Role of Genetic Variation in Cytochromes P450 in Breast Cancer Prognosis and Therapy Response
Abstract
:1. Introduction
2. Results
2.1. Evaluation Phase
2.2. Confirmation Phase
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Panel Sequencing—Evaluation Phase
4.3. Genotyping—Confirmation Phase
4.4. Quantitative Real-Time PCR
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CI | Confidence interval |
| CYP | Cytochrome P450 |
| DFS | Disease-free survival |
| FDR | False discovery rate |
| GWAS | Genome-wide association study |
| HR | Hazard ratio |
| LOF | Loss-of-function |
| MAF | Minor allele frequency |
| NACT | Neoadjuvant cytotoxic therapy |
| qPCR | Quantitative real-time PCR |
| SNP | Single nucleotide polymorphism |
| SNV | Single nucleotide variant |
| UTR | Untranslated region |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Heer, E.; Harper, A.; Escandor, N.; Sung, H.; McCormack, V.; Fidler-Benaoudia, M.M. Global burden and trends in premenopausal and postmenopausal breast cancer: A population-based study. Lancet Glob. Health 2020, 8, e1027–e1037. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Mkrtchian, S.; Zhou, Y.; Lauschke, V.M. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Genom. 2018, 12, 1–12. [Google Scholar] [CrossRef]
- Hlaváč, V.; Holý, P.; Souček, P. Pharmacogenomics to predict tumor therapy response: A focus on atp-binding cassette transporters and cytochromes P450. J. Pers. Med. 2020, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. The Cytochrome P450 Homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Gaedigk, A.; Ingelman-Sundberg, M.; Miller, N.A.; Leeder, J.S.; Whirl-Carrillo, M.; Klein, T.E. The PharmVar steering committee the pharmacogene variation (PharmVar) consortium: Incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharmacol. Ther. 2018, 103, 399–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rendic, S.P.; Guengerich, F.P. Human cytochrome P450 enzymes 5–51 as targets of drugs and natural and environmental compounds: Mechanisms, induction, and inhibition—Toxic effects and benefits. Drug Metab. Rev. 2018, 50, 256–342. [Google Scholar] [CrossRef]
- Guengerich, F.P. Human Cytochrome P450 Enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry; Ortiz de Montellano, P.R., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 523–785. ISBN 978-3-319-12108-6. [Google Scholar]
- Guengerich, F.P. A history of the roles of cytochrome P450 enzymes in the toxicity of drugs. Toxicol. Res. 2021, 37, 1–23. [Google Scholar] [CrossRef]
- Samavat, H.; Kurzer, M.S. Estrogen metabolism and breast cancer. Cancer Lett. 2015, 356, 231–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artigalás, O.; Vanni, T.; Hutz, M.H.; Ashton-Prolla, P.; Schwartz, I.V. Influence of CYP19A1 polymorphisms on the treatment of breast cancer with aromatase inhibitors: A systematic review and meta-analysis. BMC Med. 2015, 13, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hlavac, V.; Kovacova, M.; Elsnerova, K.; Brynychova, V.; Kozevnikovova, R.; Raus, K.; Kopeckova, K.; Mestakova, S.; Vrana, D.; Gatek, J.; et al. Use of germline genetic variability for prediction of chemoresistance and prognosis of breast cancer patients. Cancers 2018, 10, 511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, K.; Guengerich, F.P. Characterization of orphan human cytochromes P450. Drug Metab. Rev. 2007, 39, 627–637. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Cheng, Q. Orphans in the human cytochrome p450 superfamily: Approaches to discovering functions and relevance in pharmacology. Pharmacol. Rev. 2011, 63, 684–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, K.; Dostalek, M.; Guengerich, F.P. Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. FEBS J. 2008, 275, 3706–3717. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S. Computational identification and binding analysis of orphan human cytochrome P450 4X1 enzyme with substrates. BMC Res. Notes 2015, 8, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GeneCards® Home Page. Available online: http://www.genecards.org/ (accessed on 1 August 2012).
