Bioactive Triterpenes of Protium heptaphyllum Gum Resin Extract Display Cholesterol-Lowering Potential
Abstract
1. Introduction
2. Results and Discussion
2.1. HPLC-APCI-HRMS2(High Performance Liquid Chromatography coupled with Atmospheric Pressure Chemical Ionization and High Resolution Mass Spectrometry) and GC-FID/MS (Gas Chromatography coupled with Flame Ionization Detector and Mass Spectrometry) Analyses identified Tetra- and Pentacyclic Triterpenoic compounds as the main constituent of PHE
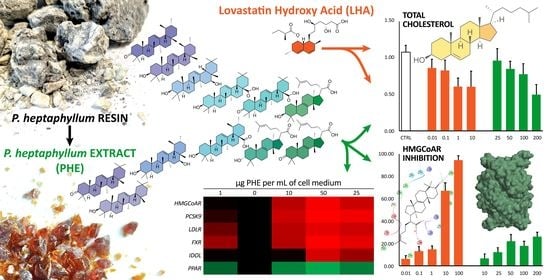
2.2. PHE Decreases Total Cholesterol in Hepatocytes
2.3. Triterpenoic Acids of PHE Modulate HMGCR Activity Assuming Similar Poses to LHE in the Active Site of the Enzyme
2.4. PHE Modulates the Expression of Cholesterol-Related Genes
3. Materials and Methods
3.1. Plant Material and Extract Preparation
3.2. Chemical Characterization Via HPLC-APCI-HRMS2
3.3. Chemical Characterization via GC-MS and GC-FID
3.4. Determination of Cholesterol Levels
3.5. Evaluation of HMGCR Activity
3.6. Molecular Docking Studies and Validation on HMGCR
3.6.1. Ligand and Protein Preparation
3.6.2. Molecular Docking
3.7. Gene Expression via qRT-PCR
3.8. Statistical Analysis
4. Conclusions
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raal, F.J.; Hovingh, G.K.; Catapano, A.L. Familial hypercholesterolemia treatments: Guidelines and new therapies. Atherosclerosis 2018, 277, 483–492. [Google Scholar] [CrossRef]
- Santos, R.D.; Gidding, S.S.; Hegele, R.A.; Cuchel, M.A.; Barter, P.J.; Watts, G.F.; Baum, S.J.; Catapano, A.L.; Chapman, M.J.; Defesche, J.C. Defining severe familial hypercholesterolaemia and the implications for clinical management: A consensus statement from the International Atherosclerosis Society Severe Familial Hypercholesterolemia Panel. Lancet Diabetes Endocrinol. 2016, 4, 850–861. [Google Scholar] [CrossRef]
- Rosso, A.; Pitini, E.; D’Andrea, E.; Massimi, A.; De Vito, C.; Marzuillo, C.; Villari, P. The cost-effectiveness of genetic screening for familial hypercholesterolemia: A systematic review. Ann. Ig. 2017, 29, 464–480. [Google Scholar] [PubMed]
- Jiang, J.; Wu, C.; Zhang, C.; Zhao, J.; Yu, L.; Zhang, H.; Narbad, A.; Chen, W.; Zhai, Q. Effects of probiotic supplementation on cardiovascular risk factors in hypercholesterolemia: A systematic review and meta-analysis of randomized clinical trial. J. Funct. Foods 2020, 74, 104177. [Google Scholar] [CrossRef]
- Yuningrum, H.; Rahmuniyati, M.E.; Sumiratsi, N.N.R. Consumption of Fried Foods as A Risk Factor for Hypercholesterolemia: Study of Eating Habits in Public Health Students. JHE J. Health Educ. 2020, 5, 78–85. [Google Scholar] [CrossRef]
- Al-Muzafar, H.M.; Amin, K.A. Efficacy of functional foods mixture in improving hypercholesterolemia, inflammatory and endothelial dysfunction biomarkers-induced by high cholesterol diet. Lipids Health Dis. 2017, 16, 194. [Google Scholar] [CrossRef]
- Stancu, C.; Sima, A. Statins: Mechanism of action and effects. J. Cell. Mol. Med. 2001, 5, 378–387. [Google Scholar] [CrossRef]
- Reiner, Ž. Management of patients with familial hypercholesterolaemia. Nat. Rev. Cardiol. 2015, 12, 565. [Google Scholar] [CrossRef]
- Joy, T.R.; Hegele, R.A. Narrative review: Statin-related myopathy. Ann. Intern. Med. 2009, 150, 858–868. [Google Scholar] [CrossRef]
- Joy, T.R.; Hegele, R.A.; Hilton-Jones, D. Statin-related myopathies. Pract. Neurol. 2018, 18, 97–105. [Google Scholar]
- Mannino, G.; Di Stefano, V.; Lauria, A.; Pitonzo, R.; Gentile, C. Vaccinium macrocarpon (Cranberry)-Based Dietary Supplements: Variation in Mass Uniformity, Proanthocyanidin Dosage and Anthocyanin Profile Demonstrates Quality Control Standard Needed. Nutrients 2020, 12, 992. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.Y.; Cooperman, T.; Obermeyer, W.; Becker, D.J. Marked variability of monacolin levels in commercial red yeast rice products: Buyer beware! Arch. Intern. Med. 2010, 170, 1722–1727. [Google Scholar] [CrossRef]
