Exogenous 1′,4′-trans-Diol-ABA Induces Stress Tolerance by Affecting the Level of Gene Expression in Tobacco (Nicotiana tabacum L.)
Abstract
1. Introduction
2. Results
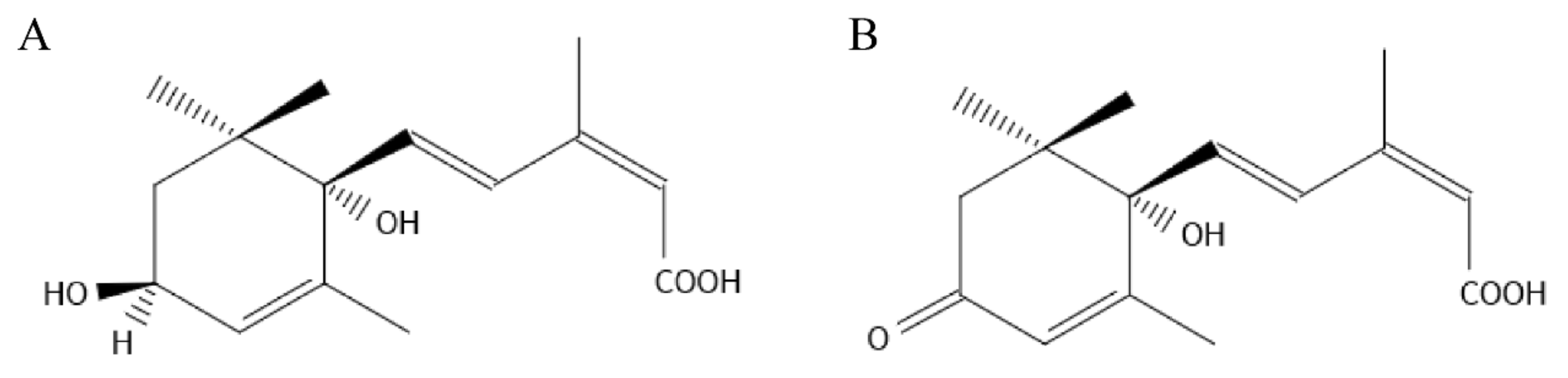
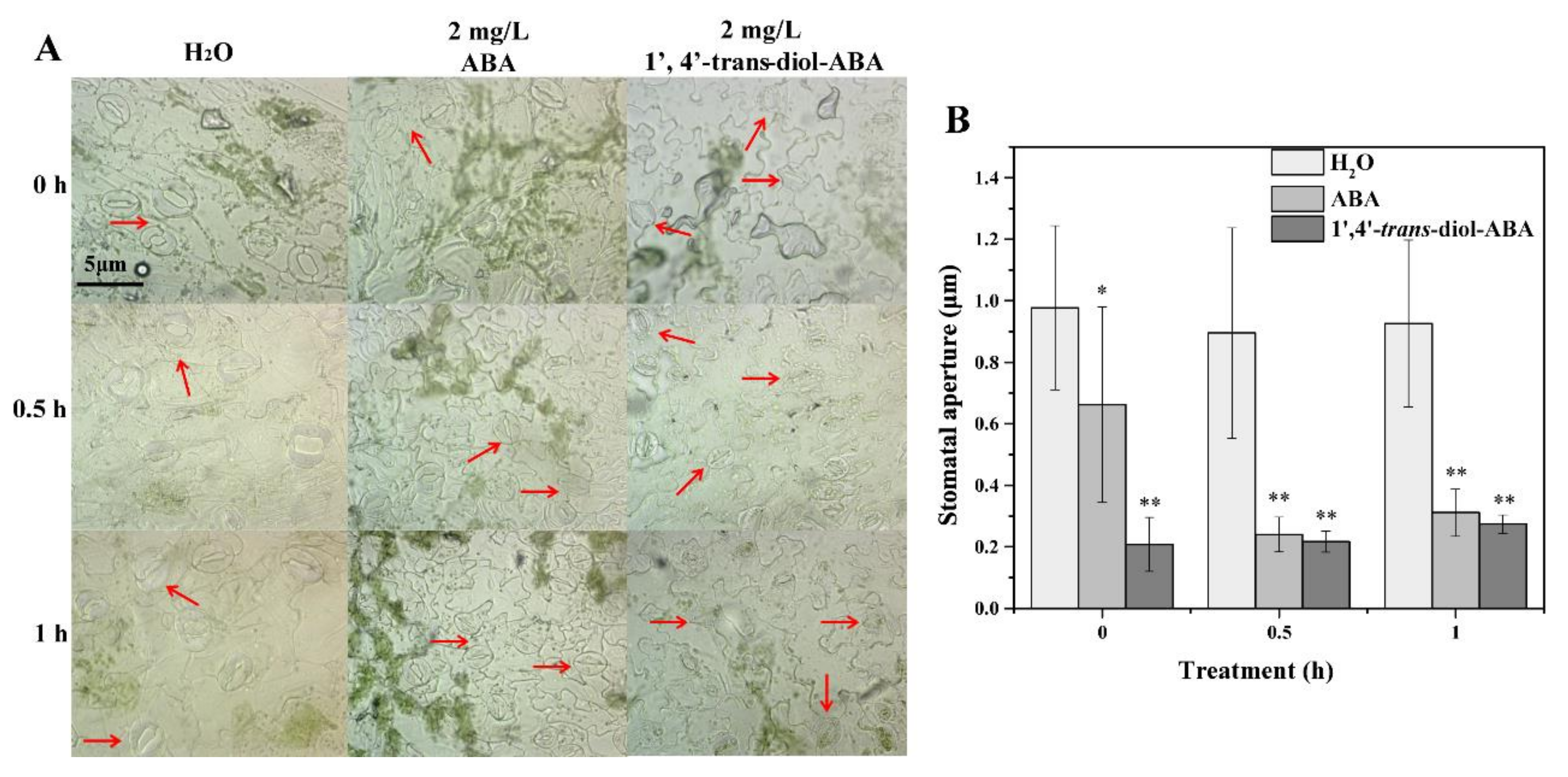
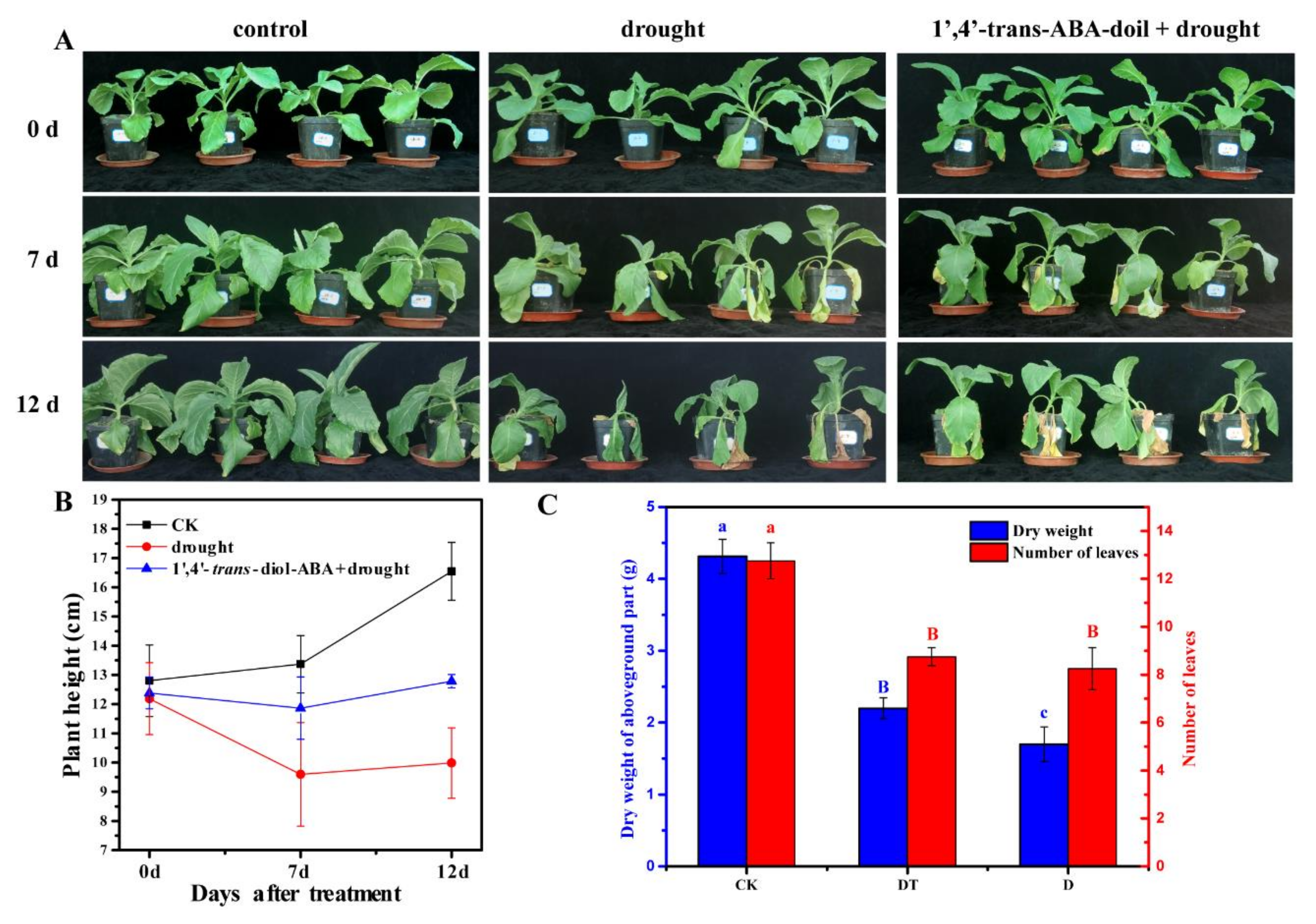
2.1. Blade Stomatal Movement and Drought Plant Morphology
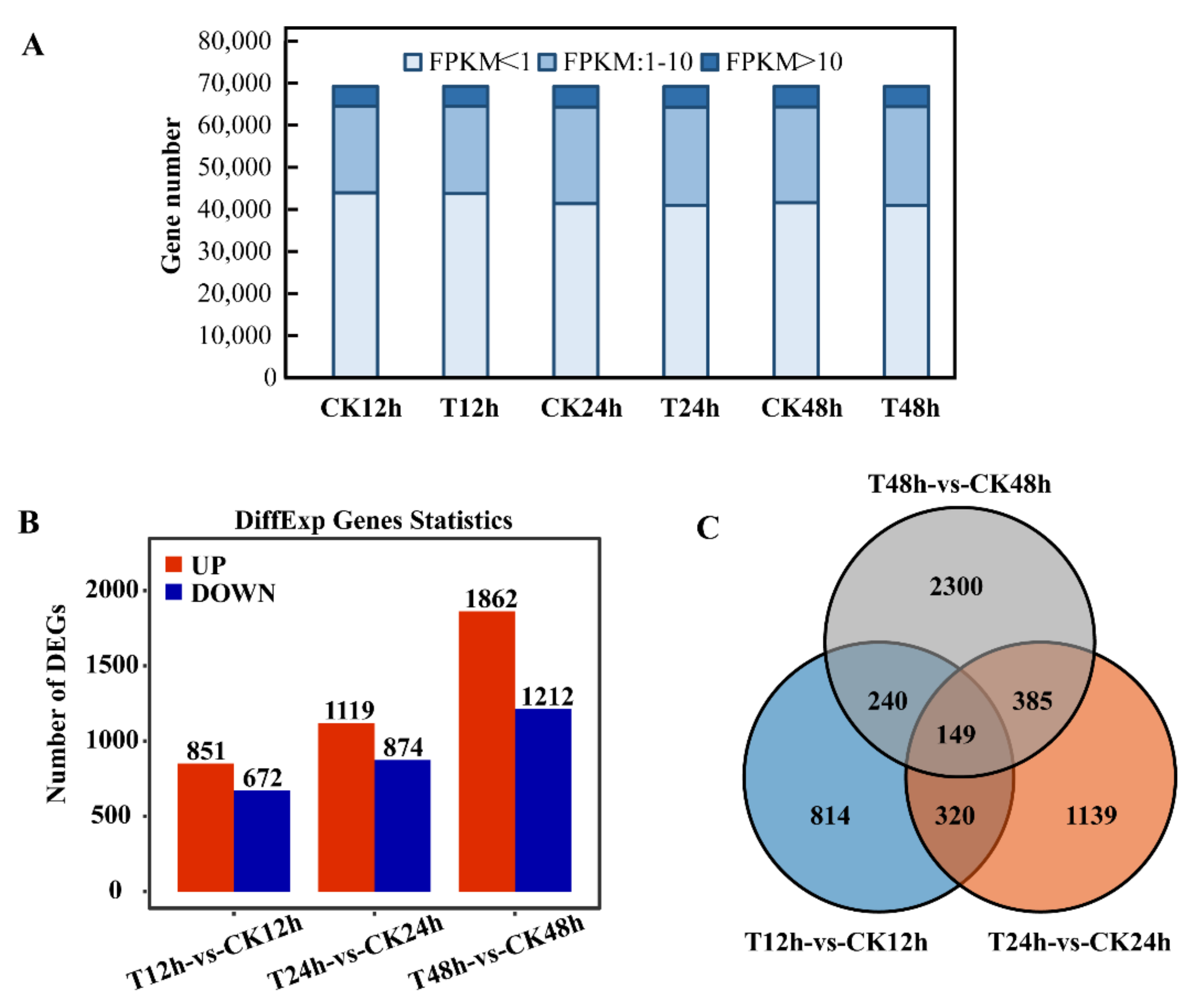
2.2. Data Analysis of RNA-Seq
2.3. Analysis of Differential Expression Genes
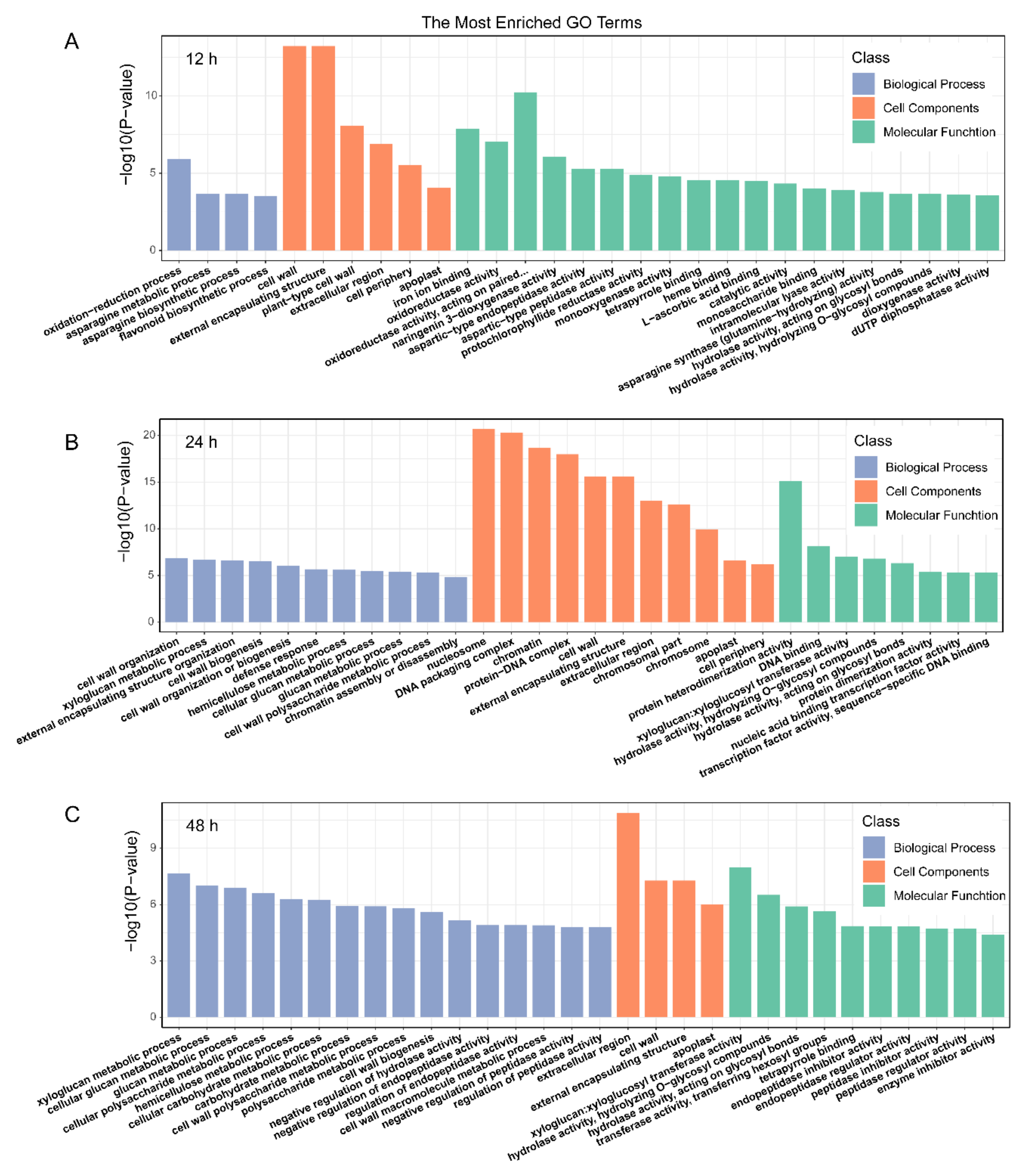
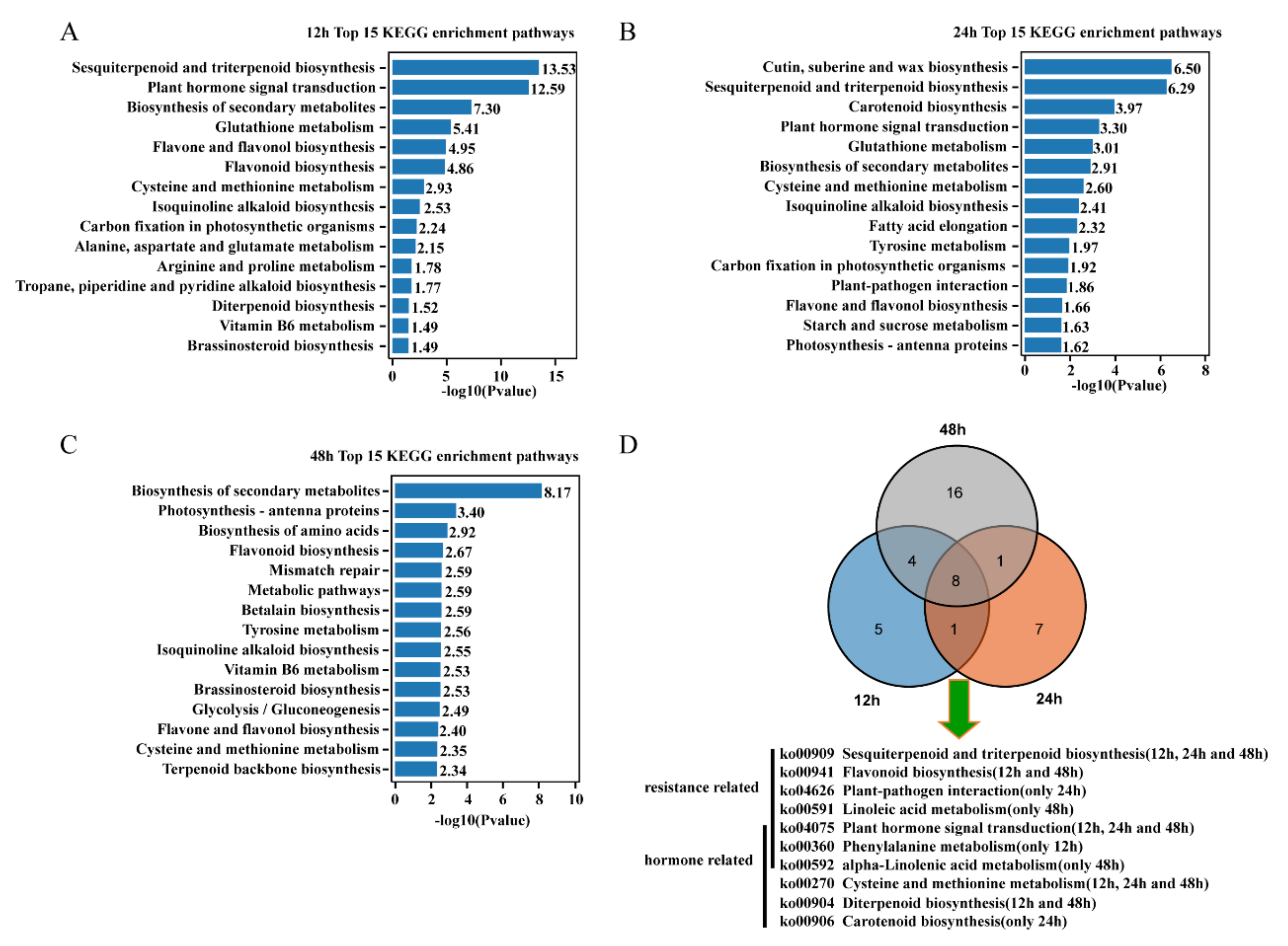
2.4. GO Function and KEGG Pathway Enrichment Analysis of DEGs
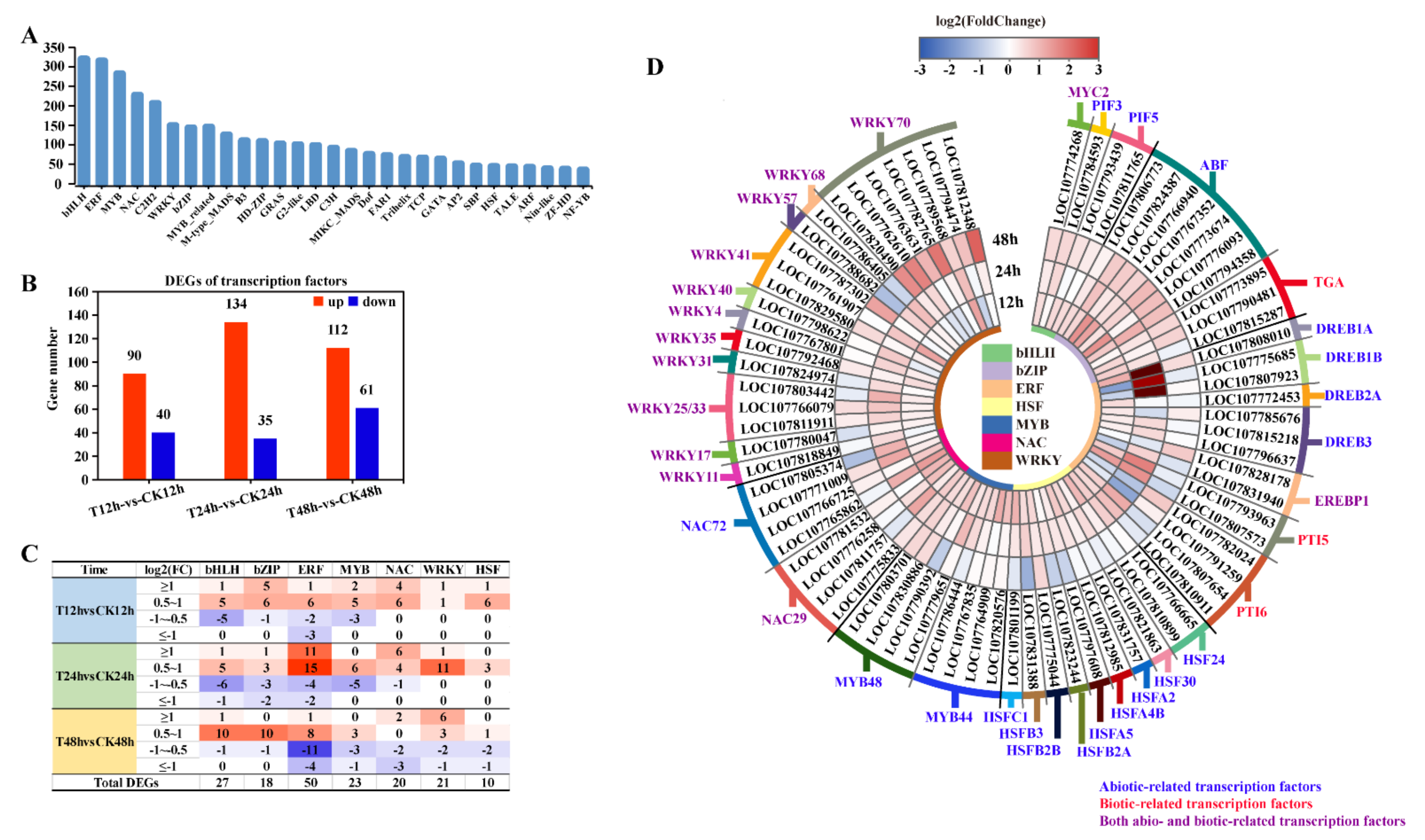
2.5. Transcription Factor Expression Analysis
2.6. Analysis of Gene Expression in the ABA Signaling Pathway
2.7. Other Hormone Signaling Pathway and Tolerance-Related Genes Expression Analysis
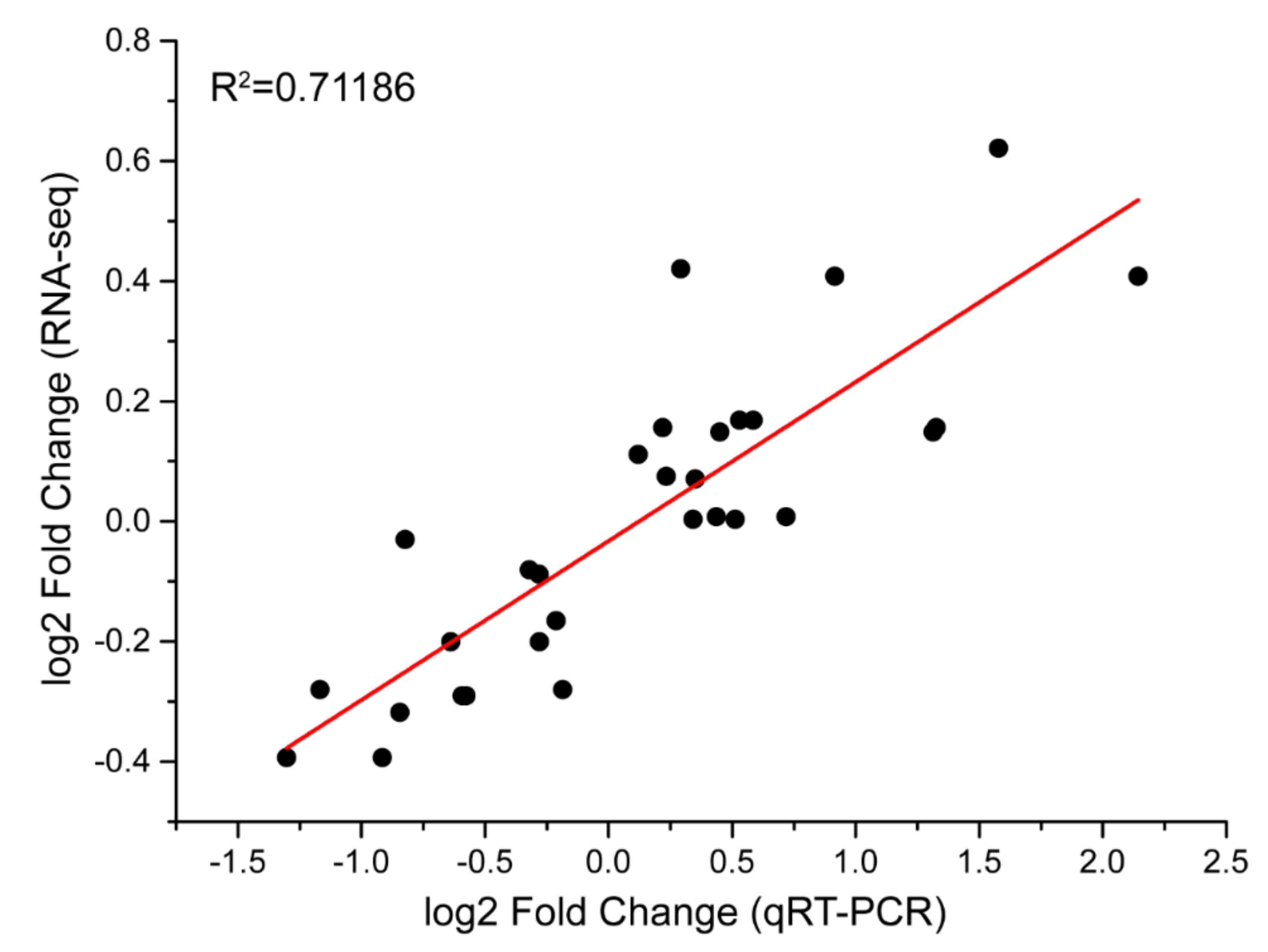
2.8. Validation of RNA-Seq Data by a qRT-PCR Analysis
3. Discussion
3.1. 1′,4′-trans-Diol-ABA Induced a Cascade Reaction of Gene Expression in Tobacco Leaves
3.2. 1′,4′-trans-Diol-ABA Induced the Expression of Transcription Factors Related to Stress Tolerance
3.3. 1′,4′-trans-Diol-ABA Induced the ABA Signaling Involved Gene Expressions
3.4. Other Hormone-Related Genes Respond to 1′,4′-trans-Diol-ABA
3.5. The Response of ROS Scavenging System and Other Defense-Related Genes to 1′,4′-trans-Diol-ABA
4. Conclusions
5. Materials and Methods
5.1. Plant Materials and Treatment
5.2. RNA Isolation and Transcriptome Sequencing
5.3. Raw Data Sorting, Filtering, and Quality Evaluation
