor-expressing T cell therapy. Biochem. Biophys. Res. Commun. 2016, 473, 73–79. [Google Scholar] [CrossRef] [PubMed]



| Components | Gene Sequences | |
|---|---|---|
| CD3ζ | STD (AA52-164) | AGAGCAAAAT TCAGCAGGAG TGCAGAGACT GCTGCCAACC TGCAGGACCC CAACCAGCTC TACAATGAGC TCAATCTAGG GCGAAGAGAG GAATATGACG TCTTGGAGAA GAAGCGGGCT CGGGATCCAG AGATGGGAGG CAAACAGCAG AGGAGGAGGA ACCCCCAGGA AGGCGTATAC AATGCACTGC AGAAAGACAA GATGGCAGAA GCCTACAGTG AGATCGGCAC AAAAGGCGAG AGGCGGAGAG GCAAGGGGCA CGATGGCCTT TACCAGGGTC TCAGCACTGC CACCAAGGAC ACCTATGATG CCCTGCATAT GCAGACCCTG GCCCCTCGC |
| CD8α | HD/TMD (AA131-217) | GTCATCAGCA ACTCGGTGAT GTACTTCAGT TCTGTCGTGC CAGTCCTTCA GAAAGTGAAC TCTACTACTA CCAAGCCAGT GCTGCGAACT CCCTCACCTG TGCACCCTAC CGGGACATCT CAGCCCCAGA GACCAGAAGA TTGTCGGCCC CGTGGCTCAG TGAAGGGGAC CGGATTGGAC TTCGCCTGTG ATATTTACAT CTGGGCACCC TTGGCCGGAA TCTGCGTGGC CCTTCTGCTG TCCTTGATCA TCACTCTCAT C |
| CD28 | HD (AA115-177) | ATTGAGTTCA TGTACCCTCC GCCTTACCTA GACAACGAGA GGAGCAATGG AACTATTATT CACATAAAAG AGAAACATCT TTGTCATACT CAGTCATCTC CTAAGCTGTT TTGGGCACTG GTCGTGGTTG CTGGAGTCCT GTTTTGTTAT GGCTTGCTAG TGACAGTGGC TCTTTGTGTG ATCTGGACA |
| STD (AA178-218) | AATAGTAGAA GGAACAGACT CCTTCAAAGT GACTACATGA ACATGACTCC CCGGAGGCCT GGGCTCACTC GAAAGCCTTA CCAGCCCTAC GCCCCTGCCA GAGACTTTGC AGCGTACCGC CCC | |
| STD (AA178-218)Mut | AATAGTAGAA GGAACAGACT CCTTCAAAGT GACTTCATGG CCATGACTCC CCGGAGGCCT GGGCTCACTC GAAAGCCTTA CCAGGCCTAC GCCGCTGCCA GAGACTTTGC AGCGTACCGC CCC | |
| STD (AA178-218)PI3K-Mut | AATAGTAGAA GGAACAGACT CCTTCAAAGT GACTTCATGG CCATGACTCC CCGGAGGCCT GGGCTCACTC GAAAGCCTTA CCAGCCCTAC GCCCCTGCCA GAGACTTTGC AGCGTACCGC CCC | |
| STD (AA178-218)Lck-Mut | AATAGTAGAA GGAACAGACT CCTTCAAAGT GACTACATGA ACATGACTCC CCGGAGGCCT GGGCTCACTC GAAAGCCTTA CCAGGCCTAC GCCGCTGCCA GAGACTTTGC AGCGTACCGC CCC | |
| CD278 | STD (AA166-200) | TCAAAAAAGA AATACGGATC CAGTGTGCAT GACCCTAATA GTGAATACAT GTTCATGGCG GCAGTCAACA CAAACAAAAA GTCTAGACTT GCAGGTGTGA CCTCA |
| STD (AA166-200)Mut | TCAAAAAAGA AATACGGATC CAGTGTGCAT GACCCTAATA GTGAATTCAT GTTCATGGCG GCAGTCAACA CAAACAAAAA GTCTAGACTT GCAGGTGTGA CCTCA | |
| CD27 | STD (AA204-250) | CAAAGAAGAA ACCACGGGCC AAATGAAGAC CGGCAGGCAG TGCCTGAAGA GCCTTGTCCT TACAGCTGCC CCAGGGAAGA GGAGGGCAGT GCTATCCCTA TCCAGGAGGA CTACCGGAAA CCCGAGCCTG CTTTCTACCC T |
| STD (AA204-250)Mut | CAAAGAAGAA ACCACGGGCC AAATGAAGAC CGGCAGGCAG TGCCTGAAGA GCCTTGTCCT TACAGCTGCC CCAGGGAAGA GGAGGGCAGT GCTATCGCTG CCGCGGCGGC CTACCGGAAA CCCGAGCCTG CTTTCTACCC T | |
| CD134 | STD (AA237-272) | CGGAAGGCTT GGAGATTGCC TAACACTCCC AAACCTTGTT GGGGAAACAG CTTCAGGACC CCGATCCAGG AGGAACACAC AGACGCACAC TTTACTCTGG CCAAGATC |
| STD (AA237-272)Mut | CGGAAGGCTT GGAGATTGCC TAACACTCCC AAACCTTGTT GGGGAAACAG CTTCAGGACC CCGATCGCGG CGGCCCACAC AGACGCACAC TTTACTCTGG CCAAGATC | |
| CD137 | STD (AA209-256) | TCTGTGCTCA AATGGATCAG GAAAAAATTC CCCCACATAT TCAAGCAACC ATTTAAGAAG ACCACTGGAG CAGCTCAAGA GGAAGATGCT TGTAGCTGCC GATGTCCACA GGAAGAAGAA GGAGGAGGAG GAGGCTATGA GCTGT |