- Cicero, A.F.G.; Fogacci, F.; Banach, M. Red yeast rice for hypercholesterolemia. Methodist Debakey Cardiovasc. J. 2019, 15, 192. [Google Scholar]
- Gerards, M.C.; Terlou, R.J.; Yu, H.; Koks, C.H.W.; Gerdes, V.E.A. Traditional Chinese lipid-lowering agent red yeast rice results in significant LDL reduction but safety is uncertain–a systematic review and meta-analysis. Atherosclerosis 2015, 240, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Kuroda, M. Citrinin, an inhibitor of cholesterol synthesis. J. Antibiot. 1976, 29, 841–843. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Du, L.-D.; Du, G.-H. Lovastatin. In Natural Small Molecule Drugs from Plants; Springer: Berlin, Germany, 2018; pp. 93–99. [Google Scholar]
- Pasha, M.K.; Muzeeb, S.; Basha, S.J.S.; Shashikumar, D.; Mullangi, R.; Srinivas, N.R. Analysis of five HMG-CoA reductase inhibitors—atorvastatin, lovastatin, pravastatin, rosuvastatin and simvastatin: Pharmacological, pharmacokinetic and analytical overview and development of a new method for use in pharmaceutical formulations analysis and. Biomed. Chromatogr. 2006, 20, 282–293. [Google Scholar] [CrossRef]
- De Oliveira Filho, J.W.G.; Islam, M.T.; Ali, E.S.; Uddin, S.J.; de Oliveira Santos, J.V.; de Alencar, M.V.O.B.; Júnior, A.L.G.; Paz, M.F.C.J.; de Brito, M.D.R.M.; de Castroe e Sousa, J.M.; et al. A comprehensive review on biological properties of citrinin. Food Chem. Toxicol. 2017, 110, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.D.; Panza, G.; Zaleski, A.; Taylor, B. Statin-associated side effects. J. Am. Coll. Cardiol. 2016, 67, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Flajs, D.; Peraica, M. Toxicological properties of citrinin. Arch. Ind. Hyg. Toxicol. 2009, 60, 457–464. [Google Scholar] [CrossRef]
- Gentile, C.; Mannino, G.; Palazzolo, E.; Gianguzzi, G.; Perrone, A.; Serio, G.; Farina, V. Pomological, Sensorial, Nutritional and Nutraceutical Profile of Seven Cultivars of Cherimoya (Annona cherimola Mill). Foods 2021, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Mannino, G.; Gentile, C.; Porcu, A.; Agliassa, C.; Caradonna, F.; Bertea, C.M. Chemical Profile and Biological Activity of Cherimoya (Annona cherimola Mill.) and Atemoya (Annona atemoya) Leaves. Molecules 2020, 25, 2612. [Google Scholar] [CrossRef]
- Hoang, L.; Beneš, F.; Fenclová, M.; Kronusová, O.; Švarcová, V.; Řehořová, K.; Švecová, E.B.; Vosátka, M.; Hajšlová, J.; Kaštánek, P. Phytochemical Composition and In Vitro Biological Activity of Iris spp. (Iridaceae): A New Source of Bioactive Constituents for the Inhibition of Oral Bacterial Biofilms. Antibiotics 2020, 9, 403. [Google Scholar] [CrossRef]
- Maheshwari, V. Phytochemicals effective in lowering Low-Density Lipoproteins. J. Biol. Eng. Res. Rev. 2020, 7, 16–23. [Google Scholar]
- Weeks, A. The Molecular Systematics and Biogeography of the Burseraceae. Ph.D. Thesis, University of Texas at Austin, Austin, TX, USA, 1969. [Google Scholar]
- Rüdiger, A.L.; Siani, A.C.; Junior, V.F.V. The chemistry and pharmacology of the South America genus Protium Burm. f. (Burseraceae). Pharmacogn. Rev. 2007, 1, 93–104. [Google Scholar]
- Da Silva, P.S.D.; Leal, I.R.; Wirth, R.; Tabarelli, M. Spatial distribution and fruiting phenology of Protium heptaphyllum (Burseraceae) determine the design of the underground foraging system of Atta sexdens L. (Hymenoptera: Formicidae). Neotrop. Entomol. 2012, 41, 257–262. [Google Scholar] [CrossRef]
- Aragão, G.F.; Carneiro, L.M.V.; Junior, A.P.F.; Vieira, L.C.; Bandeira, P.N.; Lemos, T.L.G.; Viana, G.S.d.B. A possible mechanism for anxiolytic and antidepressant effects of alpha-and beta-amyrin from Protium heptaphyllum (Aubl.) March. Pharmacol. Biochem. Behav. 2006, 85, 827–834. [Google Scholar] [CrossRef]
