Lipid Dynamics in Diisobutylene-Maleic Acid (DIBMA) Lipid Particles in Presence of Sensory Rhodopsin II
Abstract
:1. Introduction
2. Results
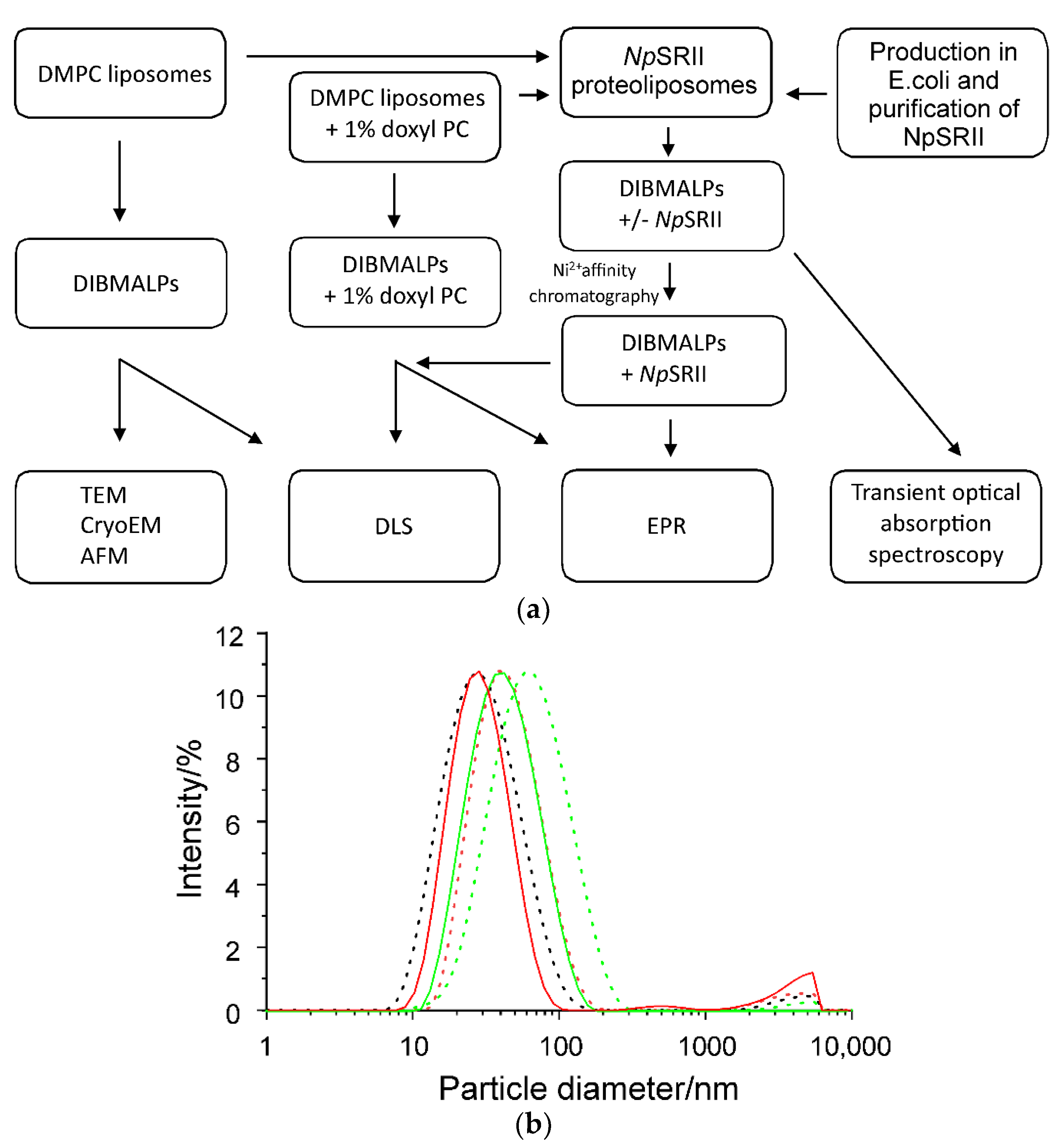
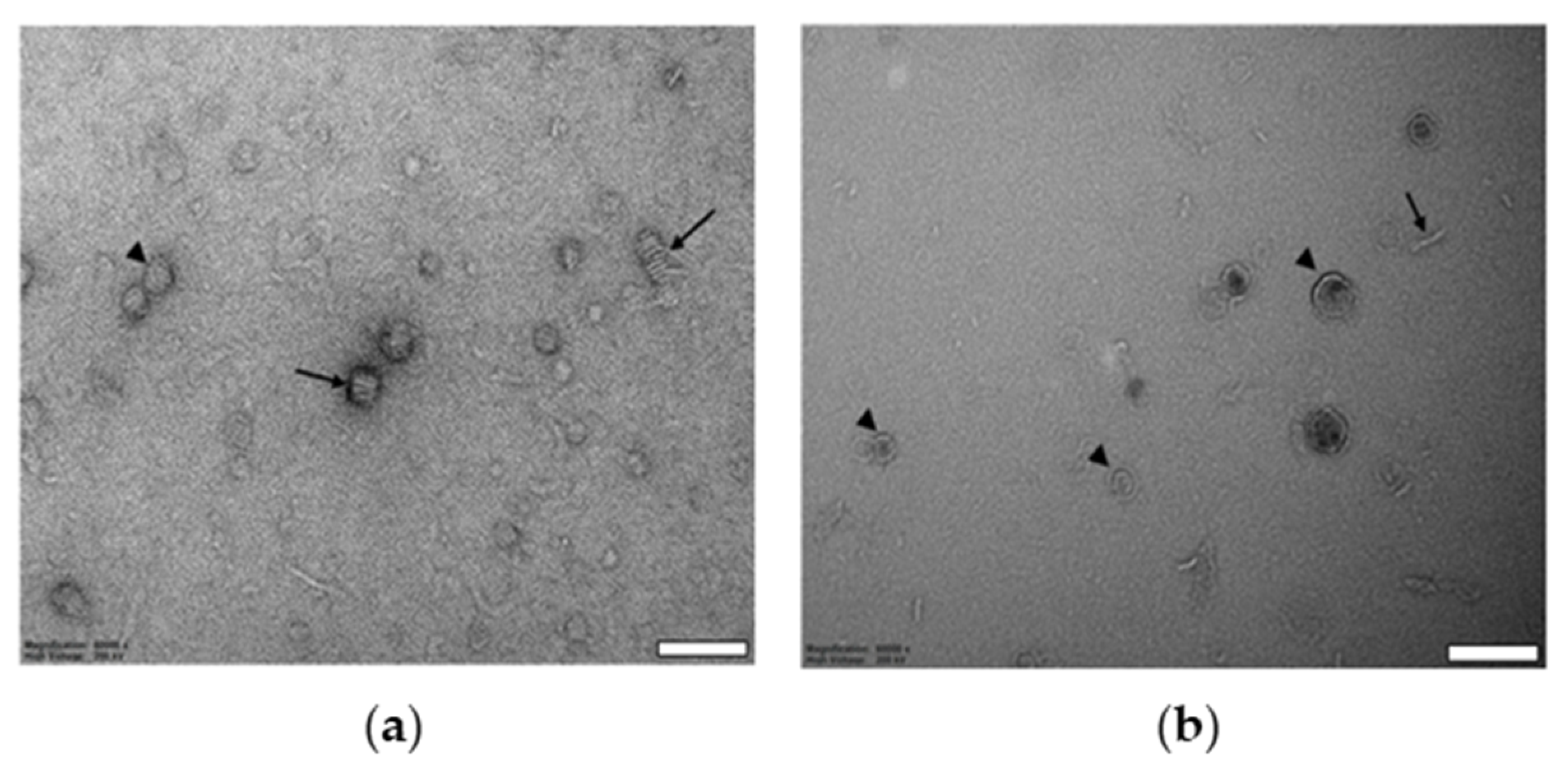
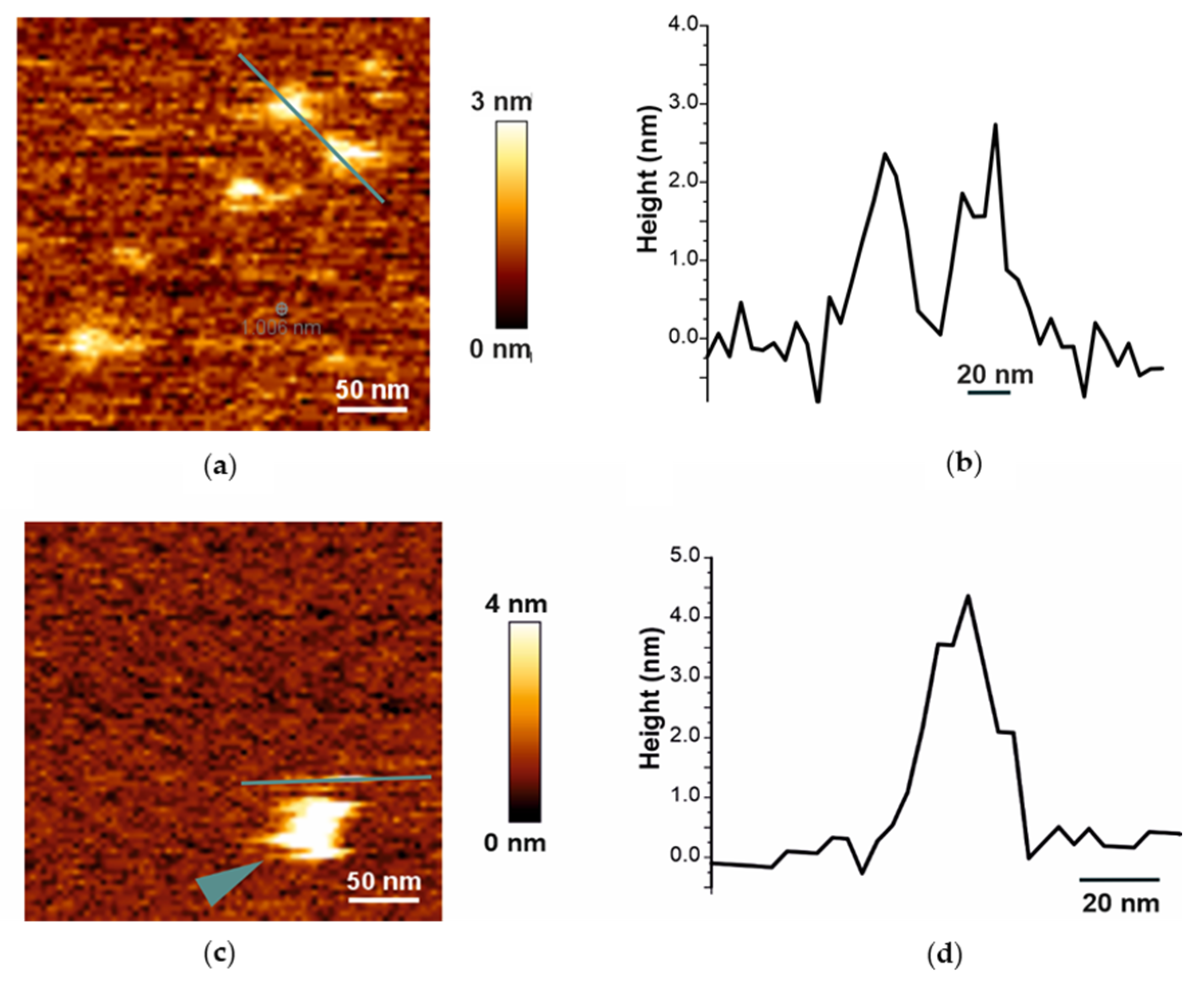
2.1. Biophysical Characterization of Empty and Protein-Containing DIBMALPs
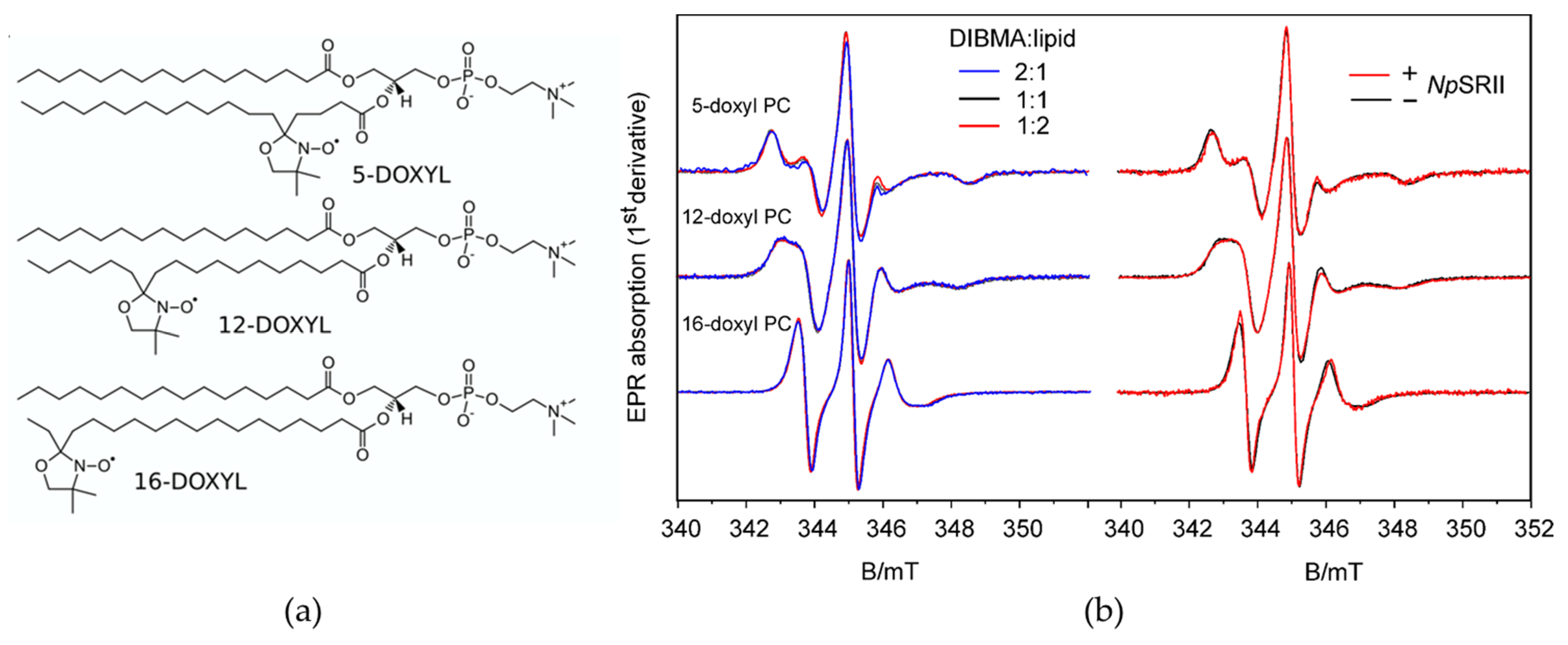
2.2. Lipid Dynamics and Ordering Are Not Affected by the Presence of NpSRII
2.3. Coarse-Grained Molecular Dynamics Simulations of Dibmalps Complement the Experimental Results
2.4. The Photocycle of NpSRII in DIBMALPs Depends on the Protein-to-Lipid Ratio
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Protein Expression and Purification
4.3. Liposome Preparations
4.4. Proteoliposome Preparation
4.5. Preparation of DIBMALPs
4.6. Preparation of DIBMALPs Containing NpSRII
4.7. Dynamic Light Scattering
4.8. Transmission Electron Microscopy
4.9. Cryo-Transmission Electron Microscopy
4.10. Atomic Force Microscopy
4.11. Electron Paramagnetic Resonance Spectroscopy
4.12. EPR Spectra Simulations
4.13. Transient Optical Absorption Spectroscopy
4.14. Molecular Dynamics Simulations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006, 5, 993–996. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Sligar, S.G. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003, 12, 2476–2481. [Google Scholar] [CrossRef] [Green Version]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef] [PubMed]
- De Zorzi, R.; Mi, W.; Liao, M.; Walz, T. Single-particle electron microscopy in the study of membrane protein structure. Microscopy 2016, 65, 81–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klare, J.P.; Bordignon, E.; Doebber, M.; Fitter, J.; Kriegsmann, J.; Chizhov, I.; Steinhoff, H.J.; Engelhard, M. Effects of solubilization on the structure and function of the sensory rhodopsin II/transducer complex. J. Mol. Biol. 2006, 356, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- Zoonens, M.; Comer, J.; Masscheleyn, S.; Pebay-Peyroula, E.; Chipot, C.; Miroux, B.; Dehez, F. Dangerous liaisons between detergents and membrane proteins. The case of mitochondrial uncoupling protein 2. J. Am. Chem. Soc. 2013, 135, 15174–15182. [Google Scholar] [CrossRef]
- Knowles, T.J.; Finka, R.; Smith, C.; Lin, Y.P.; Dafforn, T.; Overduin, M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009, 131, 7484–7485. [Google Scholar] [CrossRef] [PubMed]