5.4. Reference Genome Comparison and Quantification of Expression Level
5.5. Differentially Expressed Gene Analysis and Functional Enrichment
5.6. qRT-PCR Verification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Gong, Z.; Zhu, J.K. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J. Integr. Plant Biol. 2008, 50, 1187–1195. [Google Scholar] [CrossRef]
- Sun, Y.; Pri-Tal, O.; Michaeli, D.; Mosquna, A. Evolution of Abscisic Acid Signaling Module and Its Perception. Front Plant Sci. 2020, 11, 934. [Google Scholar] [CrossRef] [PubMed]
- Walton, D.C.; Sondheimer, E. Activity and metabolism of C-(+/−)-abscisic Acid derivatives. Plant Physiol. 1972, 49, 290–292. [Google Scholar] [CrossRef][Green Version]
- Siewers, V.; Kokkelink, L.; Smedsgaard, J.; Tudzynski, P. Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl. Environ. Microbiol. 2006, 72, 4619–4626. [Google Scholar] [CrossRef]
- Hirai, N. The 1′,4′-trans-diol of abscisic acid, a possible precursor of abscisic acid in Botrytis cinerea. Phytochemistry 1986, 25, 1865–1868. [Google Scholar] [CrossRef]
- Neill, S.; Horgan, R.; Walton, D.; Mercer, C. Biosynthesis of ABA in C-rosicola. 4. The Metabolism Of Alpha-Ionylidene Compounds By Cercospora-Rosicola. Phytochemistry 1987, 26, 2515–2519. [Google Scholar] [CrossRef]
- Okamoto, M.; Hirai, N.; Koshimizu, K. Biosynthesis of abscisic acid in Cercospora pini-densiflorae. Phytochemistry 1988, 27, 2099–2103. [Google Scholar] [CrossRef]
- Okamoto, M.; Hirai, N.; Koshimizu, K. Biosynthesis of abscisic acid from α-ionylideneethanol in Cercospora pini-densiflorae. Phytochemistry 1988, 27, 3465–3469. [Google Scholar] [CrossRef]
