Molecular Docking and Biophysical Studies for Antiproliferative Assessment of Synthetic Pyrazolo-Pyrimidinones Tethered with Hydrazide-Hydrazones
Abstract
1. Introduction
2. Results
2.1. Chemistry
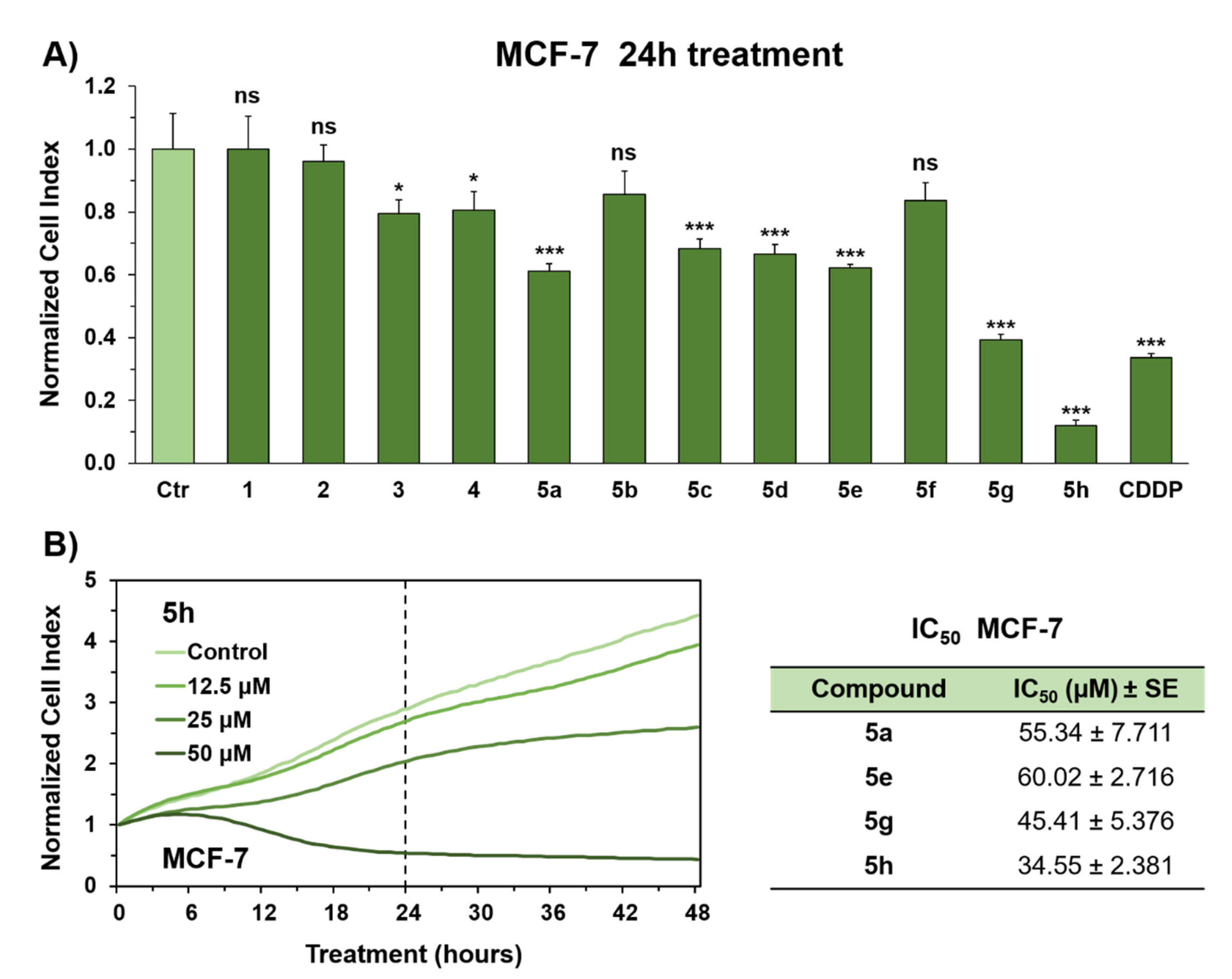
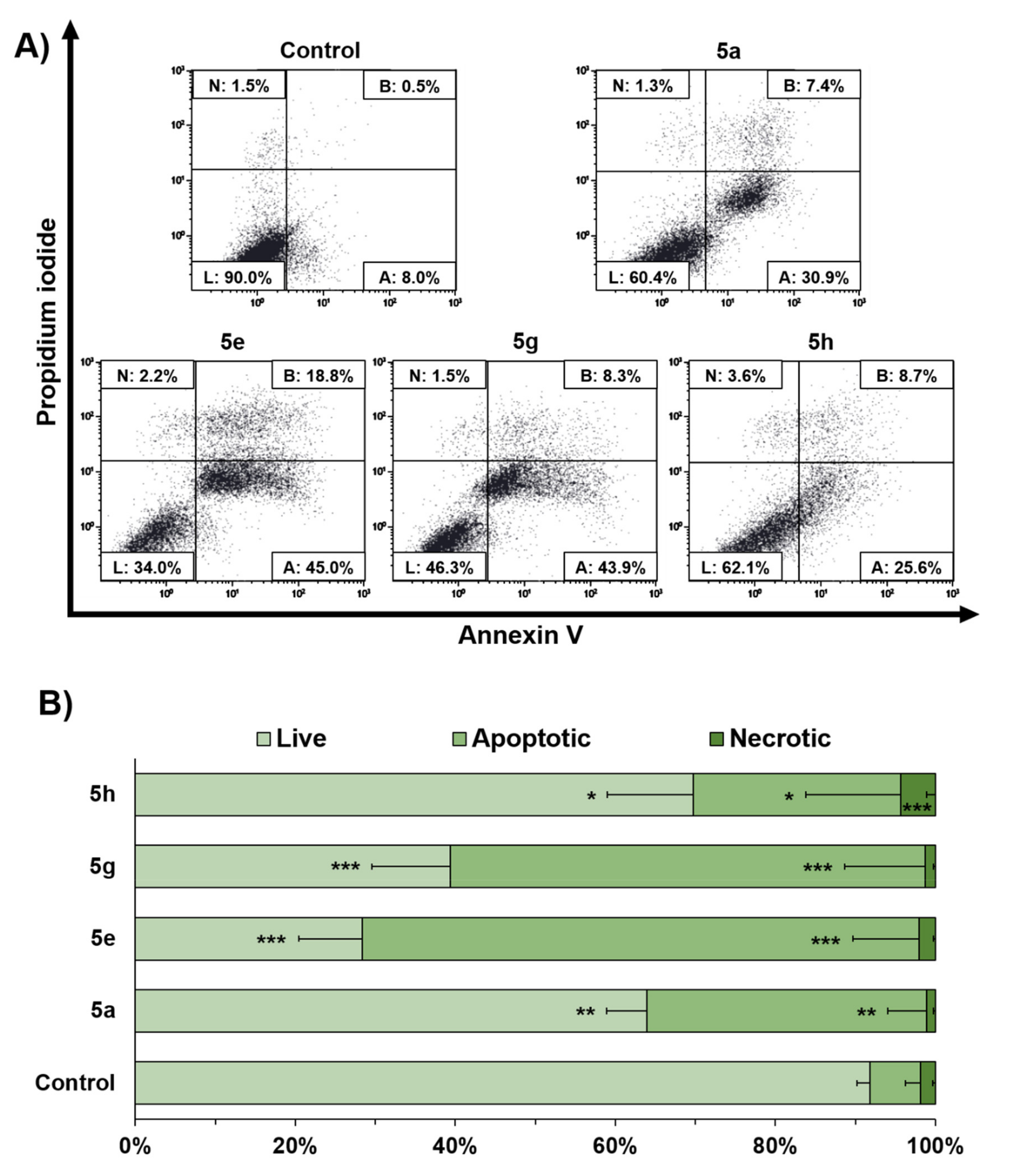
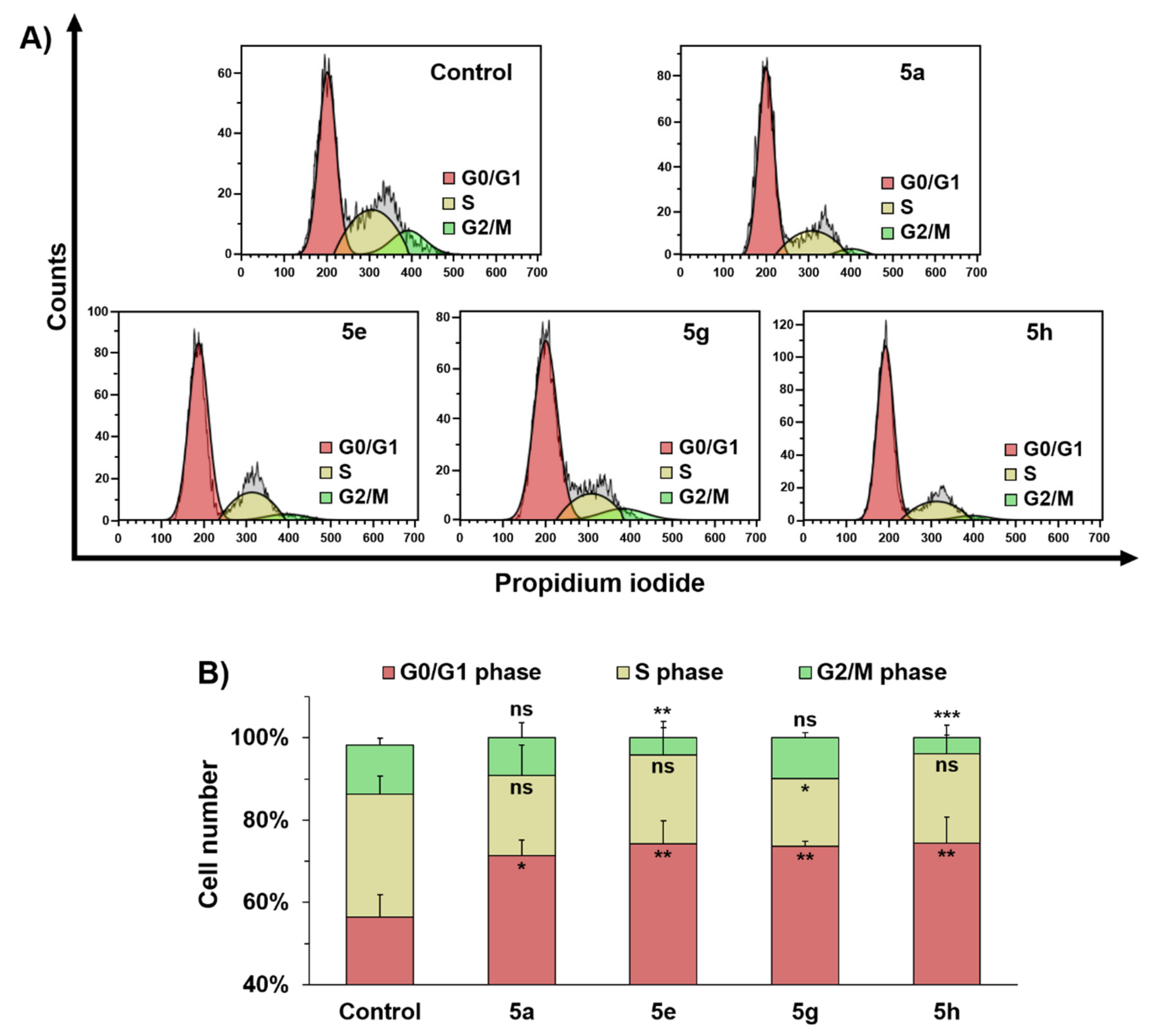
2.2. In Vitro Evaluation of Antiproliferative Activity in MCF-7 Breast Cancer Cells
2.3. Identification of Potential Anticancer Drug Targets of 5a, 5e, 5g, and 5h
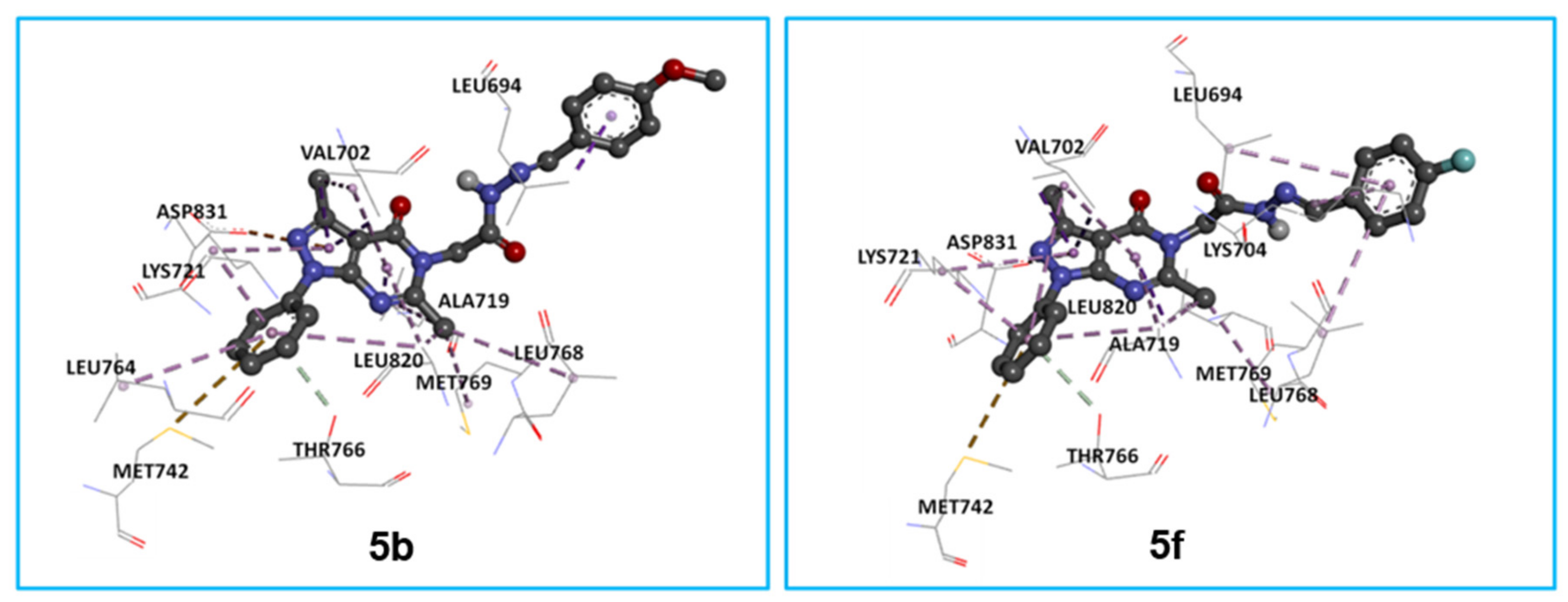