- Jamshad, M.; Lin, Y.P.; Knowles, T.J.; Parslow, R.A.; Harris, C.; Wheatley, M.; Poyner, D.R.; Bill, R.M.; Thomas, O.R.; Overduin, M.; et al. Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem. Soc. Trans. 2011, 39, 813–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orwick-Rydmark, M.; Lovett, J.E.; Graziadei, A.; Lindholm, L.; Hicks, M.R.; Watts, A. Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano Lett. 2012, 12, 4687–4692. [Google Scholar] [CrossRef]
- Orwick, M.C.; Judge, P.J.; Procek, J.; Lindholm, L.; Graziadei, A.; Engel, A.; Grobner, G.; Watts, A. Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angew. Chem. Int. Ed. 2012, 51, 4653–4657. [Google Scholar] [CrossRef] [PubMed]
- Dorr, J.M.; Scheidelaar, S.; Koorengevel, M.C.; Dominguez, J.J.; Schafer, M.; Van Walree, C.A.; Killian, J.A. The styrene-maleic acid copolymer: A versatile tool in membrane research. Eur. Biophys. J. 2016, 45, 3–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.C.; Pollock, N.L. Membrane proteins: Is the future disc shaped? Biochem. Soc. Trans. 2016, 44, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Overduin, M.; Esmaili, M. Structures and Interactions of Transmembrane Targets in Native Nanodiscs. SLAS Discov. 2019, 24, 943–952. [Google Scholar] [CrossRef]
- Ravula, T.; Hardin, N.Z.; Ramamoorthy, A. Polymer nanodiscs: Advantages and limitations. Chem. Phys. Lipids 2019, 219, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, M.J.; Overduin, M. Structure and function of proteins in membranes and nanodiscs. Biochim. Biophys. Acta Biomembr. 2020, 1863, 183445. [Google Scholar] [CrossRef] [PubMed]
- Arenas, R.C.; Klingler, J.; Vargas, C.; Keller, S. Influence of lipid bilayer properties on nanodisc formation mediated by styrene/maleic acid copolymers. Nanoscale 2016, 8, 15016–15026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez Pardo, J.J.; Dorr, J.M.; Iyer, A.; Cox, R.C.; Scheidelaar, S.; Koorengevel, M.C.; Subramaniam, V.; Killian, J.A. Solubilization of lipids and lipid phases by the styrene-maleic acid copolymer. Eur. Biophys. J. 2017, 46, 91–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, A.F.; Clark, E.E.; Sahu, I.D.; Zhang, R.; Frantz, N.D.; Al-Abdul-Wahid, M.S.; Dabney-Smith, C.; Konkolewicz, D.; Lorigan, G.A. Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies. Biochim. Biophys. Acta 2016, 1858, 2931–2939. [Google Scholar] [CrossRef] [PubMed]
