CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands
Abstract
:1. Introduction
2. Results and Discussion
2.1. G4 Prediction and Selection in the SARS-CoV-2 Positive and Negative Genome
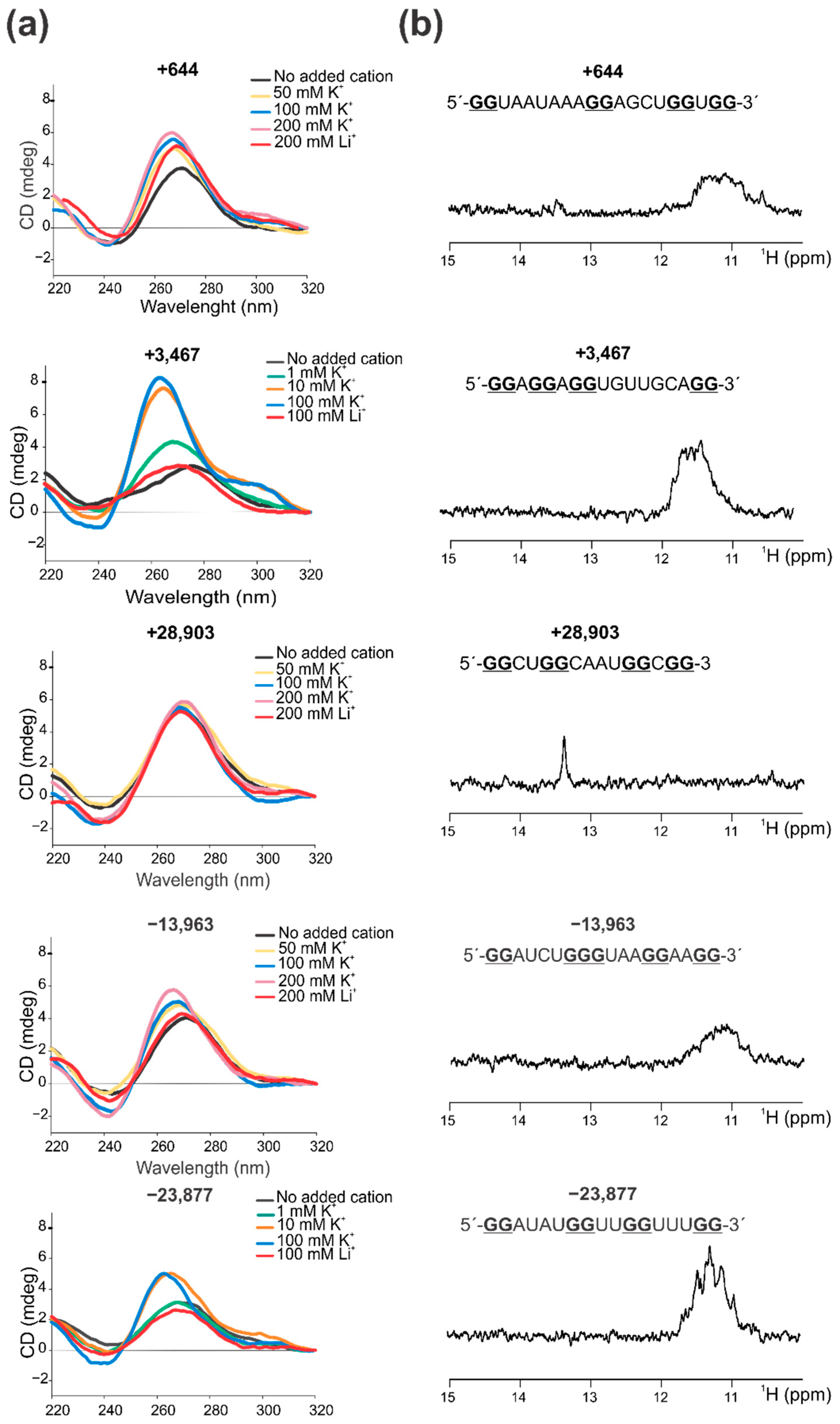
2.2. Confirmation That the Selected PQSs Fold In Vitro as G4
2.3. Cellular Nucleic Acids Binding Protein (CNBP) Interacts with Some SARS-CoV-2 G4s and Promotes Their Unfolding
2.4. Integration of Results with Putative Functions of G4 and CNBP in SARS-CoV-2 Replicative Cycle
3. Materials and Methods
3.1. Bioinformatics
3.2. Oligonucleotides
3.3. Circular Dichroism (CD) Spectroscopy
3.4. 1D 1H Nuclear Magnetic Resonance (NMR)
3.5. Thermal Difference Spectroscopy (TDS)
3.6. ThT Fluorescence Assays
3.7. CNBP Expression and Purification
3.8. Electrophoretic Mobility Shift Assay (EMSA)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Malik, Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar] [PubMed]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [Green Version]
- Ramadan, N.; Shaib, H. Middle east respiratory syndrome coronavirus (MERS-COV): A review. GERMS 2019, 9, 35–42. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Chu, H.; Chan, J.F.-W.; Yuen, T.T.-T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.C.-Y.; Tsang, J.O.-L.; Huang, X.; et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe 2020, 1, e14–e23. [Google Scholar] [CrossRef]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Coronaviruses: Methods and Protocols; Springer: New York, NY, USA, 2015; Volume 1282, pp. 1–23. ISBN 9781493924387. [Google Scholar] [CrossRef] [Green Version]
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, Y.K.; Gack, M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016, 14, 360–373. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Emanuel, W.; Kirstin, M.; Vedran, F.; Asija, D.; Theresa, G.L.; Roberto, A.; Filippos, K.; David, K.; Salah, A.; Christopher, B.; et al. Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Münch, C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020, 583, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Klann, K.; Bojkova, D.; Tascher, G.; Ciesek, S.; Münch, C.; Cinatl, J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol. Cell 2020, 80, 164–174.e4. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370. [Google Scholar] [CrossRef]
- Schmidt, N.; Lareau, C.A.; Keshishian, H.; Ganskih, S.; Schneider, C.; Hennig, T.; Melanson, R.; Werner, S.; Wei, Y.; Zimmer, M.; et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat. Microbiol. 2020. [Google Scholar] [CrossRef]
- Smyth, R.P.; Negroni, M.; Lever, A.M.; Mak, J.; Kenyon, J.C. RNA structure-a neglected puppet master for the evolution of virus and host immunity. Front. Immunol. 2018, 9, 2097. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Di Antonio, M.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [Green Version]