- Aragão, G.F.; Carneiro, L.M.V.; Rota-Junior, A.P.; Bandeira, P.N.; Lemos, T.L.G.; Viana, G.S.d.B. Alterations in brain amino acid metabolism and inhibitory effects on PKC are possibly correlated with anticonvulsant effects of the isomeric mixture of α-and β-amyrin from Protium heptaphyllum. Pharm. Biol. 2015, 53, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Lima-Júnior, R.C.P.; Oliveira, F.A.; Gurgel, L.A.; Cavalcante, Í.J.M.; Santos, K.A.; Campos, D.A.; Vale, C.A.L.; Silva, R.M.; Chaves, M.H.; Rao, V.S.N. Attenuation of visceral nociception by α-and β-amyrin, a triterpenoid mixture isolated from the resin of Protium heptaphyllum, in mice. Planta Med. 2006, 72, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, F.A.; Costa, C.L.S.; Chaves, M.H.; Almeida, F.R.C.; Cavalcante, Í.J.M.; Lima, A.F.; Lima, R.C.P., Jr.; Silva, R.M.; Campos, A.R.; Santos, F.A. Attenuation of capsaicin-induced acute and visceral nociceptive pain by α-and β-amyrin, a triterpene mixture isolated from Protium heptaphyllum resin in mice. Life Sci. 2005, 77, 2942–2952. [Google Scholar] [CrossRef]
- Siani, A.C.; Ramos, M.F.d.S.; Menezes-de-Lima, O., Jr.; Ribeiro-dos-Santos, R.; Fernadez-Ferreira, E.; Soares, R.O.A.; Rosas, E.C.; Susunaga, G.S.; Guimarães, A.C.; Zoghbi, M.d.G.B. Evaluation of anti-inflammatory-related activity of essential oils from the leaves and resin of species of Protium. J. Ethnopharmacol. 1999, 66, 57–69. [Google Scholar] [CrossRef]
- Oliveira, F.A.; Vieira-Júnior, G.M.; Chaves, M.H.; Almeida, F.R.C.; Santos, K.A.; Martins, F.S.; Silva, R.M.; Santos, F.A.; Rao, V.S.N. Gastroprotective effect of the mixture of α-and β-amyrin from Protium heptaphyllum: Role of capsaicin-sensitive primary afferent neurons. Planta Med. 2004, 70, 780–782. [Google Scholar] [CrossRef]
- Aragao, G.F.; Pinheiro, M.C.C.; Bandeira, P.N.; Lemos, T.L.G.; Viana, G.S. Analgesic and anti-inflammatory activities of the isomeric mixture of alpha-and beta-amyrin from Protium heptaphyllum (Aubl.) March. J. Herb. Pharmacother. 2008, 7, 31–47. [Google Scholar] [CrossRef]
- Pinto, L.M.S.; Helena, C.M.; Santos, F.A.; Rao, V. Anti-inflammatory effect of alpha, beta-Amyrin, a pentacyclic triterpene from Protium heptaphyllum in rat model of acute periodontitis. Inflammopharmacology 2007, 15, 1–5. [Google Scholar]
- Faustino, C.G.; de Medeiros, F.A.; Ribeiro Galardo, A.K.; Lobato Rodrigues, A.B.; Lopes Martins, R.; de Medeiros Souza Lima, Y.; Fechine Tavares, J.; Alves de Medeiros, M.A.; dos Santos Cruz, J.; Almeida, S.S.M.d.S. Larvicide Activity on Aedes aegypti of Essential Oil Nanoemulsion from the Protium heptaphyllum Resin. Molecules 2020, 25, 5333. [Google Scholar] [CrossRef] [PubMed]
- Cabral, R.S.C.; Alves, C.C.F.; Batista, H.R.F.; Sousa, W.C.; Abrahão, I.S.; Crotti, A.E.M.; Santiago, M.B.; Martins, C.H.G.; Miranda, M.L.D. Chemical composition of essential oils from different parts of Protium heptaphyllum (Aubl.) Marchand and their in vitro antibacterial activity. Nat. Prod. Res. 2020, 34, 2378–2383. [Google Scholar] [CrossRef] [PubMed]
- De Lima, E.M.; Cazelli, D.S.P.; Pinto, F.E.; Mazuco, R.A.; Kalil, I.C.; Lenz, D.; Scherer, R.; de Andrade, T.U.; Endringer, D.C. Essential oil from the resin of Protium heptaphyllum: Chemical composition, cytotoxicity, antimicrobial activity, and antimutagenicity. Pharmacogn. Mag. 2016, 12, S42. [Google Scholar] [PubMed]
- Santos, F.A.; Frota, J.T.; Arruda, B.R.; de Melo, T.S.; de Castro Brito, G.A.; Chaves, M.H.; Rao, V.S. Antihyperglycemic and hypolipidemic effects of α, β-amyrin, a triterpenoid mixture from Protium heptaphyllum in mice. Lipids Health Dis. 2012, 11, 98. [Google Scholar] [CrossRef]