- Voskoboynikova, N.; Mosslehy, W.; Colbasevici, A.; Ismagulova, T.T.; Bagrov, D.V.; Akovantseva, A.A.; Timashev, P.S.; Mulkidjanian, A.Y.; Bagratashvili, V.N.; Shaitan, K.V.; et al. Characterization of an archaeal photoreceptor/transducer complex from Natronomonas pharaonis assembled within styrene-maleic acid lipid particles. Rsc Adv. 2017, 7, 51324–51334. [Google Scholar] [CrossRef] [Green Version]
- Gulati, S.; Jamshad, M.; Knowles, T.J.; Morrison, K.A.; Downing, R.; Cant, N.; Collins, R.; Koenderink, J.B.; Ford, R.C.; Overduin, M.; et al. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J. 2014, 461, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Postis, V.; Rawson, S.; Mitchell, J.K.; Lee, S.C.; Parslow, R.A.; Dafforn, T.R.; Baldwin, S.A.; Muench, S.P. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim. Biophys. Acta 2015, 1848, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Parmar, M.; Rawson, S.; Scarff, C.A.; Goldman, A.; Dafforn, T.R.; Muench, S.P.; Postis, V.L.G. Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim. Biophys. Acta 2018, 1860, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Gennis, R.B. Single-particle cryo-EM studies of transmembrane proteins in SMA copolymer nanodiscs. Chem. Phys. Lipids 2019, 221, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Oluwole, A.O.; Danielczak, B.; Meister, A.; Babalola, J.O.; Vargas, C.; Keller, S. Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew. Chem. Int. Ed. 2017, 56, 1919–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oluwole, A.O.; Klingler, J.; Danielczak, B.; Babalola, J.O.; Vargas, C.; Pabst, G.; Keller, S. Formation of Lipid-Bilayer Nanodiscs by Diisobutylene/Maleic Acid (DIBMA) Copolymer. Langmuir 2017, 33, 14378–14388. [Google Scholar] [CrossRef] [PubMed]
- Swainsbury, D.J.K.; Scheidelaar, S.; Van Grondelle, R.; Killian, J.A.; Jones, M.R. Bacterial Reaction Centers Purified with Styrene Maleic Acid Copolymer Retain Native Membrane Functional Properties and Display Enhanced Stability. Angew. Chem. Int. Ed. 2014, 53, 11803–11807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barniol-Xicota, M.; Verhelst, S.H.L. Stable and Functional Rhomboid Proteases in Lipid Nanodiscs by Using Diisobutylene/Maleic Acid Copolymers. J. Am. Chem. Soc. 2018, 140, 14557–14561. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 2004, 1666, 62–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inagaki, S.; Ghirlando, R.; White, J.F.; Gvozdenovic-Jeremic, J.; Northup, J.K.; Grisshammer, R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 2012, 417, 95–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soubias, O.; Gawrisch, K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta 2012, 1818, 234–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rietveld, A.; Berkhout, T.A.; Roenhorst, A.; Marsh, D.; De Kruijff, B. Preferential association of apocytochrome c with negatively charged phospholipids in mixed model membranes. Biochim. Biophys. Acta 1986, 858, 38–46. [Google Scholar] [CrossRef]