2.3.1. In Silico Molecular Docking of 5a, 5e, 5g, and 5h as Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors
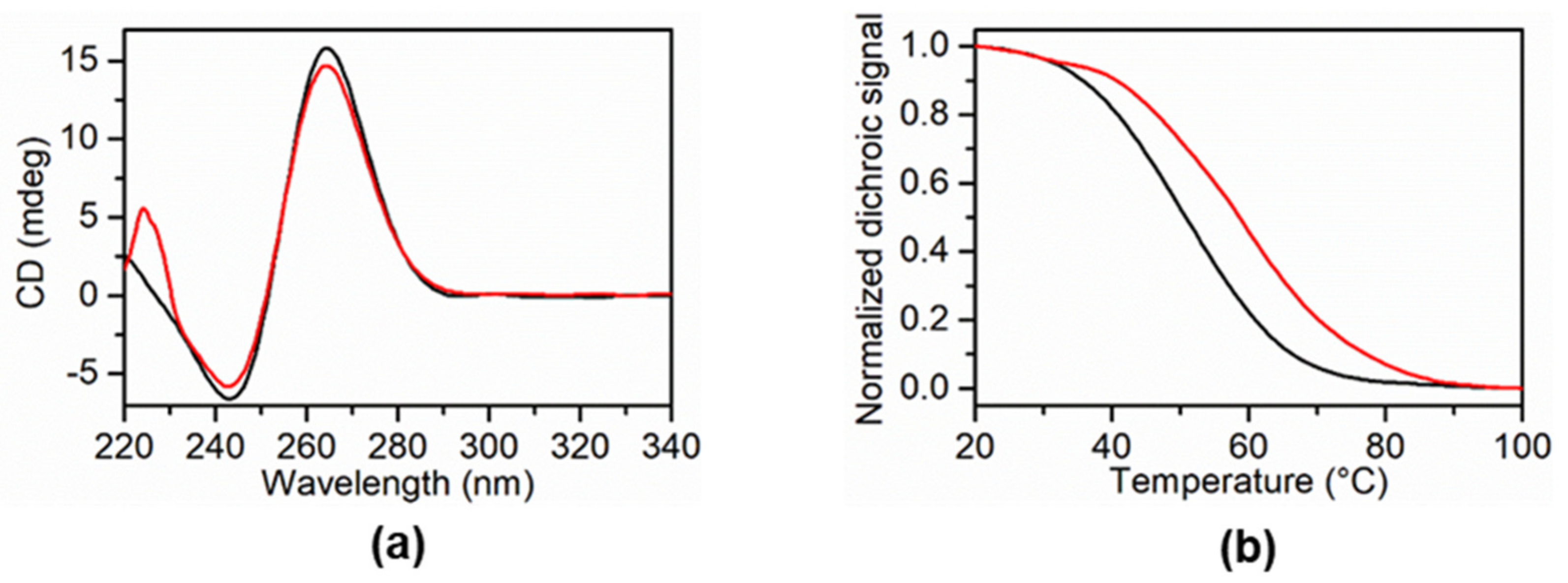
2.3.2. Physicochemical Evaluation of 5a, 5e, 5g, and 5h as G-Quadruplex DNA Stabilizers
3. Discussion
4. Materials and Methods
4.1. General Information
4.2. Chemistry
4.2.1. General Procedure for the Synthesis of 3,6-dimethyl-1-phenyl-1,5-dihydro-4H-pyrazolo [3,4-d]pyrimidin-4-one 2
4.2.2. General Procedure for the Synthesis of Ethyl 2-(3,6-dimethyl-4-oxo-1-phenyl-1,4-dihydro-5H-pyrazolo[3,4-d]pyrimidin-5-yl)acetate 3
4.2.3. General Procedure for the Synthesis of 2-(3,6-dimethyl-4-oxo-1-phenyl-1,4-dihydro-5H-pyrazolo[3,4-d]pyrimidin-5-yl)acetohydrazide 4
4.2.4. General Procedure for the Synthesis of Derivatives 5a–h
4.3. Cell Viability Assays
4.4. Apoptosis Assay and Cell Cycle Analysis
4.5. Molecular Docking Procedure
4.6. Physicochemical Studies
4.6.1. Synthesis of Oligomers
4.6.2. Circular Dichroism (CD) Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Cancer—World Health Organization. Available online: http://www.who.int/mediacentre/factsheets/fs297/en/ (accessed on 5 January 2021).