- Bartas, M.; Cutová, M.; Brázda, V.; Kaura, P.; Št’Astný, J.; Kolomazník, J.; Coufal, J.; Goswami, P.; Červeň, J.; Pečinka, P. The presence and localization of G-quadruplex forming sequences in the domain of bacteria. Molecules 2019, 24, 1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brázda, V.; Luo, Y.; Bartas, M.; Kaura, P.; Porubiaková, O.; Šťastný, J.; Pečinka, P.; Verga, D.; Da Cunha, V.; Takahashi, T.S.; et al. G-Quadruplexes in the archaea domain. Biomolecules 2020, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Seifert, S. Above and beyond watson and crick: Guanine quadruplex structures and microbes. Annu. Rev. Microbiol. 2018, 72, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More than just a kink in microbial genomes. Trends Microbiol. 2018, 27, 148–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-Quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef] [Green Version]
- Ruggiero, E.; Richter, S.N. Survey and summary G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. In Annual Reports in Medicinal Chemistry; Academic Press Inc.: San Diego, CA, USA, 2020; Volume 54, pp. 101–131. [Google Scholar] [CrossRef]
- Ji, D.; Juhas, M.; Tsang, C.M.; Kwok, C.K.; Li, Y.; Zhang, Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief. Bioinform. 2020, bbaa114. [Google Scholar] [CrossRef]
- Panera, N.; Tozzi, A.E.; Alisi, A. The G-quadruplex/helicase world as a potential antiviral approach against COVID-19. Drugs 2020, 80, 941–946. [Google Scholar] [CrossRef]
- Zhang, R.; Xiao, K.; Gu, Y.; Liu, H.; Sun, X. Whole genome identification of potential G-quadruplexes and analysis of the G-quadruplex binding domain for SARS-CoV-2. Front. Genet. 2020, 11, 587829. [Google Scholar] [CrossRef]
- Bartas, M.; Brázda, V.; Bohálová, N.; Cantara, A.; Volná, A.; Stachurová, T.; Malachová, K.; Jagelská, E.B.; Porubiaková, O.; Červeň, J.; et al. In-Depth bioinformatic analyses of nidovirales including human SARS-CoV-2, SARS-CoV, MERS-CoV viruses suggest important roles of non-canonical nucleic acid structures in their lifecycles. Front. Microbiol. 2020, 11, 1583. [Google Scholar] [CrossRef] [PubMed]
- Belmonte-Reche, E.; Serrano-Chacón, I.; Gonzalez, C.; Gallo, J.; Bañobre-López, M. Exploring G and C-quadruplex structures as potential targets against the severe acute respiratory syndrome coronavirus 2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-quadruplex in SARS-CoV-2: A promising therapeutic target for COVID-19? Angew. Chem. Int. Ed. 2021, 60, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhang, L. G-Quadruplexes are present in human coronaviruses including SARS-CoV-2. Front. Microbiol. 2020, 11, 567317. [Google Scholar] [CrossRef] [PubMed]
- Zaccaria, F.; Fonseca Guerra, C. RNA versus DNA G-quadruplex: The origin of increased stability. Chem. Eur. J. 2018, 24, 16315–16322. [Google Scholar] [CrossRef] [Green Version]
- Perrone, R.; Nadai, M.; Poe, J.A.; Frasson, I.; Palumbo, M.; Palù, G.; Smithgall, T.E.; Richter, S.N. Formation of a unique cluster of G-quadruplex structures in the HIV-1 nef coding region: Implications for antiviral activity. PLoS ONE 2013, 8, e73121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-Quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Ding, Y.; Alenko, A.; Burrows, C.J. Zika virus genomic RNA possesses conserved G-quadruplexes characteristic of the flaviviridae family. ACS Infect. Dis. 2016, 2, 674–681. [Google Scholar] [CrossRef]