- Wolfs, J.A.; Horvath, L.I.; Marsh, D.; Watts, A.; Hemminga, M.A. Spin-label ESR of bacteriophage M13 coat protein in mixed lipid bilayers. Characterization of molecular selectivity of charged phospholipids for the bacteriophage M13 coat protein in lipid bilayers. Biochemistry 1989, 28, 9995–10001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, R.C.; Ten Berge, D.; Nouwen, N.; Snel, M.M.; Tommassen, J.; Marsh, D.; De Kruijff, B. Mode of insertion of the signal sequence of a bacterial precursor protein into phospholipid bilayers as revealed by cysteine-based site-directed spectroscopy. Biochemistry 1996, 35, 3063–3071. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D.; McCarrick, R.M.; Troxel, K.R.; Zhang, R.F.; Smith, H.J.; Dunagan, M.M.; Swartz, M.S.; Rajan, P.V.; Kroncke, B.M.; Sanders, C.R.; et al. DEER EPR Measurements for Membrane Protein Structures via Bifunctional Spin Labels and Lipodisq Nanoparticles. Biochemistry 2013, 52, 6627–6632. [Google Scholar] [CrossRef] [Green Version]
- Orban-Glass, I.; Voskoboynikova, N.; Busch, K.B.; Klose, D.; Rickert, C.; Mosslehy, W.; Roder, F.; Wilkens, V.; Piehler, J.; Engelhard, M.; et al. Clustering and Dynamics of Phototransducer Signaling Domains Revealed by Site-Directed Spin Labeling Electron Paramagnetic Resonance on SRII/HtrII in Membranes and Nanodiscs. Biochemistry 2015, 54, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D.; Kroncke, B.M.; Zhang, R.; Dunagan, M.M.; Smith, H.J.; Craig, A.; McCarrick, R.M.; Sanders, C.R.; Lorigan, G.A. Structural investigation of the transmembrane domain of KCNE1 in proteoliposomes. Biochemistry 2014, 53, 6392–6401. [Google Scholar] [CrossRef] [Green Version]
- Stepien, P.; Polit, A.; Wisniewska-Becker, A. Comparative EPR studies on lipid bilayer properties in nanodiscs and liposomes. Biochim. Biophys. Acta 2015, 1848, 60–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosslehy, W.; Voskoboynikova, N.; Colbasevici, A.; Ricke, A.; Klose, D.; Klare, J.P.; Mulkidjanian, A.Y.; Steinhoff, H.J. Conformational Dynamics of Sensory Rhodopsin II in Nanolipoprotein and Styrene-Maleic Acid Lipid Particles. Photochem. Photobiol. 2019, 95, 1195–1204. [Google Scholar] [CrossRef]