- Strickaert, A.; Saiselet, M.; Dom, G.; De Deken, X.; Dumont, J.E.; Feron, O.; Sonveaux, P.; Maenhaut, C. Cancer heterogeneity is not compatible with one unique cancer cell metabolic map. Oncogene 2017, 36, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Caso, A.; Laurenzana, I.; Lamorte, D.; Trino, S.; Esposito, G.; Piccialli, V.; Costantino, V. Smenamide A Analogues. Synthesis and Biological Activity on Multiple Myeloma Cells. Mar. Drugs. 2018, 16, 206. [Google Scholar] [CrossRef]
- Caso, A.; Mangoni, A.; Piccialli, G.; Costantino, V.; Piccialli, V. Studies toward the Synthesis of Smenamide A, an Antiproliferative Metabolite from Smenospongia aurea: Total Synthesis of ent-Smenamide A and 16-epi-Smenamide A. ACS Omega 2017, 2, 1477–1488. [Google Scholar] [CrossRef]
- Esposito, G.; Della Sala, G.; Teta, R.; Caso, A.; Bourguet-Kondraki, M.L.; Pawlik, J.R.; Mangoni, A.; Costantino, V. Chlorinated Thiazole-Containing Polyketide-Peptides from the Caribbean Sponge Smenospongia conulosa: Structure Elucidation on Microgram Scale. Eur. J. Org. Chem. 2016, 2871–2875. [Google Scholar] [CrossRef]
- Della Sala, G.; Hochmuth, T.; Costantino, V.; Teta, R.; Gerwick, W.; Gerwick, L.; Piel, J.; Mangoni, A. Polyketide Genes in the Marine Sponge Plakortis simplex: A New Group of Mono-Modular Type-I Polyketide Synthases from Sponge Symbionts. Environ. Microbiol. Rep. 2013, 5, 809–818. [Google Scholar] [CrossRef]
- Costantino, V.; Fattorusso, E.; Imperatore, C.; Mangoni, A.; Teta, R. Glycolipids from Sponges. Part 21. Amphiceramide A and B, Novel Glycosphingolipids from the Marine Sponge Amphimedon compressa. Eur. J. Org. Chem. 2009, 2112–2119. [Google Scholar] [CrossRef]
- Britstein, M.; Devescovi, G.; Handley, K.; Malik, A.; Haber, M.; Saurav, K.; Teta, R.; Costantino, V.; Burgsdorf, I.; Gilbert, J.; et al. A New N-Acyl homoserine lactone synthase in an uncultured symbiont of the red sponge Theonella swinohei. Appl. Environ. Microbiol. 2016, 82, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Teta, R.; Della Sala, G.; Renga, B.; Mangoni, A.; Fiorucci, S.; Costantino, V. Chalinulasterol, a chlorinated steroid disulfate from the caribbean sponge Chalinula molitba. Evaluation of its role as PXR receptor modulator. Mar. Drugs 2012, 10, 1383–1390. [Google Scholar] [CrossRef]
- Teta, R.; Marteinsson, V.T.; Longeon, A.; Klonowski, A.M.; Groben, R.; Bourguet-Kondracki, M.-L.; Costantino, V.; Mangoni, A. Thermoactinoamide A, an antibiotic lipophilic cyclopeptide from the icelandic thermophilic bacterium thermoactinomyces vulgaris. J. Nat. Prod. 2017, 80, 2530–2535. [Google Scholar] [CrossRef]
- Rees, D.C.; Congreve, M.; Murray, C.W.; Carr, R. Fragment-based lead discovery. Nat. Rev. Drug. Discov. 2004, 3, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Hou, T.J. Drug and drug candidate building block analysis. J. Chem. Inf. Model. 2010, 50, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.A.; Dhar, K.L.; Puri, S.C.; Saxena, A.K.; Shanmugavel, M.; Qazi, G.N. Synthesis and biological evaluation of chalcones and their derived pyrazoles as potential cytotoxic agents. Bioorg. Med. Chem. Lett. 2005, 15, 3177–3180. [Google Scholar] [CrossRef]
- Nossier, E.S.; Fahmy, H.H.; Khalifa, N.M.; El-Eraky, W.I.; Baset, M.A. Design and Synthesis of Novel Pyrazole-Substituted Different Nitrogenous Heterocyclic Ring Systems as Potential Anti-Inflammatory Agents. Molecules 2017, 22, 512–528. [Google Scholar] [CrossRef] [PubMed]
- Riyadh, S.M.; Farghaly, T.A.; Abdallah, M.A.; Abdalla, M.M.; Abd El-Aziz, M.R. New Pyrazoles Incorporating Pyrazolylpyrazole Moiety: Synthesis, anti- HCV and Antitumor Activity. Eur. J. Med. Chem. 2010, 45, 1042–1050. [Google Scholar] [CrossRef]