- Milborrow, B.V.; Lee, H.S. Endogenous biosynthetic precursors of (+)-abscisic acid. VII. The 1′,4′-trans-diol is formed from ABA, it is not a precursor. Funct. Plant Biol. 1998, 25, 729–737. [Google Scholar] [CrossRef]
- Rock, C.D.; Zeevaart, J.A.D. Abscisic (ABA)-Aldehyde Is a Precursor to, and 1′,4′-trans-ABA-Diol a Catabolite of, ABA in Apple. Plant Physiol. 1990, 93, 915–923. [Google Scholar] [CrossRef]
- Sindhu, R.K.; Walton, D.C. Xanthoxin Metabolism in Cell-free Preparations from Wild Type and Wilty Mutants of Tomato. Plant Physiol. 1988, 88, 178–182. [Google Scholar] [CrossRef]
- Wang, T.; Zhou, J.; Bai, B.; Yang, J.; Tan, H. Stereochemistry And Biological Activity of 1′,4′-trans-diol of ABA. Chin. J. Appl. Environ. Biol. 2005, 11, 699–701. [Google Scholar]
- Wang, T.S.; Zhou, J.Y.; Tan, H. Isolation and Crystal Structure of 1′,4′-Trans-diol of Abscisic Acid. Chin. J. Struc. Chem. 2006, 25, 1085–1089. [Google Scholar]
- Finkelstein, R. Abscisic Acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Ali, A.; Pardo, J.M.; Yun, D.J. Desensitization of ABA-Signaling: The Swing From Activation to Degradation. Front. Plant Sci. 2020, 11, 379. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Hu, R.S.; Liu, D.; Liu, X.; Wang, J.; Xiang, X.H.; Li, Y.Y. The AP2 transcription factor NtERF172 confers drought resistance by modifying NtCAT. Plant Biotechnol. J. 2020, 18, 2444–2455. [Google Scholar] [CrossRef]