- Bali, A.P.; Sahu, I.D.; Craig, A.F.; Clark, E.E.; Burridge, K.M.; Dolan, M.T.; Dabney-Smith, C.; Konkolewicz, D.; Lorigan, G.A. Structural characterization of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using EPR spectroscopy. Chem. Phys. Lipids 2019, 220, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, W.L.; McConnell, H.M. Molecular motion in spin-labeled phospholipids and membranes. J. Am. Chem. Soc. 1971, 93, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Subczynski, W.K.; Raguz, M.; Widomska, J. Studying lipid organization in biological membranes using liposomes and EPR spin labeling. Methods Mol. Biol. 2010, 606, 247–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subczynski, W.K.; Wisniewska, A.; Yin, J.J.; Hyde, J.S.; Kusumi, A. Hydrophobic barriers of lipid bilayer membranes formed by reduction of water penetration by alkyl chain unsaturation and cholesterol. Biochemistry 1994, 33, 7670–7681. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Cheng, L.; Faustino, I.; Guo, W.; Marrink, S.J. Molecular Mechanism of Lipid Nanodisk Formation by Styrene-Maleic Acid Copolymers. Biophys. J. 2018, 115, 494–502. [Google Scholar] [CrossRef] [Green Version]
- Orekhov, P.S.; Bozdaganyan, M.E.; Voskoboynikova, N.; Mulkidjanian, A.Y.; Steinhoff, H.J.; Shaitan, K.V. Styrene/Maleic Acid Copolymers Form SMALPs by Pulling Lipid Patches out of the Lipid Bilayer. Langmuir 2019, 35, 3748–3758. [Google Scholar] [CrossRef] [PubMed]
- Colbasevici, A.; Voskoboynikova, N.; Orekhov, P.S.; Bozdaganyan, M.E.; Karlova, M.G.; Sokolova, O.S.; Klare, J.P.; Mulkidjanian, A.Y.; Shaitan, K.V.; Steinhoff, H.J. Lipid dynamics in nanoparticles formed by maleic acid-containing copolymers: EPR spectroscopy and molecular dynamics simulations. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183207. [Google Scholar] [CrossRef]
- Bagrov, D.V.; Voskoboynikova, N.; Armeev, G.A.; Mosslehy, W.; Gluhov, G.S.; Ismagulova, T.T.; Mulkidjanian, A.Y.; Kirpichnikov, M.P.; Steinhoff, H.J.; Shaitan, K.V. Characterization of lipodisc nanoparticles containing sensory rhodopsin ii and its cognate transducer from Natronomonas pharaonis. Biophysics 2016, 61, 942–949. [Google Scholar] [CrossRef]
- Freed, J.H. Theory of slow tumbling ESR spectra for nitroxides. In Spin Labeling: Theory and Applications; Springer: Berlin/Heidelberg, Germany, 1976; Volume 1, pp. 53–132. [Google Scholar]
- Marsh, D.; Watts, A.; Pates, R.D.; Uhl, R.; Knowles, P.F.; Esmann, M. ESR spin-label studies of lipid-protein interactions in membranes. Biophys. J. 1982, 37, 265–274. [Google Scholar] [CrossRef] [Green Version]