- Carvalho, K.M.M.B.; de Melo, T.S.; de Melo, K.M.; Quinderé, A.L.G.; de Oliveira, F.T.B.; Viana, A.F.S.C.; Nunes, P.I.G.; da Silva Quetz, J.; de Araújo Viana, D.; Havt, A. Amyrins from Protium heptaphyllum reduce high-fat diet-induced obesity in mice via modulation of enzymatic, hormonal and inflammatory responses. Planta Med. 2017, 83, 285–291. [Google Scholar] [CrossRef]
- Carvalho, K.M.M.B.; Marinho Filho, J.D.B.; de Melo, T.S.; Araújo, A.J.; Quetz, J.d.S.; da Cunha, M.d.P.S.S.; de Melo, K.M.; da Silva, A.A.d.C.A.; Tomé, A.R.; Havt, A. The resin from protium heptaphyllum prevents high-fat diet-induced obesity in mice: Scientific evidence and potential mechanisms. Evid. Based Complement. Altern. Med. 2015, 2015. [Google Scholar] [CrossRef]
- de Melo, K.M.; de Oliveira, F.T.B.; Silva, R.A.C.; Quinderé, A.L.G.; Marinho Filho, J.D.B.; Araújo, A.J.; Pereira, E.D.B.; Carvalho, A.A.; Chaves, M.H.; Rao, V.S. α, β-Amyrin, a pentacyclic triterpenoid from Protium heptaphyllum suppresses adipocyte differentiation accompanied by down regulation of PPARγ and C/EBPα in 3T3-L1 cells. Biomed. Pharmacother. 2019, 109, 1860–1866. [Google Scholar] [CrossRef]
- Serajuddin, A.T.M.; Ranadive, S.A.; Mahoney, E.M. Relative lipophilicities, solubilities, and structure-pharmacological considerations of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors pravastatin, lovastatin, mevastatin, and simvastatin. J. Pharm. Sci. 1991, 80, 830–834. [Google Scholar] [CrossRef]
- McGraw, G.W.; Hemingway, R.W.; Ingram, L.L.; Canady, C.S.; McGraw, W.B. Thermal degradation of terpenes: Camphene, Δ3-carene, limonene, and α-terpinene. Environ. Sci. Technol. 1999, 33, 4029–4033. [Google Scholar] [CrossRef]
- Chern, L.Y. Monoterpenes in plants-a mini review. Asian J. Plant. Biol. 2013, 1, 15–19. [Google Scholar]
- Nogueira, A.O.; Oliveira, Y.I.S.; Adjafre, B.L.; de Moraes, M.E.A.; Aragao, G.F. Pharmacological effects of the isomeric mixture of alpha and beta amyrin from Protium heptaphyllum: A literature review. Fundam. Clin. Pharmacol. 2019, 33, 4–12. [Google Scholar] [CrossRef]
- Büchele, B.; Zugmaier, W.; Simmet, T. Analysis of pentacyclic triterpenic acids from frankincense gum resins and related phytopharmaceuticals by high-performance liquid chromatography. Identification of lupeolic acid, a novel pentacyclic triterpene. J. Chromatogr. B 2003, 791, 21–30. [Google Scholar] [CrossRef]
- Singh, B.; Kumar, R.; Bhandari, S.; Pathania, S.; Lal, B. Volatile constituents of natural Boswellia serrata oleo-gum-resin and commercial samples. Flavour Fragr. J. 2007, 22, 145–147. [Google Scholar] [CrossRef]
- Mahajan, B.; Taneja, S.C.; Sethi, V.K.; Dhar, K.L. Two triterpenoids from Boswellia serrata gum resin. Phytochemistry 1995, 39, 453–455. [Google Scholar] [CrossRef]
- López-Hortas, L.; Pérez-Larrán, P.; González-Muñoz, M.J.; Falqué, E.; Domínguez, H. Recent developments on the extraction and application of ursolic acid. A review. Food Res. Int. 2018, 103, 130–149. [Google Scholar] [CrossRef]
- Volkman, J.K. Sterols and other triterpenoids: Source specificity and evolution of biosynthetic pathways. Org. Geochem. 2005, 36, 139–159. [Google Scholar] [CrossRef]
- Khan, M.S.Y.; Bano, S.; Javed, K.; Mueed, M.A. A comprehensive review on the chemistry and pharmacology of Corchorus species—a source of cardiac glycosides, triterpenoids, ionones, flavonoids, coumarins, steroids and some other compounds. J. Sci. Ind. Res. 2006, 65, 283–298. [Google Scholar]
- Kimura, Y.; Taniguchi, M.; Baba, K. Antitumor and antimetastatic effects on liver of triterpenoid fractions of Ganoderma lucidum: Mechanism of action and isolation of an active substance. Anticancer Res. 2002, 22, 3309–3318. [Google Scholar]
- Alberts, A.W. Discovery, biochemistry and biology of lovastatin. Am. J. Cardiol. 1988, 62, J10–J15. [Google Scholar] [CrossRef]