- Zhou, S.; Zheng, W.J.; Liu, B.H.; Zheng, J.C.; Dong, F.S.; Liu, Z.F.; Wen, Z.Y.; Yang, F.; Wang, H.B.; Xu, Z.S.; et al. Characterizing the Role of TaWRKY13 in Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 5712. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Nguyen, N.C.; Nguyen, Y.H.; Watanabe, Y.; Tran, L.P.; Thao, N.P. The Soybean GmNAC019 Transcription Factor Mediates Drought Tolerance in Arabidopsis in an Abscisic Acid-Dependent Manner. Int. J. Mol. Sci. 2019, 21, 286. [Google Scholar] [CrossRef]
- Zhang, B.; Su, L.; Hu, B.; Li, L. Expression of AhDREB1, an AP2/ERF Transcription Factor Gene from Peanut, Is Affected by Histone Acetylation and Increases Abscisic Acid Sensitivity and Tolerance to Osmotic Stress in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1441. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Blakeslee, J.J.; Jones, M.L.; Chapin, L.J.; Dami, I.E. Exogenous abscisic acid enhances physiological, metabolic, and transcriptional cold acclimation responses in greenhouse-grown grapevines. Plant Sci. 2020, 293, 110437. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liang, C.J. Enhancing tolerance of rice (Oryza sativa) to simulated acid rain by exogenous abscisic acid. Environ. Sci. Pollut. R. 2017, 24, 4860–4870. [Google Scholar] [CrossRef]
- An, Y.; Zhou, P.; Liang, J.F. Effects of exogenous application of abscisic acid on membrane stability, osmotic adjustment, photosynthesis and hormonal status of two lucerne (Medicago sativa L.) genotypes under high temperature stress and drought stress. Crop Pasture Sci. 2014, 65, 274–286. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef]