- Zhang, W.; Holyoke, C.W.; Barry, J.; Cordova, D.; Leighty, R.M.; Tong, M.T.; Hughes, K.A.; Pahutski, T.F.; Xu, M.; Briddell, T.A.; et al. Mesoionicpyrido[1,2-a]pyrimidinones: Discovery of triflumezopyrim as a potent hopper insecticide. Bioorg. Med. Chem. Lett. 2017, 27, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chai, W.M.; Yang, Q.; Wei, M.K.; Peng, Y. 2-(4-Fluorophenyl)-quinazolin- 4(3H)-one as a novel tyrosinase inhibitor: Synthesis, inhibitory activity, and mechanism. Bioorg. Med. Chem. 2016, 24, 4620–4625. [Google Scholar] [CrossRef]
- Patel, N.B.; Patel, J.C. Synthesis and antimicrobial activity of Schiff bases and 2-azetidinones derived from quinazolin-4(3H)-one. Arab. J. Chem. 2011, 4, 403–411. [Google Scholar] [CrossRef]
- Nagwa, M.A.G.; Hanan, H.G.; Riham, M.Y.; Nehad, A.E.S. Synthesis and antitumor activity of some 2,3-disubstituted quinazolin-4(3H)- ones and 4,6-disubstituted-1,2,3,4-tetrahydroquinazolin-2H-ones. Eur. J. Med. Chem. 2010, 45, 6058–6067. [Google Scholar] [CrossRef]
- El-Mekabaty, A. Synthesis and Antioxidant Activity of Some New Heterocycles Incorporating the Pyrazolo[3,4-d]pyrimidin-4-one Moiety. Chem. Heterocycl. Compd. 2015, 50, 1698–1706. [Google Scholar] [CrossRef]
- De Vita, D.; Pandolfi, F.; Cirilli, R.; Scipione, L.; Di Santo, R.; Friggeri, L.; Mori, M.; Fiorucci, D.; Maccari, G.; Christopher, R.S.A.; et al. Discovery of in vitro antitubercular agents through in silico ligand-based approaches. Eur. J. Med. Chem. 2016, 121, 169–180. [Google Scholar] [CrossRef]
- Gong, P.; Zhao, Y.F.; Wang, D. Synthesis and vasodilatory activities of new pyrazolo[3,4- d]pyrimidin-4-one derivatives. Chin. Chem. Lett. 2002, 13, 613–616. [Google Scholar]
- Kumar, R.; Joshi, Y.C. Synthesis, antimicrobial and antifungal activities of novel 1H-1,4-diazepines containing pyrazolopyrimidinone moiety. J. Chem. Sci. 2009, 121, 497–502. [Google Scholar] [CrossRef]
- Baviskar, A.T.; Banerjee, U.C.; Gupta, M.; Singh, R.; Kumar, S.; Gupta, M.K.; Kumar, S.; Raut, S.K.; Khullar, M.; Singh, S.; et al. Synthesis of imine-pyrazolopyrimidinones and their mechanistic interventions on anticancer activity. Bio. Org. Med. Chem. 2013, 21, 5782–5793. [Google Scholar] [CrossRef] [PubMed]
- El-Mekabaty, A.; Etman, H.A.; Mosbah, A.; Fadda, A.A. Synthesis, In Vitro Cytotoxicity and Bleomycin-Dependent DNA Damage Evaluation of Some Heterocyclic-Fused Pyrimidinone Derivatives. ChemistrySelect 2020, 5, 4856–4861. [Google Scholar] [CrossRef]
- Hassan, G.S.; Kadry, H.H.; Abou-Seri, S.M.; Ali, M.M.; Mahmoud, A.E.E. Synthesis and in vitro cytotoxic activity of novel pyrazolo[3,4-d]pyrimidines and related pyrazolehydrazones toward breast adenocarcinoma MCF-7 cell line. Bioorg. Med. Chem. 2011, 19, 6808–6817. [Google Scholar] [CrossRef]
- Reddy, G.L.; Guru, S.K.; Srinivas, M.; Pathania, A.S.; Mahajan, P.; Nargotra, A.; Bhushan, S.; Vishwakarma, R.A.; Sawant, S.D. Synthesis of 5-substituted-1H-pyrazolo[4,3-d]pyrimidin-7(6H)-one analogs and their biological evaluation as anticancer agents: mTOR inhibitors. Eur. J. Med. Chem. 2014, 80, 201–208. [Google Scholar] [CrossRef]
- Salar, U.; Taha, M.; Khan, K.M.; Ismail, N.H.; Imran, S.; Perveen, S.; Gul, S.; Wadood, A. Syntheses of new 3-thiazolyl coumarin derivatives, in vitro α-glucosidase inhibitory activity, and molecular modeling studies. Eur. J. Med. Chem. 2016, 122, 196–204. [Google Scholar] [CrossRef]