- Seenivasan, A.; Gummadi, S.N.; Panda, T. Comparison of the elution characteristics of individual forms of lovastatin in both isocratic and gradient modes and HPLC-PDA method development for pure and fermentation-derived lovastatin. Prep. Biochem. Biotechnol. 2017, 47, 901–908. [Google Scholar] [CrossRef]
- Klingelhöfer, I.; Morlock, G.E. Lovastatin in lactone and hydroxy acid forms and citrinin in red yeast rice powders analyzed by HPTLC-UV/FLD. Anal. Bioanal. Chem. 2019, 411, 6655–6665. [Google Scholar] [CrossRef]
- Beltrán, D.; Frutos-Lisón, M.D.; Espín, J.C.; García-Villalba, R. Re-examining the role of the gut microbiota in the conversion of the lipid-lowering statin monacolin K (lovastatin) into its active β-hydroxy acid metabolite. Food Funct. 2019, 10, 1787–1791. [Google Scholar] [CrossRef]
- Tang, J.-J.; Li, J.-G.; Qi, W.; Qiu, W.-W.; Li, P.-S.; Li, B.-L.; Song, B.-L. Inhibition of SREBP by a Small Molecule, Betulin, Improves Hyperlipidemia and Insulin Resistance and Reduces Atherosclerotic Plaques. Cell Metab. 2011, 13, 44–56. [Google Scholar] [CrossRef]
- Prabhakar, P.; Reeta, K.; Maulik, S.K.; Dinda, A.K.; Gupta, Y.K. α-Amyrin attenuates high fructose diet-induced metabolic syndrome in rats. Appl. Physiol. Nutr. Metab. 2017, 42, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Somova, L.O.; Nadar, A.; Rammanan, P.; Shode, F.O. Cardiovascular, antihyperlipidemic and antioxidant effects of oleanolic and ursolic acids in experimental hypertension. Phytomedicine 2003, 10, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Nam, G.W.; Kim, S.H.; Lee, S.H. Phytocomponents of triterpenoids, oleanolic acid and ursolic acid, regulated differently the processing of epidermal keratinocytes via PPAR-alpha pathway. Exp. Dermatol. 2006, 15, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef]
- Tobert, J.A. Lovastatin and beyond: The history of the HMG-CoA reductase inhibitors. Nat. Rev. Drug Discov. 2003, 2, 517–526. [Google Scholar] [CrossRef]
- Schachter, M. Chemical, pharmacokinetic and pharmacodynamic properties of statins: An update. Fundam. Clin. Pharmacol. 2005, 19, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Zolkiflee, N.F.; Majeed, A.B.A. Lovastatin: History, physicochemistry, pharmacokinetics and enhanced solubility. Int. J. Res. Pharm. Sci. 2017, 8, 90–102. [Google Scholar]
- Gesto, D.S.; Pereira, C.; Cerqueira, N.M.F.S.; Sousa, S.F. An Atomic-Level Perspective of HMG-CoA-Reductase: The Target Enzyme to Treat Hypercholesterolemia. Molecules 2020, 25, 3891. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.; Sharma, A.; Tuli, H.S.; Punia, S.; Sharma, A.K. Ursolic acid and oleanolic acid: Pentacyclic terpenoids with promising anti-inflammatory activities. Recent Pat. Inflamm. Allergy Drug Discov. 2016, 10, 21–33. [Google Scholar] [CrossRef]
- Lateef, T.; Naeem, S.; Qureshi, S.A. In-silico studies of HMG-Co A reductase inhibitors present in fruits of Withania coagulans Dunal (Solanaceae). Trop. J. Pharm. Res. 2020, 19, 305–312. [Google Scholar] [CrossRef]
- Marahatha, R.; Basnet, S.; Bhattarai, B.R.; Budhathoki, P.; Aryal, B.; Adhikari, B.; Lamichhane, G.; Poudel, D.K.; Parajuli, N. Potential natural inhibitors of xanthine oxidase and HMG-CoA reductase in cholesterol regulation: In silico analysis. BMC Complement. Med. Ther. 2021, 21, 1–11. [Google Scholar] [CrossRef]
- Goswami, S.; Vidyarthi, A.S.; Bhunia, B.; Mandal, T. A review on lovastatin and its production. J. Biochem. Technol. 2013, 4, 581–587. [Google Scholar]
- Alberts, A.W. Lovastatin and simvastatin-inhibitors of HMG CoA reductase and cholesterol biosynthesis. Cardiology 1990, 77, 14–21. [Google Scholar] [CrossRef]