- Wang, S.R.; Zhang, Q.Y.; Wang, J.Q.; Ge, X.Y.; Song, Y.Y.; Wang, Y.F.; Li, X.D.; Fu, B.S.; Xu, G.H.; Shu, B.; et al. Chemical targeting of a G-quadruplex RNA in the Ebola Virus L. Gene. Cell Chem. Biol. 2016, 23, 1113–1122. [Google Scholar] [CrossRef] [Green Version]
- Zahin, M.; Dean, W.L.; Ghim, S.; Joh, J.; Gray, R.D.; Khanal, S.; Bossart, G.D.; Mignucci-Giannoni, A.A.; Rouchka, E.C.; Jenson, A.B.; et al. Identification of G-quadruplex forming sequences in three manatee papillomaviruses. PLoS ONE 2018, 13, e0195625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majee, P.; Kumar Mishra, S.; Pandya, N.; Shankar, U.; Pasadi, S.; Muniyappa, K.; Nayak, D.; Kumar, A. Identification and characterization of two conserved G-quadruplex forming motifs in the Nipah virus genome and their interaction with G-quadruplex specific ligands. Sci. Rep. 2020, 10, 1477. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, S.; Jiang, H.; Deng, H.; Dong, C.; Shen, W.; Chen, H.; Gao, C.; Xiao, S.; Liu, Z.F.; et al. G2-Quadruplex in the 3’UTR of IE180 regulates pseudorabies virus replication by enhancing gene expression. RNA Biol. 2020, 17, 816–827. [Google Scholar] [CrossRef]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef] [PubMed]
- Labudova, D.; Hon, J.; Lexa, M.; Lexa, M. Pqsfinder web: G-quadruplex prediction using optimized pqsfinder algorithm. Bioinformatics 2020, 36, 2584–2586. [Google Scholar] [CrossRef] [PubMed]
- Garant, J.M.; Perreault, J.P.; Scott, M.S. G4RNA screener web server: User focused interface for RNA G-quadruplex prediction. Biochimie 2018, 151, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.D.; Jodoin, R.; Perreault, J.P. New scoring system to identify RNA G-quadruplex folding. Nucleic Acids Res. 2014, 42, 1209–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-Evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Garant, J.M.; Perreault, J.P.; Scott, M.S. Motif independent identification of potential RNA G-quadruplexes by G4RNA screener. Bioinformatics 2017, 33, 3532–3537. [Google Scholar] [CrossRef] [Green Version]
- Rangan, R.; Zheludev, I.N.; Hagey, R.J.; Pham, E.A.; Wayment-Steele, H.K.; Glenn, J.S.; Das, R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: A first look. RNA 2020, 26, 937–959. [Google Scholar] [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Nakken, S.; Rognes, T.; Hovig, E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009, 37, 5749–5756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baral, A.; Kumar, P.; Halder, R.; Mani, P.; Yadav, V.K.; Singh, A.; Das, S.K.; Chowdhury, S. Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res. 2012, 40, 3800–3811. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, H.; Ota, S.; Nishizawa, H.; Miyamura, H.; Nakahira, K.; Suzuki, M.; Nishiyama, S.; Kato, T.; Yanagihara, I.; Kurahashi, H. Obstetric complication-associated ANXA5 promoter polymorphisms may affect gene expression via DNA secondary structures. J. Hum. Genet. 2019, 64, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.D.; Perreault, J.P. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010, 38, 7022–7036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeraati, M.; Moye, A.L.; Wong, J.W.H.; Perera, D.; Cowley, M.J.; Christ, D.U.; Bryan, T.M.; Dinger, M.E. Cancer-Associated noncoding mutations affect RNA G-quadruplex-mediated regulation of gene expression. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Lee, D.S.M.; Ghanem, L.R.; Barash, Y. Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T.; Mergny, J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002, 30, 4618–4625. [Google Scholar] [CrossRef]