- Garkani-Nejad, Z.; Ahmadi-Roudi, B. Modeling the antileishmanial activity screening of 5-nitro-2-heterocyclic benzylidene hydrazides using different chemometrics methods. Eur. J. Med. Chem. 2010, 45, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Walcourt, A.; Loyevsky, M.; Lovejoy, D.B.; Gordeuk, V.R.; Richardson, D.R. Novel aroylhydrazone and thiosemicarbazone iron chelators with anti-malarial activity against chloroquine-resistant and -sensitive parasites. Int. J. Biochem. Cell Biol. 2004, 36, 401–407. [Google Scholar] [CrossRef]
- Mohareb, R.M.; Fleita, D.H.; Sakka, O.K. Novel synthesis of hydrazide-hydrazone derivatives and their utilization in the synthesis of coumarin, pyridine, thiazole and thiophene derivatives with antitumor activity. Molecules 2010, 16, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Bernardino, A.; Gomes, A.; Charret, K.; Freitas, A.; Machado, G.; Canto-Cavalheiro, M.; Leon, L.; Amaral, V. Synthesis and leishmanicidal activities of 1-(4-X-phenyl)-N′-[(4-Y-phenyl)methylene]-1H-pyrazole-4-carbohydrazides. Eur. J. Med. Chem. 2006, 41, 80–87. [Google Scholar] [CrossRef]
- Gürsoy, A.; Karali, N. Synthesis and primary cytotoxicity evaluation of 3-[[(3-phenyl-4(3H)-quinazolinone-2-yl)mercaptoacetyl]hydrazono]-1H-2-indolinones. Eur. J. Med. Chem. 2003, 38, 633–643. [Google Scholar] [CrossRef]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Caterino, M.; D’Aria, F.; Kustov, A.V.; Belykh, D.V.; Khudyaeva, I.S.; Starseva, O.M.; Berezin, D.B.; Pylina, Y.I.; Usacheva, T.; Amato, J.; et al. Selective binding of a bioactive porphyrin-based photosensitizer to the G-quadruplex from the KRAS oncogene promoter. Int. J. Biol. Macromol. 2020, 145, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Pagano, B.; Fotticchia, I.; De Tito, S.; Mattia, C.A.; Mayol, L.; Novellino, E.; Randazzo, A.; Giancola, C. Selective binding of distamycin a derivative to G-quadruplex structure [d(TGGGGT)]4. J. Nucleic Acids 2010. [Google Scholar] [CrossRef]
- Maennling, A.E.; Tur, M.K.; Niebert, M.; Klockenbring, T.; Zeppernick, F.; Gattenlöhner, S.; Meinhold-Heerlein, I.; Hussain, A.F. Molecular Targeting Therapy against EGFR Family in Breast Cancer: Progress and Future Potentials. Cancers 2019, 11, 1826. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009, 28 (Suppl. 1), S24–S31. [Google Scholar] [CrossRef]
- Choi, J.; Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. [Google Scholar] [CrossRef]
- Zhao, J.; Bacolla, A.; Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010, 67, 43–62. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Zorzet, S.; Rapozzi, V.; Géci, I.; Pedersen, E.B.; Xodo, L.E. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013, 41, 4049–4064. [Google Scholar] [CrossRef]
- Adams, J.M. The Bcl-2 Protein Family: Arbiters of Cell Survival. Science 1998, 281, 1322–1326. [Google Scholar] [CrossRef]
- Yip, K.W.; Reed, J.C. Bcl-2 family proteins and cancer. Oncogene 2008, 27, 6398–6406. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Gue’rin, O.; Fischel, J.L.; Ferrero, J.M.; Bozec, A.; Milano, G. EGFR targeting in hormone-refractory prostate cancer: Current appraisal and prospects for treatment. Pharmaceuticals 2010, 3, 2238–2247. [Google Scholar] [CrossRef] [PubMed]
- Abdelgawad, M.A.; Bakr, R.B.; Alkhoja, O.A.; Mohamed, W.R. Design, synthesis and antitumor activity of novel pyrazolo[3,4-d]pyrimidine derivatives as EGFR-TK inhibitors. Bioorg. Chem. 2016, 66, 88–96. [Google Scholar] [CrossRef]
- Holbro, T.; Hynes, N.E. ErbB receptors: Directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Ducray, R.; Ballard, P.; Barlaam, B.C.; Hickinson, M.D.; Kettle, J.G.; Ogilvie, D.J.; Trigwell, C.B. Novel 3-alkoxy-1H-pyrazolo[3,4-d]pyrimidines as EGFR and erbB2 receptor tyrosine kinase inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 959–962. [Google Scholar] [CrossRef]