- Liu, J.L.; Du, H.T.; Ding, X.; Zhou, Y.D.; Xie, P.F.; Wu, J.C. Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stal (Hemiptera: Delphacidae). Pest Manag. Sci. 2017, 73, 2559–2568. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Ma, X.; Tan, H.; Zhou, J. Abscisic acid enhances resistance to Alternaria solani in tomato seedlings. Plant Physiol. Biochem. Ppb 2011, 49, 693. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, A.S.; Helander, J.D.M.; Peterson, F.C.; Elzinga, D.; Cutler, S.R. Dynamic control of plant water use using designed ABA receptor agonists. Science 2019, 366, eaaw8848. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.J.; Zhang, Y.L.; Liu, X.; Huang, H.; Zhou, X.E.; Wang, W.L.; Zeng, A.; Zhao, C.Z.; Si, T.; Du, J.; et al. Combining chemical and genetic approaches to increase drought resistance in plants. Nat. Commun. 2017, 8, 1183. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.; Barros, R.; Bright, J.; Desikan, R.; Hancock, J.; Harrison, J.; Morris, P.; Ribeiro, D.; Wilson, I. Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot 2008, 59, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Hajihashemi, S. Stomatal Regulation as a Drought-tolerance Mechanism; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019. [Google Scholar]
- Wilkinson, S.; Davies, W.J. ABA-based chemical signalling: The co-ordination of responses to stress in plants. Plant Cell Environ. 2002, 25, 195–210. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Heidarvand, L.; Amiri, R.M. What happens in plant molecular responses to cold stress? Acta Physiol. Plant. 2010, 32, 419–431. [Google Scholar] [CrossRef]