| STD Δ20 (AA229-256) | ACCACTGGAG CAGCTCAAGC GGCGGCCGCT TGTAGCTGCC GATGTCCACA GGAAGAAGAA GGAGGAGGAG GAGGCTATGA GCTGT | |
| STD Δ20 (AA229-256)Mut | ACCACTGGAG CAGCTCAAGA GGAAGATGCT TGTAGCTGCC GATGTCCACA GGCGGCGGCC GGAGGAGGAG GAGGCTATGA GCTGT | |
| Components | Amino Acid Sequences | |
|---|---|---|
| CD3ζ | STD (AA52-164) | RAKFSRSAET AANLQDPNQL YNELNLGRRE EYDVLEKKRA RDPEMGGKQQ RRRNPQEGVY NALQKDKMAE AYSEIGTKGE RRRGKGHDGL YQGLSTATKD TYDALHMQTL APR |
| CD8α | HD/TMD (AA131-217) | VISNSVMYFS SVVPVLQKVN STTTKPVLRT PSPVHPTGTS QPQRPEDCRP RGSVKGTGLD FACDIYIWAP LAGICVALLL SLIITLI |
| CD28 | HD (AA115-177) | IEFMYPPPYL DNERSNGTII HIKEKHLCHT QSSPKLFWAL VVVAGVLFCY GLLVTVALCV IWT |
| STD (AA178-218) | NSRRNRLLQS DYMNMTPRRP GLTRKPYQPY APARDFAAYR P | |
| STD (AA178-218)Mut | NSRRNRLLQS DFMAMTPRRP GLTRKPYQAY AAARDFAAYR P | |
| STD (AA178-218)PI3K-Mut | NSRRNRLLQS DFMAMTPRRP GLTRKPYQPY APARDFAAYR P | |
| STD (AA178-218)Lck-Mut | NSRRNRLLQS DYMNMTPRRP GLTRKPYQAY AAARDFAAYR P | |
| CD278 | STD (AA166-200) | SKKKYGSSVH DPNSEYMFMA AVNTNKKSRL AGVTS |
| STD (AA166-200)Mut | SKKKYGSSVH DPNSEFMFMA AVNTNKKSRL AGVTS | |
| CD27 | STD (AA204-250) | QRRNHGPNED RQAVPEEPCP YSCPREEEGS AIPIQEDYRK PEPAFYP |
| STD (AA204-250)Mut | QRRNHGPNED RQAVPEEPCP YSCPREEEGS AIAAAAAYRK PEPAFYP | |
| CD134 | STD (AA237-272) | RKAWRLPNTP KPCWGNSFRT PIQEEHTDAH FTLAKI |
| STD (AA237-272)Mut | RKAWRLPNTP KPCWGNSFRT PIAAAHTDAH FTLAKI | |
| CD137 | STD (AA209-256) | SVLKWIRKKF PHIFKQPFKK TTGAAQEEDA CSCRCPQEEE GGGGGYEL |
| STD Δ20 (AA229-256) | TTGAAQEEDA CSCRCPQEEE GGGGGYEL | |
| STD Δ20 (AA229-256)Mut | TTGAAQAAAA CSCRCPQAAA GGGGGYEL | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujiwara, K.; Kitaura, M.; Tsunei, A.; Kusabuka, H.; Ogaki, E.; Okada, N. Structure of the Signal Transduction Domain in Second-Generation CAR Regulates the Input Efficiency of CAR Signals. Int. J. Mol. Sci. 2021, 22, 2476. https://doi.org/10.3390/ijms22052476
Fujiwara K, Kitaura M, Tsunei A, Kusabuka H, Ogaki E, Okada N. Structure of the Signal Transduction Domain in Second-Generation CAR Regulates the Input Efficiency of CAR Signals. International Journal of Molecular Sciences. 2021; 22(5):2476. https://doi.org/10.3390/ijms22052476
Chicago/Turabian StyleFujiwara, Kento, Masaki Kitaura, Ayaka Tsunei, Hotaka Kusabuka, Erika Ogaki, and Naoki Okada. 2021. "Structure of the Signal Transduction Domain in Second-Generation CAR Regulates the Input Efficiency of CAR Signals" International Journal of Molecular Sciences 22, no. 5: 2476. https://doi.org/10.3390/ijms22052476
APA StyleFujiwara, K., Kitaura, M., Tsunei, A., Kusabuka, H., Ogaki, E., & Okada, N. (2021). Structure of the Signal Transduction Domain in Second-Generation CAR Regulates the Input Efficiency of CAR Signals. International Journal of Molecular Sciences, 22(5), 2476. https://doi.org/10.3390/ijms22052476