- Bakr, R.B.; Mehany, A.B.M.; Abdellatif, K.R.A. Synthesis, EGFR Inhibition and Anti-cancer Activity of New 3,6-dimethyl-1-phenyl-4-(substituted-methoxy)pyrazolo[3,4-d] pyrimidine Derivatives. Anticancer Agents Med. Chem. 2017, 17, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.E.; Aly, E.I.; Awadallah, F.M.; Mahmoud, W.R. 4-Substituted-1-phenyl-1H-pyrazolo[3,4-d]pyrimidine derivatives: Design, synthesis, antitumor and EGFR tyrosine kinase inhibitory activity. Chem. Biol. Drug Des. 2015, 85, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Chen, Y.; Wang, X.Y.; Lu, R.F.; Zhang, S.Y.; Tian, M.; Xie, T.; Liu, B.; He, G. Polygonatum odoratum lectin induces apoptosis and autophagy via targeting EGFR-mediated Ras-Raf-MEK-ERK pathway in human MCF-7 breast cancer cells. Phytomedicine 2014, 21, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, S.; Mukherjee, S.; Das, S.; Bhat, F.A.; Raja Singh, P.; Patra, C.R.; Arunakaran, J. Gold nanoparticles-conjugated quercetin induces apoptosis via inhibition of EGFR/PI3K/Akt-mediated pathway in breast cancer cell lines (MCF-7 and MDA-MB-231). Cell Biochem. Funct. 2017, 35, 217–231. [Google Scholar] [CrossRef]
- Moiseeva, E.P.; Heukers, R.; Manson, M.M. EGFR and Src are involved in indole-3-carbinol-induced death and cell cycle arrest of human breast cancer cells. Carcinogenesis 2007, 28, 435–445. [Google Scholar] [CrossRef]
- Mahernia, S.; Hassanzadeh, M.; Sharifi, N.; Mehravi, B.; Paytam, F.; Adib, M.; Amanlou, M. Structure-based pharmacophore design and virtual screening for novel potential inhibitors of epidermal growth factor receptor as an approach to breast cancer chemotherapy. Mol. Divers. 2018, 22, 173–181. [Google Scholar] [CrossRef]
- Singh, P.; Bast, F. In silico molecular docking study of natural compounds on wild and mutated epidermal growth factor receptor. Med. Chem. Res. 2014, 23, 5074–5085. [Google Scholar] [CrossRef]
- Ayaz, M.; Sadiq, A.; Wadood, A.; Junaid, M.; Ullah, F.; Khan, N.Z. Cytotoxicity and molecular docking studies on Phytosterols isolated from Polygonum hydropiper L. Steroids 2018, 141, 30–35. [Google Scholar] [CrossRef]
- Huether, A.; Höpfner, M.; Sutter, A.P.; Schuppan, D.; Scherübl, H. Erlotinib induces cell cycle arrest and apoptosis in hepatocellular cancer cells and enhances chemosensitivity towards cytostatics. J. Hepatol. 2005, 43, 661–669. [Google Scholar] [CrossRef]
- Shan, F.; Shao, Z.; Jiang, S.; Cheng, Z. Erlotinib induces the human non-small-cell lung cancer cells apoptosis via activating ROS-dependent JNK pathways. Cancer Med. 2016, 5, 3166–3175. [Google Scholar] [CrossRef]
- Saleh, M.M.; Laughton, C.A.; Bradshaw, T.D.; Moody, C.J. Development of a series of bis-triazoles as G-quadruplex ligands. RSC Adv. 2017, 7, 47297–47308. [Google Scholar] [CrossRef]
- Horchani, M.; Hajlaoui, A.; Harrath, A.H.; Mansour, L.; Ben Jannet, H.; Romdhane, A. New pyrazolo-triazolo-pyrimidine derivatives as antibacterial agents: Design and synthesis, molecular docking and DFT studies. J. Mol. Struct. 2020, 1199, 127007. [Google Scholar] [CrossRef]
- Lopes, A.B.; Miguez, E.; Kümmerle, A.E.; Rumjanek, V.M.; Fraga, C.A.; Barreiro, E.J. Characterization of amide bond conformers for a novel heterocyclic template of N-acylhydrazone derivatives. Molecules 2013, 18, 11683–11704. [Google Scholar] [CrossRef]
- Patorski, P.; Wyrzykiewicz, E.; Bartkowiak, G. Synthesis and conformational assignment of N-(E)-stilbenyloxymethylenecarbonyl-substituted hydrazones of acetone and o- (m-and p-) chloro- (nitro-) benzaldehydes by means of and NMR spectroscopy. J. Spectrosc. 2013, 1–12. [Google Scholar] [CrossRef]