- Zhu, J.; Dong, C.H.; Zhu, J.K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007, 10, 290–295. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef]
- Ramirez, S.R.; Basu, C. Comparative Analyses of Plant Transcription Factor Databases. Curr. Genom. 2009, 10, 10–17. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Besseau, S.; Toronen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 455–472. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.L.; Ba, L.J.; Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Involvement of WRKY Transcription Factors in Abscisic-Acid-Induced Cold Tolerance of Banana Fruit. J. Agric. Food Chem. 2017, 65, 3627–3635. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Kim, S.Y. The role of ABF family bZIP class transcription factors in stress response. Physiol. Plant. 2010, 126, 519–527. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.M.; Zhu, S.; Kachroo, P.; Kachroo, A. Signal regulators of systemic acquired resistance. Front. Plant Sci. 2015, 6, 228. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tao, X.; Tang, X.M.; Xiao, L.; Sun, J.L.; Yan, X.F.; Li, D.; Deng, H.Y.; Ma, X.R. Comparative transcriptome analysis of tomato (Solanum lycopersicum) in response to exogenous abscisic acid. BMC Genom. 2013, 14, 841. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Wirthmueller, L. Mapping FLS2 function to structure: LRRs, kinase and its working bits. Protoplasma 2013, 250, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Pattison, R.J.; Csukasi, F.; Zheng, Y.; Fei, Z.; van der Knaap, E.; Catala, C. Comprehensive Tissue-Specific Transcriptome Analysis Reveals Distinct Regulatory Programs during Early Tomato Fruit Development. Plant Physiol. 2015, 168, 1684–1701. [Google Scholar] [CrossRef]