- Mergny, J.L.; Li, J.; Lacroix, L.; Amrane, S.; Chaires, J.B. Thermal difference spectra: A specific signature for nucleic acid structures. Nucleic Acids Res. 2005, 33, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Alfajaro, M.M.; DeWeirdt, P.C.; Hanna, R.E.; Lu-Culligan, W.J.; Cai, W.L.; Strine, M.S.; Zhang, S.M.; Graziano, V.R.; Schmitz, C.O.; et al. Genome-Wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 2020, 184, 76–91.e13. [Google Scholar] [CrossRef]
- Calcaterra, N.B.; Armas, P.; Weiner, A.M.J.; Borgognone, M. CNBP: A multifunctional nucleic acid chaperone involved in cell death and proliferation control. IUBMB Life 2010, 62, 707–714. [Google Scholar] [CrossRef]
- Armas, P.; Margarit, E.; Mouguelar, V.S.; Allende, M.L.; Calcaterra, N.B. Beyond the binding site: In vivo identification of tbx2, smarca5 and wnt5b as molecular targets of CNBP during embryonic development. PLoS ONE 2013, 8, e63234. [Google Scholar] [CrossRef] [Green Version]
- Benhalevy, D.; Gupta, S.K.; Danan, C.H.; Ghosal, S.; Sun, H.-W.; Kazemier, H.G.; Paeschke, K.; Hafner, M.; Juranek, S.A. The human CCHC-type zinc finger nucleic acid-binding protein binds g-rich elements in target mRNA coding sequences and promotes translation. Cell Rep. 2017, 18, 2979–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, A.P.; Pipier, A.; Pascutti, F.; Binolfi, A.; Weiner, A.M.J.; Challier, E.; Heckel, S.; Calsou, P.; Gomez, D.; Calcaterra, N.B.; et al. CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Lee, T.A.; Kim, J.H.; Park, A.; Ra, E.A.; Kang, S.; Choi, H.; Choi, J.L.; Huh, H.D.; Lee, J.E.; et al. CNBP acts as a key transcriptional regulator of sustained expression of interleukin-6. Nucleic Acids Res. 2017, 45, 3280–3296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Sharma, S.; Assis, P.A.; Jiang, Z.; Elling, R.; Olive, A.J.; Hang, S.; Bernier, J.; Huh, J.R.; Sassetti, C.M.; et al. CNBP controls IL-12 gene transcription and Th1 immunity. J. Exp. Med. 2018, 215, 3136–3150. [Google Scholar] [CrossRef]
- Tan, J.; Vonrhein, C.; Smart, O.S.; Bricogne, G.; Bollati, M.; Kusov, Y.; Hansen, G.; Mesters, J.R.; Schmidt, C.L.; Hilgenfeld, R. The SARS-Unique Domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009, 5, e1000428. [Google Scholar] [CrossRef]
- Endoh, T.; Kawasaki, Y.; Sugimoto, N. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew. Chem. Int. Ed. 2013, 52, 5522–5526. [Google Scholar] [CrossRef]
- Wang, S.R.; Min, Y.Q.; Wang, J.Q.; Liu, C.X.; Fu, B.S.; Wu, F.; Wu, L.Y.; Qiao, Z.X.; Song, Y.Y.; Xu, G.H.; et al. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti–hepatitis C target. Sci. Adv. 2016, 2, e1501535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontier, D.B.; Kruisselbrink, E.; Guryev, V.; Tijsterman, M. Isolation of deletion alleles by G4 DNA-induced mutagenesis. Nat. Methods 2009, 6, 655–657. [Google Scholar] [CrossRef]