- Murray, G.; Patimalla, S.; Stewart, K.N.; Miller, I.D.; Heys, S.D. Profiling the expression of cytochrome P450 in breast cancer. Histopathology 2010, 57, 202–211. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, Z.; Shen, R.; Jiang, Y.; Xu, W.; Gu, M.; Gu, X. Identification of biomarkers predicting the chemotherapeutic outcomes of capecitabine and oxaliplatin in patients with gastric cancer. Oncol. Lett. 2020, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Poleggi, A.; Van Der Lee, S.; Capellari, S.; Puopolo, M.; Ladogana, A.; De Pascali, E.; Lia, D.; Formato, A.; Bartoletti-Stella, A.; Parchi, P.; et al. Age at onset of genetic (E200K) and sporadic Creutzfeldt-Jakob diseases is modulated by the CYP4X1 gene. J. Neurol. Neurosurg. Psychiatry 2018, 89, 1243–1249. [Google Scholar] [CrossRef]
- Guindon, J.; Hohmann, A.G. The endocannabinoid system and cancer: Therapeutic implication. Br. J. Pharmacol. 2011, 163, 1447–1463. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Zhong, R.; Tian, J.; Li, J.; Zhai, K.; Ke, J.; Lou, J.; Chen, W.; Zhu, B.; Shen, N.; et al. Exome-wide analyses identify low-frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat. Genet. 2018, 50, 338–343. [Google Scholar] [CrossRef]
- Trubicka, J.; Grabowska-Kłujszo, E.; Suchy, J.; Masojć, B.; Serrano-Fernández, P.; Kurzawski, G.; Cybulski, C.; Górski, B.; Huzarski, T.; Byrski, T.; et al. Variant alleles of the CYP1B1 gene are associated with colorectal cancer susceptibility. BMC Cancer 2010, 10, 420. [Google Scholar] [CrossRef] [Green Version]
- Kamiza, A.B.; You, J.-F.; Wang, W.-C.; Tang, R.; Chang, C.-Y.; Chien, H.-T.; Lai, C.-H.; Chiu, L.-L.; Lo, T.-P.; Hung, K.-Y.; et al. Polymorphisms of xenobiotic-metabolizing genes and colorectal cancer risk in patients with lynch syndrome: A retrospective cohort study in Taiwan. Environ. Mol. Mutagen. 2018, 59, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Reding, K.W.; Weiss, N.S.; Chen, C.; Li, C.I.; Carlson, C.S.; Wilkerson, H.-W.; Farin, F.M.; Thummel, K.E.; Daling, J.R.; Malone, K.E. Genetic polymorphisms in the catechol estrogen metabolism pathway and breast cancer risk. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1461–1467. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, M.; Li, L.; Sun, H.; Lin, X. Association of single nucleotide polymorphisms in the CYP1B1 gene with the risk of primary open-angle glaucoma: A meta-analysis. Genet. Mol. Res. 2015, 14, 17262–17272. [Google Scholar] [CrossRef]