- Liu, J.; Shi, M.; Wang, J.; Zhang, B.; Li, Y.; Wang, J.; El-Sappah, A.H.; Liang, Y. Comparative Transcriptomic Analysis of the Development of Sepal Morphology in Tomato (Solanum Lycopersicum L.). Int. J. Mol. Sci. 2020, 21, 16. [Google Scholar]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools—An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
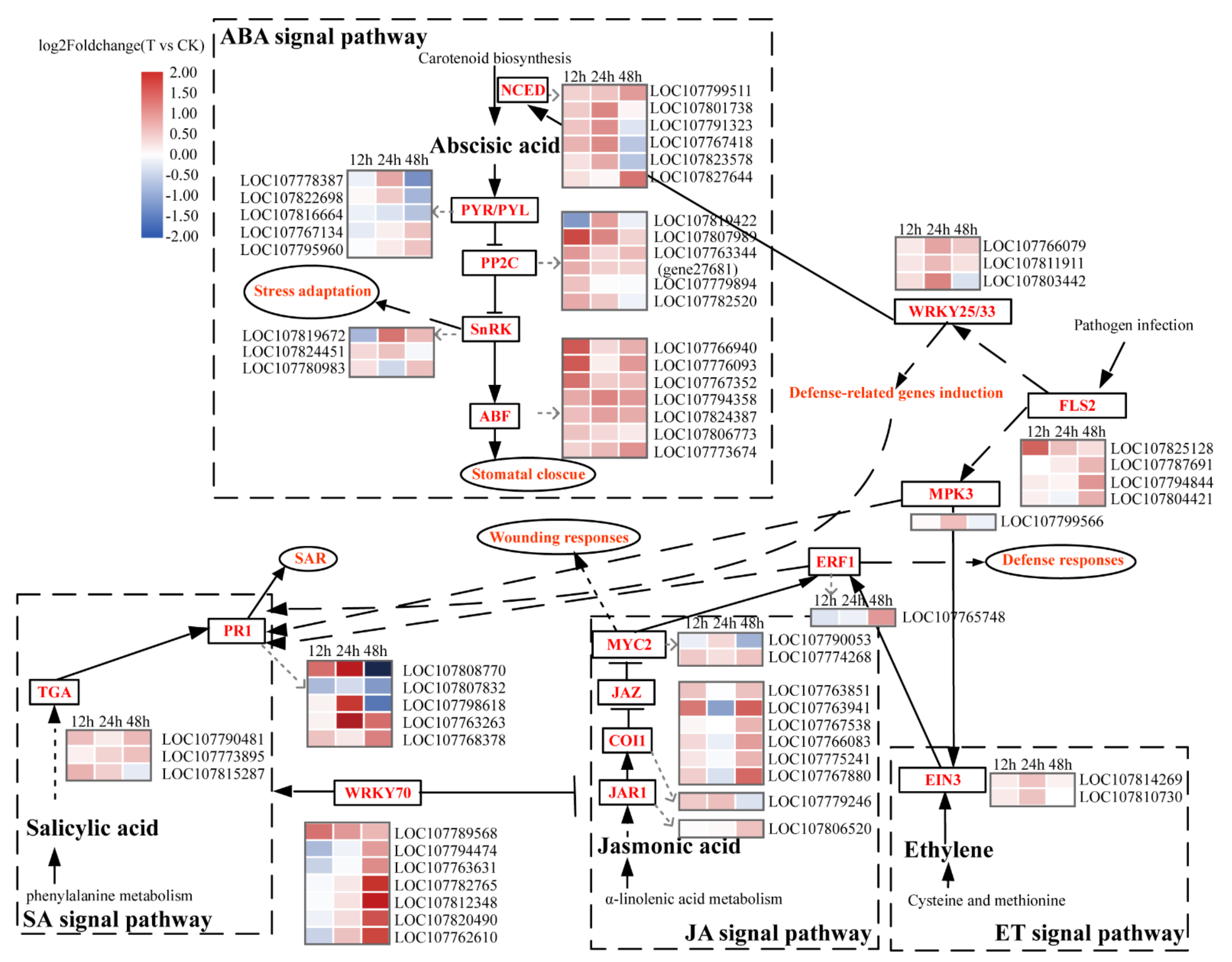

| Trait | Description | 12 h | 24 h | 48 h | Total DEGs | Sum |
|---|---|---|---|---|---|---|
| up(down) * | up(down) | up(down) | ||||
| Abscisic Acid | NCED | 2(0) | 5(0) | 2(2) | 6 | 6 |
| PYR/PYL | 0(0) | 1(0) | 2(3) | 5 | 21 | |
| PP2C | 5(0) | 2(0) | 1(0) | 6 | 13 | |
| SnRK2 | 0(1) | 2(0) | 2(0) | 3 | 19 | |
| ABF | 6(0) | 3(0) | 6(0) | 7 | 7 | |
| Salicylic Acid | TGA | 2(0) | 0(0) | 2(0) | 3 | 9 |
| PR1 | 1(0) | 3(0) | 2(3) | 5 | 5 | |
| Jasmonic Acid | JAR1 | 0(0) | 0(0) | 1(0) | 1 | 7 |
| COI1 | 0(0) | 1(0) | 0(0) | 1 | 4 | |
| JAZ | 2(0) | 0(1) | 6(0) | 6 | 9 | |
| MYC2 | 0(0) | 0(0) | 1(1) | 4 | 9 | |
| Ethylene | EIN3 | 0(0) | 2(0) | 0(0) | 2 | 11 |
| ERF1 | 0(0) | 0(0) | 0(1) | 1 | 2 | |
| ROS Scavenging System | CAT | 0(0) | 1(1) | 0(1) | 2 | 6 |
| POD | 2(2) | 4(2) | 5(5) | 14 | 34 | |
| SOD | 1(0) | 2(0) | 0(0) | 3 | 16 | |
| GST | 14(2) | 9(3) | 3(9) | 25 | 54 | |
| GLR | 1(1) | 1(1) | 9(0) | 12 | 40 | |
| APX | 0(1) | 0(0) | 1(1) | 2 | 15 | |
| Trx | 0(0) | 2(4) | 1(1) | 6 | 30 | |
| PrxR | 0(1) | 0(0) | 0(0) | 1 | 9 | |