- Mannino, G.; Perrone, A.; Campobenedetto, C.; Schittone, A.; Margherita Bertea, C.; Gentile, C. Phytochemical profile and antioxidative properties of Plinia trunciflora fruits: A new source of nutraceuticals. Food Chem. 2020, 307, 125515. [Google Scholar] [CrossRef]
- Rohit Singh, T.; Ezhilarasan, D. Ethanolic extract of Lagerstroemia Speciosa (L.) Pers., induces apoptosis and cell cycle arrest in HepG2 cells. Nutr. Cancer 2020, 72, 146–156. [Google Scholar] [CrossRef]
- Mannino, G.; Caradonna, F.; Cruciata, I.; Lauria, A.; Perrone, A.; Gentile, C. Melatonin reduces inflammatory response in human intestinal epithelial cells stimulated by interleukin-1β. J. Pineal Res. 2019, 67. [Google Scholar] [CrossRef]
- Caradonna, F.; Consiglio, O.; Luparello, C.; Gentile, C. Science and Healthy Meals in the World: Nutritional Epigenomics and Nutrigenetics of the Mediterranean Diet. Nutrients 2020, 12, 1748. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.H.; Van Vliet, A.; Roodenburg, L.; Jansen, L.M.C.; Griffigen, M. Pravastatin inhibited the cholesterol synthesis in human hepatoma cell line Hep G2 less than simvastatin and lovastatin, which is reflected in the upregulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and squalene synthase. Biochem. Pharmacol. 1993, 45, 2203–2208. [Google Scholar] [CrossRef]
- Yu, X.-H.; Zheng, X.-L.; Tang, C.-K. Peroxisome proliferator-activated receptor α in lipid metabolism and atherosclerosis. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2015; Volume 71, pp. 171–203. ISBN 0065-2423. [Google Scholar]
- Choi, Y.-J.; Lee, S.J.; Kim, H.I.; Lee, H.J.; Kang, S.J.; Kim, T.Y.; Cheon, C.; Ko, S.-G. Platycodin D enhances LDLR expression and LDL uptake via down-regulation of IDOL mRNA in hepatic cells. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Zhang, C.; Tian, Y.; Zhang, M.; Tuo, Q.; Chen, J.; Liao, D. IDOL, inducible degrader of low-density lipoprotein receptor, serves as a potential therapeutic target for dyslipidemia. Med. Hypotheses 2016, 86, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, M.; Long, S.; Tuo, Q.; Tian, Y.; Chen, J.; Zhang, C.; Liao, D. Cholesterol in LDL receptor recycling and degradation. Clin. Chim. Acta 2020, 500, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, E.H.; Kato, H.; Nardone, G.A.; Usukura, J. A prospective mechanism and source of cholesterol uptake by Plasmodium falciparum-infected erythrocytes co-cultured with HepG2 cells. Parasitol. Int. 2020, 80, 102179. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, F.; Li, C.; Lin, M.; Briggs, M.R. Synergistic activation of human LDL receptor expression by SCAP ligand and cytokine oncostatin M. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 90–96. [Google Scholar] [CrossRef][Green Version]
- Dong, B.; Wu, M.; Cao, A.; Li, H.; Liu, J. Suppression of Idol expression is an additional mechanism underlying statin-induced up-regulation of hepatic LDL receptor expression. Int. J. Mol. Med. 2011, 27, 103–110. [Google Scholar]
- Hageman, J.; Herrema, H.; Groen, A.K.; Kuipers, F. A role of the bile salt receptor FXR in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1519–1528. [Google Scholar] [CrossRef]
- Tacer, K.F. Oxysterols and bile acid act as signaling molecules that regulate cholesterol homeostasis: Nuclear receptors LXR, FXR, and fibroblast growth factor 15/19. In Mammalian Sterols; Springer: Berlin, Germany, 2020; pp. 117–143. [Google Scholar]
- Lu, J.; Huang, G.; Hu, S.; Wang, Z.; Guan, S. 1, 3-Dichloro-2-propanol induced hyperlipidemia in C57BL/6J mice via AMPK signaling pathway. Food Chem. Toxicol. 2014, 64, 403–409. [Google Scholar] [CrossRef]
- Cao, Y.; Bei, W.; Hu, Y.; Cao, L.; Huang, L.; Wang, L.; Luo, D.; Chen, Y.; Yao, X.; He, W. Hypocholesterolemia of Rhizoma Coptidis alkaloids is related to the bile acid by up-regulated CYP7A1 in hyperlipidemic rats. Phytomedicine 2012, 19, 686–692. [Google Scholar] [CrossRef]