- Shalaeva, D.N.; Galperin, M.Y.; Mulkidjanian, A.Y. Eukaryotic G protein-coupled receptors as descendants of prokaryotic sodium-translocating rhodopsins. Biol. Direct 2015, 10, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordeliy, V.I.; Labahn, J.; Moukhametzianov, R.; Efremov, R.; Granzin, J.; Schlesinger, R.; Buldt, G.; Savopol, T.; Scheidig, A.J.; Klare, J.P.; et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 2002, 419, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Chizhov, I.; Schmies, G.; Seidel, R.; Sydor, J.R.; Luttenberg, B.; Engelhard, M. The photophobic receptor from Natronobacterium pharaonis: Temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 1998, 75, 999–1009. [Google Scholar] [CrossRef] [Green Version]
- Wegener, A.A.; Klare, J.P.; Engelhard, M.; Steinhoff, H.J. Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. Embo J. 2001, 20, 5312–5319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klare, J.P.; Bordignon, E.; Engelhard, M.; Steinhoff, H.J. Sensory rhodopsin II and bacteriorhodopsin: Light activated helix F movement. Photochem. Photobiol. Sci. 2004, 3, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Schmies, G.; Luttenberg, B.; Chizhov, I.; Engelhard, M.; Becker, A.; Bamberg, E. Sensory rhodopsin II from the haloalkaliphilic natronobacterium pharaonis: Light-activated proton transfer reactions. Biophys. J. 2000, 78, 967–976. [Google Scholar] [CrossRef] [Green Version]
- Wegener, A.A.; Chizhov, I.; Engelhard, M.; Steinhoff, H.J. Time-resolved detection of transient movement of helix F in spin-labelled pharaonis sensory rhodopsin II. J. Mol. Biol. 2000, 301, 881–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forte, T.M.; Nichols, A.V.; Gong, E.L.; Levy, R.I.; Lux, S. Electron microscopic study on reassembly of plasma high density apoprotein with various lipids. Biochim. Biophys. Acta 1971, 248, 381–386. [Google Scholar] [CrossRef]
- Brouillette, C.G.; Jones, J.L.; Ng, T.C.; Kercret, H.; Chung, B.H.; Segrest, J.P. Structural studies of apolipoprotein A-I/phosphatidylcholine recombinants by high-field proton NMR, nondenaturing gradient gel electrophoresis, and electron microscopy. Biochemistry 1984, 23, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.F.; Walker, M.; Siebert, C.A.; Muench, S.P.; Ranson, N.A. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods 2016, 100, 3–15. [Google Scholar] [CrossRef]