- Tateishi-Karimata, H.; Isono, N.; Sugimoto, N. New insights into transcription fidelity: Thermal stability of non-canonical structures in template DNA regulates transcriptional arrest, pause, and slippage. PLoS ONE 2014, 9, e90580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagihara, M.; Yoneda, K.; Yabuuchi, H.; Okuno, Y.; Nakatani, K. A reverse transcriptase stop assay revealed diverse quadruplex formations in UTRs in mRNA. Bioorg. Med. Chem. Lett. 2010, 20, 2350–2353. [Google Scholar] [CrossRef]
- Jaubert, C.; Bedrat, A.; Bartolucci, L.; Di Primo, C.; Ventura, M.; Mergny, J.L.; Amrane, S.; Andreola, M.L. RNA synthesis is modulated by G-quadruplex formation in Hepatitis C virus negative RNA strand. Sci. Rep. 2018, 8, 8120. [Google Scholar] [CrossRef]
- Agarwala, P.; Pandey, S.; Mapa, K.; Maiti, S. The G-quadruplex augments translation in the 5’ untranslated region of transforming growth factor β2. Biochemistry 2013, 52, 1528–1538. [Google Scholar] [CrossRef]
- Kusov, Y.; Tan, J.; Alvarez, E.; Enjuanes, L.; Hilgenfeld, R. A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology 2015, 484, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Hognon, C.; Miclot, T.; Garcĺa-Iriepa, C.; Francés-Monerris, A.; Grandemange, S.; Terenzi, A.; Marazzi, M.; Barone, G.; Monari, A. Role of RNA guanine quadruplexes in favoring the dimerization of SARS unique domain in coronaviruses. J. Phys. Chem. Lett. 2020, 11, 5661–5667. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Juhas, M.; Zhang, Y. G-quadruplex based biosensor: A potential tool for SARS-CoV-2 detection. Biosens. Bioelectron. 2020, 167, 112494. [Google Scholar] [CrossRef]
- Chen, J.Y.; Chen, W.N.; Poon, K.M.V.; Zheng, B.J.; Lin, X.; Wang, Y.X.; Wen, Y.M. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch. Virol. 2009, 154, 507–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Gray, R.D.; Chaires, J.B. Characterization of quadruplex DNA structure by circular dichroism. Curr. Protoc. Nucleic Acid Chem. 2017, 68, 17.8.1–17.8.16. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Petraccone, L.; Erra, E.; Esposito, V.; Randazzo, A.; Galeone, A.; Barone, G.; Giancola, C.; Scienze, D.; Cintia, V.; Chimica, D. Biophysical properties of quadruple helices of modified human telomeric DNA. Biopolymers 2005, 77, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.L.; Shaka, A.J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- David, A.P.; Margarit, E.; Domizi, P.; Banchio, C.; Armas, P.; Calcaterra, N.B. G-Quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 2016, 44, 4163–4173. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Bugaut, A.; Huppert, J.L.; Balasubramanian, S. An RNA G-quadruplex in the 5’ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007, 3, 218–221. [Google Scholar] [CrossRef] [Green Version]
- Challier, E.; Lisa, M.-N.; Nerli, B.B.; Calcaterra, N.B.; Armas, P. Novel high-performance purification protocol of recombinant CNBP suitable for biochemical and biophysical characterization. Protein Expr. Purif. 2014, 93, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Armas, P.; Nasif, S.; Calcaterra, N.B. Cellular nucleic acid binding protein binds G-rich single-stranded nucleic acids and may function as a nucleic acid chaperone. J. Cell. Biochem. 2008, 103, 1013–1036. [Google Scholar] [CrossRef]