- Iwayanagi, Y.; Takada, T.; Tomura, F.; Yamanashi, Y.; Terada, T.; Inui, K.; Suzuki, H. Human NPC1L1 expression is positively regulated by PPARα. Pharm. Res. 2011, 28, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Geeraert, B.; Arnould, T.; Tsatsanis, C.; Holvoet, P. PPAR agonist-induced reduction of Mcp1 in atherosclerotic plaques of obese, insulin-resistant mice depends on adiponectin-induced Irak3 expression. PLoS ONE 2013, 8, e62253. [Google Scholar] [CrossRef]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. [Google Scholar] [CrossRef]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [Google Scholar] [CrossRef]
- Kellici, T.F.; Ntountaniotis, D.; Liapakis, G.; Tzakos, A.G.; Mavromoustakos, T. The dynamic properties of angiotensin II type 1 receptor inverse agonists in solution and in the receptor site. Arab. J. Chem. 2019, 12, 5062–5078. [Google Scholar] [CrossRef]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef]
- Rose, P.W.; Bi, C.; Bluhm, W.F.; Christie, C.H.; Dimitropoulos, D.; Dutta, S.; Green, R.K.; Goodsell, D.S.; Prlić, A.; Quesada, M. The RCSB Protein Data Bank: New resources for research and education. Nucleic Acids Res. 2012, 41, D475–D482. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Zhong, H.; Tran, L.M.; Stang, J.L. Induced-fit docking studies of the active and inactive states of protein tyrosine kinases. J. Mol. Graph. Model. 2009, 28, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Hasanat, A.; Chowdhury, T.A.; Kabir, M.S.H.; Chowdhury, M.S.; Chy, M.; Uddin, N.; Barua, J.; Chakrabarty, N.; Paul, A. Antinociceptive activity of Macaranga denticulata Muell. Arg.(Family: Euphorbiaceae): In vivo and in silico studies. Medicines 2017, 4, 88. [Google Scholar] [CrossRef]
- Lauria, A.; Mannino, S.; Gentile, C.; Mannino, G.; Martorana, A.; Peri, D. DRUDIT: Web-based DRUgs DIscovery Tools to design small molecules as modulators of biological targets. Bioinformatics 2019, 36, 1562–1569. [Google Scholar] [CrossRef]
- Wang, H.; Aslanian, R.; Madison, V.S. Induced-fit docking of mometasone furoate and further evidence for glucocorticoid receptor 17α pocket flexibility. J. Mol. Graph. Model. 2008, 27, 512–521. [Google Scholar] [CrossRef]
- Luo, H.-J.; Wang, J.-Z.; Deng, W.-Q.; Zou, K. Induced-fit docking and binding free energy calculation on furostanol saponins from Tupistra chinensis as epidermal growth factor receptor inhibitors. Med. Chem. Res. 2013, 22, 4970–4979. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinform. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef]
- Campobenedetto, C.; Mannino, G.; Agliassa, C.; Acquadro, A.; Contartese, V.; Garabello, C.; Bertea, C.M. Transcriptome Analyses and Antioxidant Activity Profiling Reveal the Role of a Lignin-Derived Biostimulant Seed Treatment in Enhancing Heat Stress Tolerance in Soybean. Plants 2020, 9, 1308. [Google Scholar] [CrossRef] [PubMed]
- Campobenedetto, C.; Grange, E.; Mannino, G.; Van Arkel, J.; Beekwilder, J.; Karlova, R.; Garabello, C.; Contartese, V.; Bertea, C.M. A Biostimulant Seed Treatment Improved Heat Stress Tolerance During Cucumber Seed Germination by Acting on the Antioxidant System and Glyoxylate Cycle. Front. Plant. Sci. 2020, 11, 836. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Siani, A.C.; Moraes, R.; Veiga Junior, V.F. Toward Establishing the Productive Chain for Triterpene-Based Amazonian Oleoresins as Valuable Non-Timber Forest Products. Open J. For. 2017, 7, 188–208. [Google Scholar] [CrossRef]









| # | CAS ID | MW | Chemical Formula | Class | Name | Composition (%) |
|---|---|---|---|---|---|---|
| 1 | 13466-78-9 | 136.23 | C10H16 | VOCs | Δ3-carene | 0.70 ± 0.08% |
| 2 | 99-87-6 | 134.22 | C10H14 | p-cymene | 0.90 ± 0.02% | |