- Dobro, M.J.; Melanson, L.A.; Jensen, G.J.; McDowall, A.W. Plunge freezing for electron cryomicroscopy. Methods Enzymol. 2010, 481, 63–82. [Google Scholar] [CrossRef] [PubMed]
- Markvoort, A.J.; Van Santen, R.A.; Hilbers, P.A. Vesicle shapes from molecular dynamicssimulations. J. Phys. Chem. B 2006, 110, 22780–22785. [Google Scholar] [CrossRef] [PubMed]
- Chng, C.-P. Effect of simulation temperature on phospholipid bilayer–vesicle transition studied by coarse-grained molecular dynamics simulations. Soft Matter 2013, 9, 7294–7301. [Google Scholar] [CrossRef]
- Shillcock, J.C. Spontaneous vesicle self-assembly: A mesoscopic view of membrane dynamics. Langmuir 2012, 28, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, R.; Souza, P.C.T.; Thallmair, S.; Melo, M.N.; De Vries, A.H.; Marrink, S.J. Pitfalls of the Martini Model. J. Chem. Theory Comput. 2019, 15, 5448–5460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, J.; Spudich, J.L. The transducer protein HtrII modulates the lifetimes of sensory rhodopsin II photointermediates. Biophys. J. 1998, 75, 2435–2440. [Google Scholar] [CrossRef] [Green Version]
- Shibata, M.; Inoue, K.; Ikeda, K.; Konno, M.; Singh, M.; Kataoka, C.; Abe-Yoshizumi, R.; Kandori, H.; Uchihashi, T. Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci. Rep. 2018, 8, 8262. [Google Scholar] [CrossRef] [PubMed]
- Hohenfeld, I.P.; Wegener, A.A.; Engelhard, M. Purification of histidine tagged bacteriorhodopsin, pharaonis halorhodopsin and pharaonis sensory rhodopsin II functionally expressed in Escherichia coli. FEBS Lett. 1999, 442, 198–202. [Google Scholar] [CrossRef] [Green Version]
- Mennes, N.; Klare, J.P.; Chizhov, I.; Seidel, R.; Schlesinger, R.; Engelhard, M. Expression of the halobacterial transducer protein HtrII from Natronomonas pharaonis in Escherichia coli. FEBS Lett. 2007, 581, 1487–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klose, D.; Voskoboynikova, N.; Orban-Glass, I.; Rickert, C.; Engelhard, M.; Klare, J.P.; Steinhoff, H.J. Light-induced switching of HAMP domain conformation and dynamics revealed by time-resolved EPR spectroscopy. FEBS Lett. 2014, 588, 3970–3976. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Wassenaar, T.A.; Ingolfsson, H.I.; Bockmann, R.A.; Tieleman, D.P.; Marrink, S.J. Computational Lipidomics with insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory Comput. 2015, 11, 2144–2155. [Google Scholar] [CrossRef]