- Teta, R.; Della Sala, G.; Esposito, G.; Via, C.W.; Mazzoccoli, C.; Piccoli, C.; Bertin, M.J.; Costantino, V.; Mangoni, A. A joint molecular networking study of a Smenospongia sponge and a cyanobacterial bloom revealed new antiproliferative chlorinated polyketides. Org. Chem. Front. 2019, 6, 1762–1774. [Google Scholar] [CrossRef] [PubMed]
- Imperatore, C.; Della Sala, G.; Casertano, M.; Luciano, P.; Aiello, A.; Laurenzana, I.; Piccoli, C.; Menna, M. In Vitro Antiproliferative Evaluation of Synthetic Meroterpenes Inspired by Marine Natural Products. Mar. Drugs 2019, 17, 684. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef]
- AutoDock—Scripps Research. Available online: http://autodock.scripps.edu/ (accessed on 15 January 2021).
- The PyMOL Molecular Graphics System, Version 1.5.0.4; Schrödinger, LLC.: New York, NY, USA, 2015.
- Oliviero, G.; Errico, S.D.; Borbone, N.; Amato, J.; Piccialli, V.; Varra, M.; Piccialli, G.; Mayol, L. A solid-phase approach to the synthesis of N -1-alkyl analogues of cyclic inosine-diphosphate-ribose (cIDPR). Tetrahedron 2010, 66, 1931–1936. [Google Scholar] [CrossRef]
- Cantor, C.R.; Warshaw, M.M.; Shapiro, H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleolides. Biopolymers 1970, 9, 1059–1077. [Google Scholar] [CrossRef]

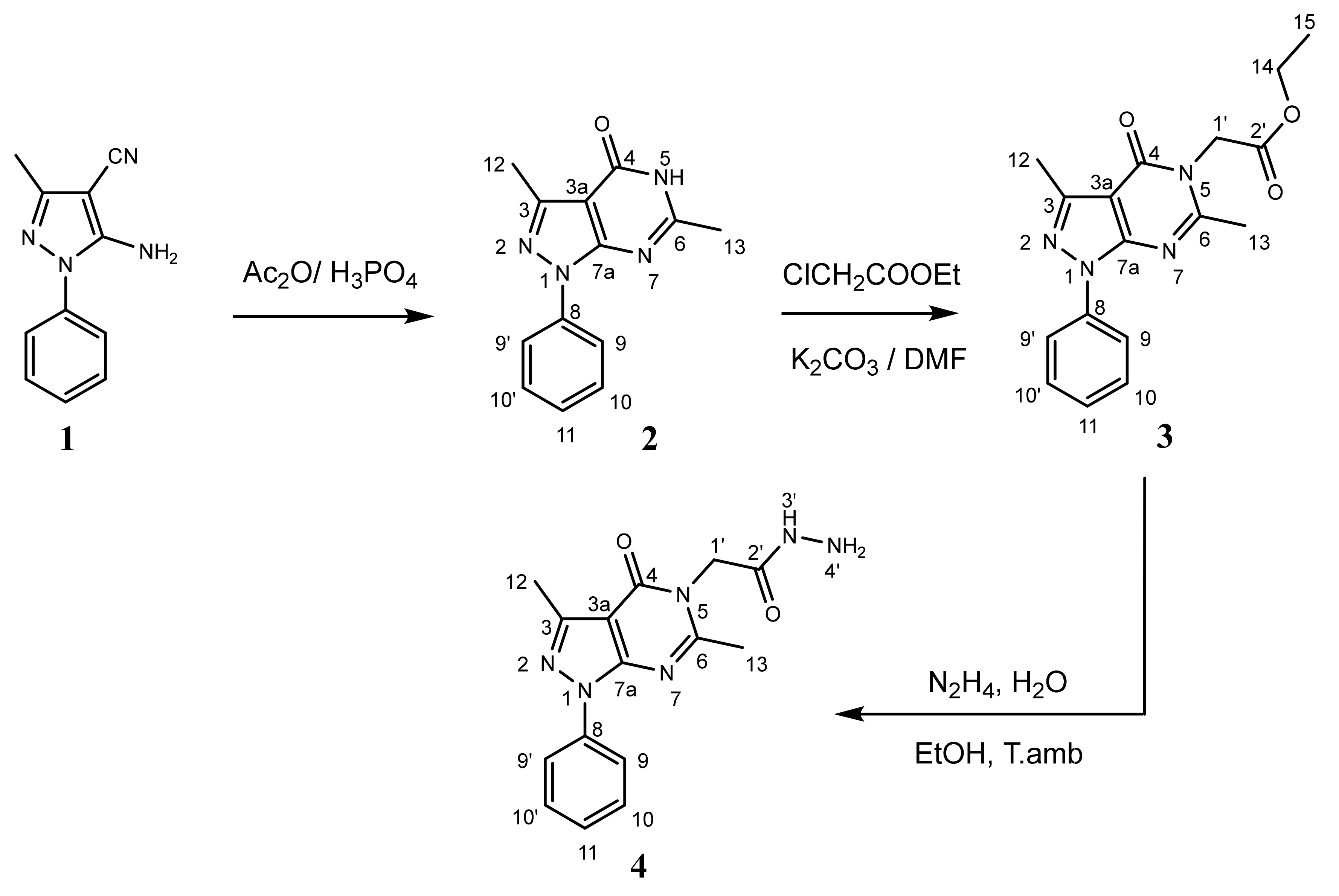


| Compound | R | Yield (%) |
|---|---|---|
| 5a | 4-N(CH3)2 | 60 |
| 5b | 4-OCH3 | 75 |
| 5c | 4-NO2 | 78 |
| 5d | 3,4-diOCH3 | 70 |
| 5e | 3-OC2H5,4-OH | 62 |
| 5f | 4-F | 68 |
| 5g | 2-OH,5-Br | 64 |
| 5h | 4-Br | 72 |
| Compound | Binding Energy (kcal/mol) | |||||
|---|---|---|---|---|---|---|
| Erlotinib | −7.1 | −6.9 | 6.8 | 6.8 | 6.8 | −6.6 |
| 3 | −8.0 | −7.6 | −7.5 | −7.4 | −7.3 | −7.1 |
| 5a | −8.9 | −8.5 | −8.4 | −8.4 | −8.0 | −7.9 |
| 5e | −8.9 | −8.7 | −8.6 | −8.4 | −8.2 | −8.1 |
| 5g | −9.1 | −8.8 | −8.2 | −8.1 | −8.1 | −8.1 |
| 5h | −8.9 | −8.7 | −8.7 | −8.5 | −8.3 | −8.3 |
| Ligand | KRAS 22RT ΔTm (°C) 1 | BCL2-G4 ΔTm (°C) 1 | Tel 23 Δ Tm (°C) 1 | Hairpin Duplex Δ Tm (°C) 1 |
|---|---|---|---|---|
| 5a | +7.0 | −3.0 | −3.2 | +1.0 |
| 5e | 0 | 0 | 0 | +5.4 |
| 5g | 0 | 0 | −2.0 | 0 |
| 5h | 0 | +1.0 | +1.0 | +3.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horchani, M.; Della Sala, G.; Caso, A.; D’Aria, F.; Esposito, G.; Laurenzana, I.; Giancola, C.; Costantino, V.; Jannet, H.B.; Romdhane, A. Molecular Docking and Biophysical Studies for Antiproliferative Assessment of Synthetic Pyrazolo-Pyrimidinones Tethered with Hydrazide-Hydrazones. Int. J. Mol. Sci. 2021, 22, 2742. https://doi.org/10.3390/ijms22052742
Horchani M, Della Sala G, Caso A, D’Aria F, Esposito G, Laurenzana I, Giancola C, Costantino V, Jannet HB, Romdhane A. Molecular Docking and Biophysical Studies for Antiproliferative Assessment of Synthetic Pyrazolo-Pyrimidinones Tethered with Hydrazide-Hydrazones. International Journal of Molecular Sciences. 2021; 22(5):2742. https://doi.org/10.3390/ijms22052742
Chicago/Turabian StyleHorchani, Mabrouk, Gerardo Della Sala, Alessia Caso, Federica D’Aria, Germana Esposito, Ilaria Laurenzana, Concetta Giancola, Valeria Costantino, Hichem Ben Jannet, and Anis Romdhane. 2021. "Molecular Docking and Biophysical Studies for Antiproliferative Assessment of Synthetic Pyrazolo-Pyrimidinones Tethered with Hydrazide-Hydrazones" International Journal of Molecular Sciences 22, no. 5: 2742. https://doi.org/10.3390/ijms22052742
APA StyleHorchani, M., Della Sala, G., Caso, A., D’Aria, F., Esposito, G., Laurenzana, I., Giancola, C., Costantino, V., Jannet, H. B., & Romdhane, A. (2021). Molecular Docking and Biophysical Studies for Antiproliferative Assessment of Synthetic Pyrazolo-Pyrimidinones Tethered with Hydrazide-Hydrazones. International Journal of Molecular Sciences, 22(5), 2742. https://doi.org/10.3390/ijms22052742