| 3 | 638-95-9 | 556.9 | C39H56O2 | NATs | α-amyrin | 12.40 ± 1.39% |
| 4 | 559-70-6 | 426.7 | C30H50O | β-amyrin | 9.50 ± 1.17% | |
| 5 | 638-96-0 | 424.7 | C30H48O | α-amyron | 2.20 ± 0.08% | |
| 6 | 638-97-1 | 424.7 | C30H48O | β-amyron | 1.70 ± 0.61% | |
| 7 | 465-08-7 | 442.7 | C30H50O2 | Brein | 1.30 ± 0.08% | |
| 8 | 595-17-5 | 442.7 | C30H50O | Maniladiol | 0.90 ± 0.06% | |
| 9 | 508-02-1 | 456.7 | C30H48O3 | ATs | Oleanolic acid | 0.05 ± 0.01% |
| 10 | 77-52-1 | 456.7 | C30H48O3 | Ursolic acid | 0.06 ± 0.01% | |
| 11 | 28282-25-9 | 454.7 | C30H46O3 | Elemonic acid | 15.60 ± 0.06% | |
| 12 | 28282-27-1 | 456.7 | C30H48O3 | α-elemolic acid | 10.57 ± 0.76% | |
| 13 | 28282-54-4 | 456.7 | C30H48O3 | β-elemolic acid | 12.80 ± 1.38% |
| # | Compound Name | Docking Score | Prime Energy | IFD Score |
|---|---|---|---|---|
| Lovastatin | −6.247 | −29124.9 | −1462.494 | |
| Lovastatin Hydroxy Acid | −14.667 | −29097.0 | −1469.500 | |
| 1 | Δ3-carene | −3.220 | −29015.2 | −1453.979 |
| 2 | p-cymene | −4.009 | −29030.2 | −1455.521 |
| 3 | α-amyrin | −4.637 | −29073.5 | −1458.313 |
| 4 | β-amyrin | −4.405 | −29079.4 | −1458.376 |
| 5 | α-amyron | −5.196 | −29066.6 | −1458.528 |
| 6 | β-amyron | −4.245 | −29030.2 | −1455.753 |
| 7 | Brein | −5.307 | −29066.2 | −1458.666 |
| 8 | Maniladiol | −5.356 | −29151.0 | −1462.869 |
| 9 | Oleanolic acid | −9.835 | −29059.2 | −1462.795 |
| 10 | Ursolic acid | −8.008 | −29074.3 | −1461.721 |
| 11 | Elemonic acid | −6.786 | −29212.4 | −1467.408 |
| 12 | α-elemolic acid | −4.724 | −29220.5 | −1465.750 |
| 13 | β-elemolic acid | −6.828 | −29166.3 | −1465.142 |
| Gene Acronym | Sequence (5′−3′) | Accession Number | |
|---|---|---|---|
| HMGCR | F | TGATTGACCTTTCCAGAGCAAG | NM_000859.2 |
| R | CTAAAATTGCCATTCCACGAGC | ||
| FXR | F | TCTCCTGGGTCGCCTGACT | NM_005123 |
| R | ACTGCACGTCCCAGATTTCAC | ||
| LDLR | F | AGTTGGCTGCGTTAATGTGA | NM_000527.4 |
| R | TGATGGGTTCATCTGACCAGT | ||
| IDOL | F | AAGTTCTTCGTGGAGCCTCA | NM_013262 |
| R | ACTGAGTTCCACTGCCTGCT | ||
| PCSK9 | F | GCTGAGCTGCTCCAGTTTCT | NM_174936.3 |
| R | AATGGCGTAGACACCCTCAC | ||
| PPARα | F | CTGGAAGCTTTGGCTTTACG | NM_005036.4 |
| R | GTTGTGTGACATCCCGACAG | ||
| βACT | F | CGGGAAATCGTGCGTGACAT | NM_001101.3 |
| R | GGACTCCATGCCCAGGAAGG | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mannino, G.; Iovino, P.; Lauria, A.; Genova, T.; Asteggiano, A.; Notarbartolo, M.; Porcu, A.; Serio, G.; Chinigò, G.; Occhipinti, A.; et al. Bioactive Triterpenes of Protium heptaphyllum Gum Resin Extract Display Cholesterol-Lowering Potential. Int. J. Mol. Sci. 2021, 22, 2664. https://doi.org/10.3390/ijms22052664
Mannino G, Iovino P, Lauria A, Genova T, Asteggiano A, Notarbartolo M, Porcu A, Serio G, Chinigò G, Occhipinti A, et al. Bioactive Triterpenes of Protium heptaphyllum Gum Resin Extract Display Cholesterol-Lowering Potential. International Journal of Molecular Sciences. 2021; 22(5):2664. https://doi.org/10.3390/ijms22052664
Chicago/Turabian StyleMannino, Giuseppe, Piera Iovino, Antonino Lauria, Tullio Genova, Alberto Asteggiano, Monica Notarbartolo, Alessandra Porcu, Graziella Serio, Giorgia Chinigò, Andrea Occhipinti, and et al. 2021. "Bioactive Triterpenes of Protium heptaphyllum Gum Resin Extract Display Cholesterol-Lowering Potential" International Journal of Molecular Sciences 22, no. 5: 2664. https://doi.org/10.3390/ijms22052664
APA StyleMannino, G., Iovino, P., Lauria, A., Genova, T., Asteggiano, A., Notarbartolo, M., Porcu, A., Serio, G., Chinigò, G., Occhipinti, A., Capuzzo, A., Medana, C., Munaron, L., & Gentile, C. (2021). Bioactive Triterpenes of Protium heptaphyllum Gum Resin Extract Display Cholesterol-Lowering Potential. International Journal of Molecular Sciences, 22(5), 2664. https://doi.org/10.3390/ijms22052664