- De Jong, D.H.; Baoukina, S.; Ingolfsson, H.I.; Marrink, S.J. Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 2016, 199, 1–7. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kholina, E.G.; Kovalenko, I.B.; Bozdaganyan, M.E.; Strakhovskaya, M.G.; Orekhov, P.S. Cationic Antiseptics Facilitate Pore Formation in Model Bacterial Membranes. J. Phys. Chem. B 2020, 124, 8593–8600. [Google Scholar] [CrossRef] [PubMed]
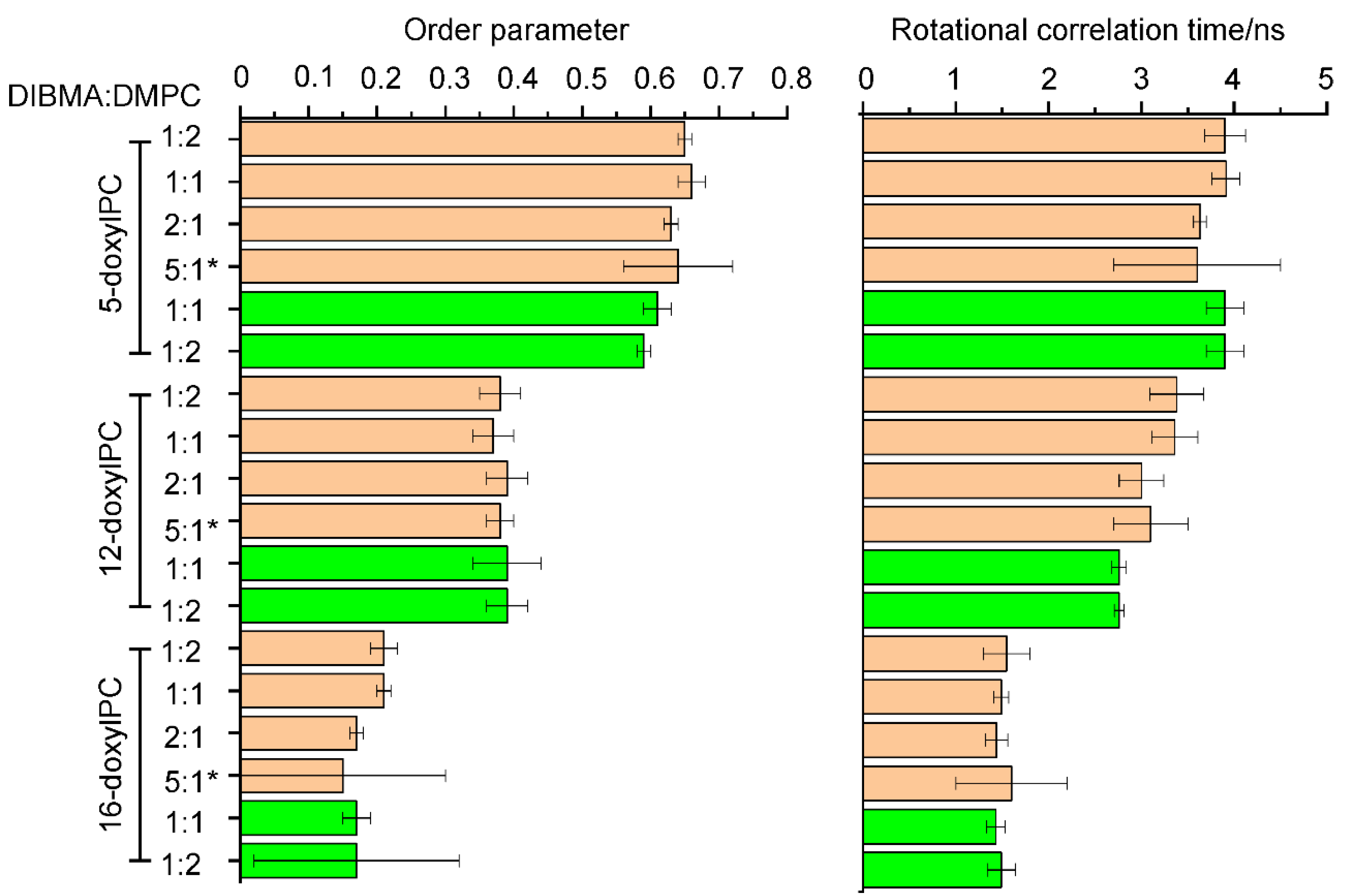
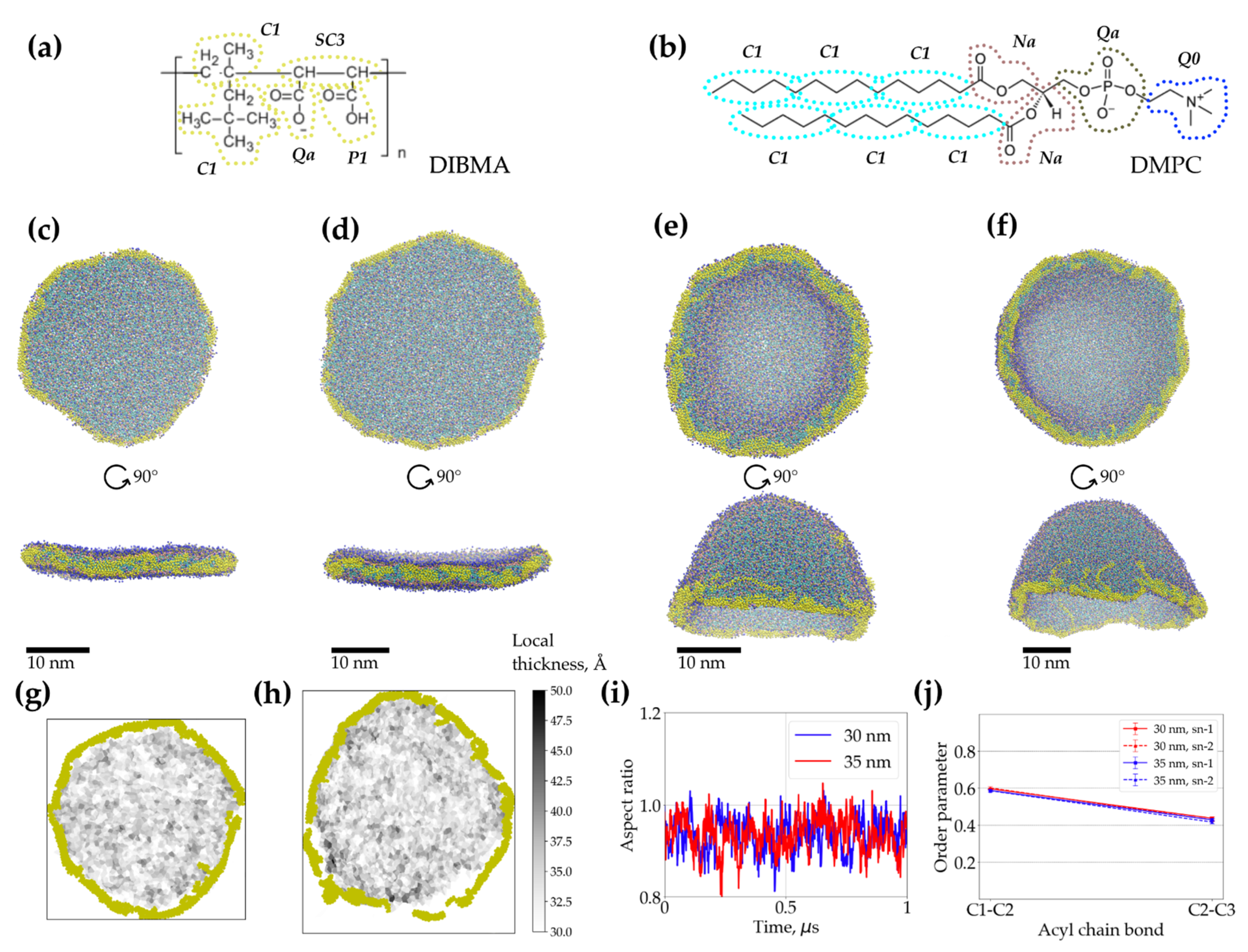

| DIBMA/DMPC w/w | Z-Average Hydrodynamic Diameter [nm] | |
|---|---|---|
| −NpSRII | +NpSRII | |
| 1:2 | 53.9 ± 0.4 | 36.3 ± 4.6 |
| 1:1 | 41.5 ± 2.0 | 25.9 ± 1.0 |
| 2:1 | 26.5 ± 1.6 | - |
| 5:1 * | 26.2 ± 3.0 | - |
| DIBMALP Diameter, nm | Number of DMPC | Number of DIBMA | Box Size, nm × nm × nm |
|---|---|---|---|
| 30 | 2322 | 26 | 39.5 × 39.5 × 39.5 |
| 35 | 3162 | 30 | 45.7 × 45.7 × 45.7 |
| 40 | 4138 | 35 | 51.7 × 51.7 × 51.7 |
| 50 | 6520 | 44 | 60.8 × 60.8 × 60.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voskoboynikova, N.; Orekhov, P.; Bozdaganyan, M.; Kodde, F.; Rademacher, M.; Schowe, M.; Budke-Gieseking, A.; Brickwedde, B.; Psathaki, O.-E.; Mulkidjanian, A.Y.; et al. Lipid Dynamics in Diisobutylene-Maleic Acid (DIBMA) Lipid Particles in Presence of Sensory Rhodopsin II. Int. J. Mol. Sci. 2021, 22, 2548. https://doi.org/10.3390/ijms22052548
Voskoboynikova N, Orekhov P, Bozdaganyan M, Kodde F, Rademacher M, Schowe M, Budke-Gieseking A, Brickwedde B, Psathaki O-E, Mulkidjanian AY, et al. Lipid Dynamics in Diisobutylene-Maleic Acid (DIBMA) Lipid Particles in Presence of Sensory Rhodopsin II. International Journal of Molecular Sciences. 2021; 22(5):2548. https://doi.org/10.3390/ijms22052548
Chicago/Turabian StyleVoskoboynikova, Natalia, Philipp Orekhov, Marine Bozdaganyan, Felix Kodde, Malte Rademacher, Maurice Schowe, Annette Budke-Gieseking, Britta Brickwedde, Olympia-Ekaterini Psathaki, Armen Y. Mulkidjanian, and et al. 2021. "Lipid Dynamics in Diisobutylene-Maleic Acid (DIBMA) Lipid Particles in Presence of Sensory Rhodopsin II" International Journal of Molecular Sciences 22, no. 5: 2548. https://doi.org/10.3390/ijms22052548
APA StyleVoskoboynikova, N., Orekhov, P., Bozdaganyan, M., Kodde, F., Rademacher, M., Schowe, M., Budke-Gieseking, A., Brickwedde, B., Psathaki, O.-E., Mulkidjanian, A. Y., Cosentino, K., Shaitan, K. V., & Steinhoff, H.-J. (2021). Lipid Dynamics in Diisobutylene-Maleic Acid (DIBMA) Lipid Particles in Presence of Sensory Rhodopsin II. International Journal of Molecular Sciences, 22(5), 2548. https://doi.org/10.3390/ijms22052548