- Shimada, T.; Hayes, C.L.; Yamazaki, H.; Amin, S.; Hecht, S.S.; Guengerich, F.P.; Sutter, T.R. Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res. 1996, 56, 2979–2984. [Google Scholar]
- Watkins, G.; Douglas-Jones, A.; Mansel, R.E.; Jiang, W.G. Expression of thromboxane synthase, TBXAS1 and the thromboxane A2 receptor, TBXA2R, in human breast cancer. Int. Semin. Surg. Oncol. 2005, 2, 23. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Jin, F.; Fan, W.; Liu, F.; Zou, Y.; Hu, X.; Xu, H.; Han, P. Gene expression meta-analysis in diffuse low-grade glioma and the corresponding histological subtypes. Sci. Rep. 2017, 7, 11741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wang, G.; Hong, X.; Wang, D.; Tsai, H.-J.; Zhang, S.; Arguelles, L.; Kumar, R.; Wang, H.; Liu, R.; et al. Gene-vitamin D interactions on food sensitization: A prospective birth cohort study. Allergy 2011, 66, 1442–1448. [Google Scholar] [CrossRef] [Green Version]
- Fuhrman, B.J.; Freedman, D.M.; Bhatti, P.; Doody, M.M.; Fu, Y.P.; Chang, S.C.; Linet, M.S.; Sigurdson, A.J. Sunlight, Poly-morphisms of vitamin d-related genes and risk of breast cancer. Anticancer. Res. 2013, 33, 543–551. [Google Scholar]
- Yang, W.; Ma, F.; Wang, L.; He, X.; Zhang, H.; Zheng, J.; Wang, Y.; Jin, T.; Yuan, D.; He, Y. The association analysis between CYP24A1 genetic polymorphisms and the risk of ischemic stroke in Chinese Han population. Brain Behav. 2019, 10, e01503. [Google Scholar] [CrossRef] [Green Version]
- Bao, Q.; Wang, D.; Zhang, Y.; Bao, L.; Jia, H. The impact of CYP24A1 polymorphisms on hypertension susceptibility. Kidney Blood Press. Res. 2020, 45, 28–37. [Google Scholar] [CrossRef]
- Hlaváč, V.; Brynychová, V.; Václavíková, R.; Ehrlichová, M.; Vrána, D.; Pecha, V.; Koževnikovová, R.; Trnková, M.; Gatěk, J.; Kopperová, D.; et al. The expression profile of ATP-binding cassette transporter genes in breast carcinoma. Pharmacogenomics 2013, 14, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.H.; Litière, S.; De Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Update and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 2014, 30, 2843–2851. [Google Scholar] [CrossRef] [Green Version]