| Other Stress Tolerance Related Genes | PAL | 0(1) | 0(1) | 0(2) | 3 | 8 |
| PPO | 1(1) | 1(3) | 5(1) | 6 | 6 | |
| GLU | 0(0) | 2(0) | 0(0) | 2 | 2 | |
| FLS2 | 1(0) | 1(0) | 3(0) | 4 | 7 | |
| chitinase | 3(0) | 1(0) | 3(1) | 6 | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Li, C.-X.; Zhong, J.; Shu, D.; Luo, D.; Li, Z.-M.; Zhou, J.-Y.; Yang, J.; Tan, H.; Ma, X.-R. Exogenous 1′,4′-trans-Diol-ABA Induces Stress Tolerance by Affecting the Level of Gene Expression in Tobacco (Nicotiana tabacum L.). Int. J. Mol. Sci. 2021, 22, 2555. https://doi.org/10.3390/ijms22052555
Liu T, Li C-X, Zhong J, Shu D, Luo D, Li Z-M, Zhou J-Y, Yang J, Tan H, Ma X-R. Exogenous 1′,4′-trans-Diol-ABA Induces Stress Tolerance by Affecting the Level of Gene Expression in Tobacco (Nicotiana tabacum L.). International Journal of Molecular Sciences. 2021; 22(5):2555. https://doi.org/10.3390/ijms22052555
Chicago/Turabian StyleLiu, Teng, Cai-Xia Li, Juan Zhong, Dan Shu, Di Luo, Zhe-Min Li, Jin-Yan Zhou, Jie Yang, Hong Tan, and Xin-Rong Ma. 2021. "Exogenous 1′,4′-trans-Diol-ABA Induces Stress Tolerance by Affecting the Level of Gene Expression in Tobacco (Nicotiana tabacum L.)" International Journal of Molecular Sciences 22, no. 5: 2555. https://doi.org/10.3390/ijms22052555
APA StyleLiu, T., Li, C.-X., Zhong, J., Shu, D., Luo, D., Li, Z.-M., Zhou, J.-Y., Yang, J., Tan, H., & Ma, X.-R. (2021). Exogenous 1′,4′-trans-Diol-ABA Induces Stress Tolerance by Affecting the Level of Gene Expression in Tobacco (Nicotiana tabacum L.). International Journal of Molecular Sciences, 22(5), 2555. https://doi.org/10.3390/ijms22052555