- Tuma, R.S.; Beaudet, M.P.; Jin, X.; Jones, L.J.; Cheung, C.; Yue, S.; Singer, V.L. Characterization of SYBR gold nucleic acid gel stain: A dye optimized for use with 300-nm ultraviolet transilluminators. Anal. Biochem. 1999, 288, 278–288. [Google Scholar] [CrossRef] [PubMed]



| Betacoronavirus | Genome Accession Number | % G | Number of PQSs in QGRS Mapper | Selected PQSs | % of Selected over Predicted PQSs | |||
|---|---|---|---|---|---|---|---|---|
| Low Probability to Form G4 | Medium Probability to Form G4 | High Probability to Form G4 | Total | |||||
| +gRNA | ||||||||
| SARS-CoV-2 | NC_045512.2 | 19.6 | 37 | 1 | 1 | 1 | 3 | 8.1 |
| RaTG13 | MN996532.2 | 19.6 | 37 | 1 | 2 | 1 | 4 | 10.8 |
| Bat-SL-CoVZXC21 | MG772934.1 | 20.1 | 32 | 2 | 2 | 4 | 8 | 25.0 |
| Bat-SL-CoVZC45 | MG772933.1 | 20.2 | 29 | 1 | 1 | 3 | 5 | 17.2 |
| SARS-CoV | NC_004718.3 | 20.8 | 49 | 2 | 2 | 2 | 6 | 12.2 |
| MERS-CoV | NC_019843.3 | 20.9 | 40 | 3 | 1 | 2 | 6 | 15.0 |
| −gRNA | ||||||||
| SARS-CoV-2 | NC_045512.2 | 18.4 | 19 | - | 3 | 2 | 5 | 26.3 |
| RaTG13 | MN996532.2 | 18.4 | 20 | - | 1 | 3 | 4 | 20.0 |
| Bat-SL-CoVZXC21 | MG772934.1 | 18.7 | 18 | 2 | 2 | - | 4 | 22.2 |
| Bat-SL-CoVZC45 | MG772933.1 | 18.7 | 16 | 2 | 2 | - | 4 | 25.0 |
| SARS-CoV | NC_004718.3 | 20.0 | 29 | 2 | - | 3 | 5 | 17.2 |
| MERS-CoV | NC_019843.3 | 20.3 | 38 | 2 | 2 | 5 | 9 | 23.7 |
| PQS Name | Genome | Length | Sequence | Prediction Scores | Reference of Previous Prediction | Reference of Experimental Evidence of G4 Formation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| G4RNA Screener | QGRS Mapper | PQS Finder | ||||||||
| cGcC | G4H | G4NN | ||||||||
| +644 | ORF1ab nsp1 | 20 | GGUAAUAAAGGAGCUGGUGG C G G C | 17 | 0.8 | 0.69 | 30 | 20 | [30,31,32,33,34,36] | [36] |
| +3467 | ORF1ab nsp3 | 17 | GGAGGAGGUGUUGCAGGA A A G A A GAAG U U | 18 | 1 | 0.87 | 30 | 24 | [30,31,32,33,34,36] | [34] |
| +28,903 | N | 15 | GGCUGGCAAUGGCGG UAU UU UU UUUAU AU ----- C | 5.33 | 0.87 | 0.57 | 33 | 27 | [30,31,32,33,34,36] | [34,35,36] |
| −13,963 | ORF1ab nsp12 | 18 | GGAUCUGGGUAAGGAAGG G G G AA | 22 | 1.11 | 0.97 | 34 | 23 | [32,33,36] | - |
| −23,877 | ORF1ab nsp3 | 17 | GGAUAUGGUUGGUUUGG AA C C A A | 160 | 0.94 | 0.02 | 34 | 24 | [32,33,36] | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bezzi, G.; Piga, E.J.; Binolfi, A.; Armas, P. CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands. Int. J. Mol. Sci. 2021, 22, 2614. https://doi.org/10.3390/ijms22052614
Bezzi G, Piga EJ, Binolfi A, Armas P. CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands. International Journal of Molecular Sciences. 2021; 22(5):2614. https://doi.org/10.3390/ijms22052614
Chicago/Turabian StyleBezzi, Georgina, Ernesto J. Piga, Andrés Binolfi, and Pablo Armas. 2021. "CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands" International Journal of Molecular Sciences 22, no. 5: 2614. https://doi.org/10.3390/ijms22052614
APA StyleBezzi, G., Piga, E. J., Binolfi, A., & Armas, P. (2021). CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands. International Journal of Molecular Sciences, 22(5), 2614. https://doi.org/10.3390/ijms22052614