- Van Der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
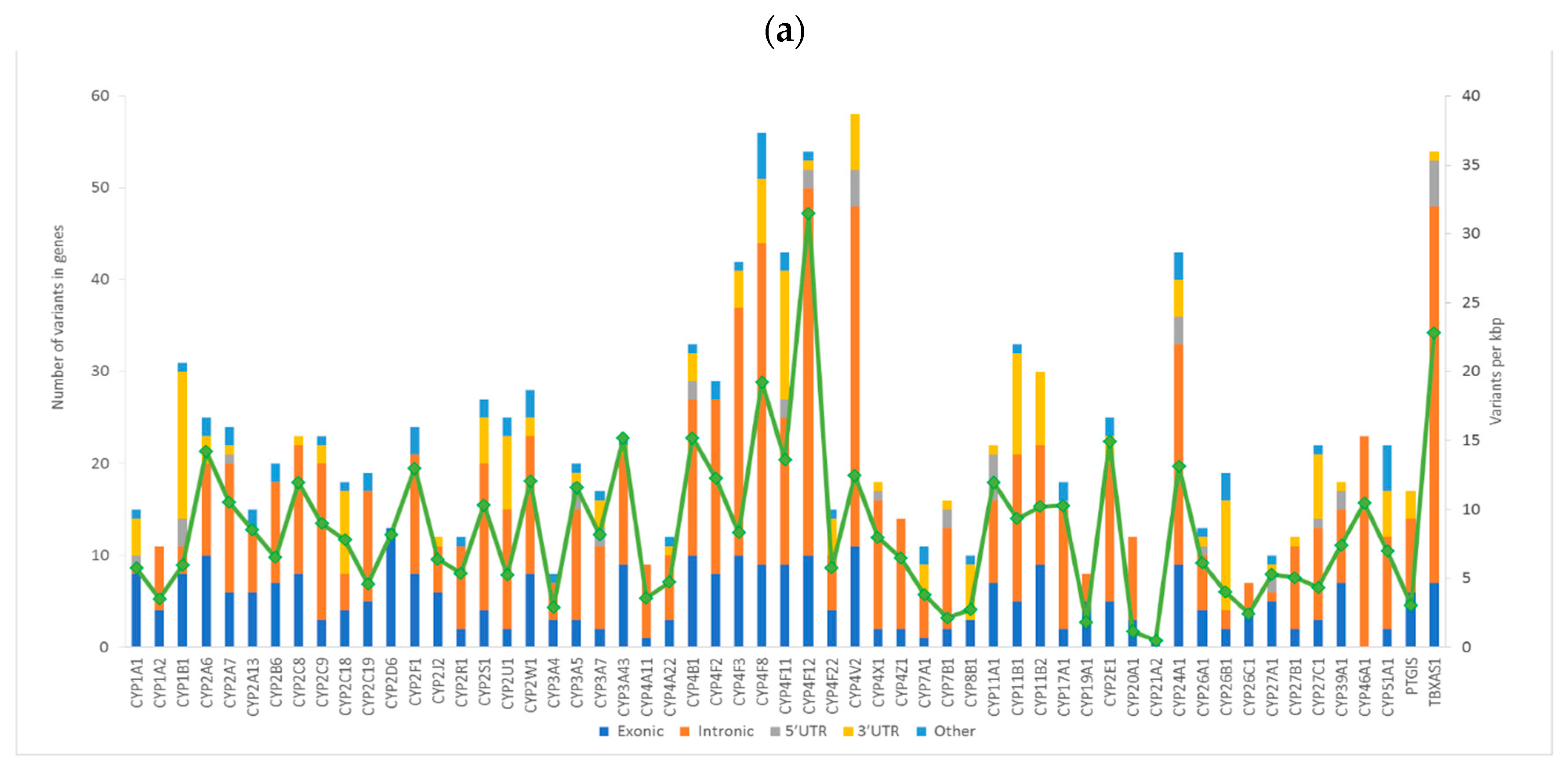



| Function | Total | Percentage |
|---|---|---|
| Intronic | 685 | 53.8 |
| Exonic (coding) | 302 | 23.7 |
| 3′UTR | 167 | 13.1 |
| 5′UTR | 43 | 3.4 |
| Upstream 1 | 45 | 3.5 |
| Downstream 1 | 19 | 1.5 |
| Intergenic | 10 | 0.8 |
| Splicing 2 | 3 | 0.2 |
| Classification | Total | Percentage |
|---|---|---|
| Non-synonymous SNV | 178 | 58.9 |
| Synonymous SNV | 99 | 32.8 |
| Stop-gain | 6 | 2.0 |
| Frameshift deletion | 4 | 1.3 |
| Frameshift insertion | 4 | 1.3 |
| Non-frameshift deletion | 2 | 0.7 |
| Unknown | 9 | 3.0 |
| Gene | SNP ID 1 | Genotype Distribution 2 | Minor Allele Frequency | |||
|---|---|---|---|---|---|---|
| Common Homozygotes | Heterozygotes | Rare Homozygotes | Confirmation Set | Evaluation Set | ||
| CYP1B1 | rs1056827 | 362 | 354 | 77 | 0.32 | 0.34 |
| CYP2S1 | rs184623 | 308 | 379 | 100 | 0.37 | 0.38 |
| CYP2W1 | rs3808348 | 538 | 237 | 23 | 0.18 | 0.20 |
| CYP2W1 | rs12701220 | 533 | 239 | 25 | 0.18 | 0.11 |
| CYP4A11 | rs3890011 | 459 | 291 | 46 | 0.24 | 0.27 |
| CYP4F2 | rs2074900 | 367 | 343 | 83 | 0.32 | 0.32 |
| CYP4F2 | rs3093198 | 398 | 325 | 73 | 0.30 | 0.29 |
| CYP4F8 | rs714772 | 506 | 258 | 35 | 0.21 | 0.25 |
| CYP4F8 | rs4646522 | 225 | 401 | 158 | 0.46 | 0.42 |
| CYP4F12 | rs593421 | 416 | 308 | 54 | 0.27 | 0.29 |
| CYP4F12 | rs593818 | 230 | 373 | 187 | 0.47 | 0.43 |
| CYP4F12 | rs2074568 | 518 | 211 | 23 | 0.17 | 0.21 |
| CYP4V2 | rs62350517 | 693 | 104 | 4 | 0.07 | 0.08 |
| CYP4X1 | rs17102977 | 653 | 125 | 8 | 0.09 | 0.10 |
| CYP24A1 | rs2259735 | 246 | 365 | 155 | 0.44 | 0.39 |
| CYP24A1 | rs2762934 | 549 | 231 | 17 | 0.17 | 0.17 |
| CYP24A1 | rs6022999 | 496 | 251 | 50 | 0.22 | 0.21 |
| CYP24A1 | rs10623012 | 294 | 382 | 105 | 0.38 | 0.32 |
| CYP26B1 | rs61138718 | 606 | 183 | 12 | 0.13 | 0.11 |
| CYP26B1 | rs62150087 | 661 | 132 | 6 | 0.09 | 0.07 |
| CYP27C1 | rs12476709 | 236 | 379 | 174 | 0.46 | 0.47 |
| TBXAS1 | rs3819733 | 590 | 195 | 14 | 0.14 | 0.15 |
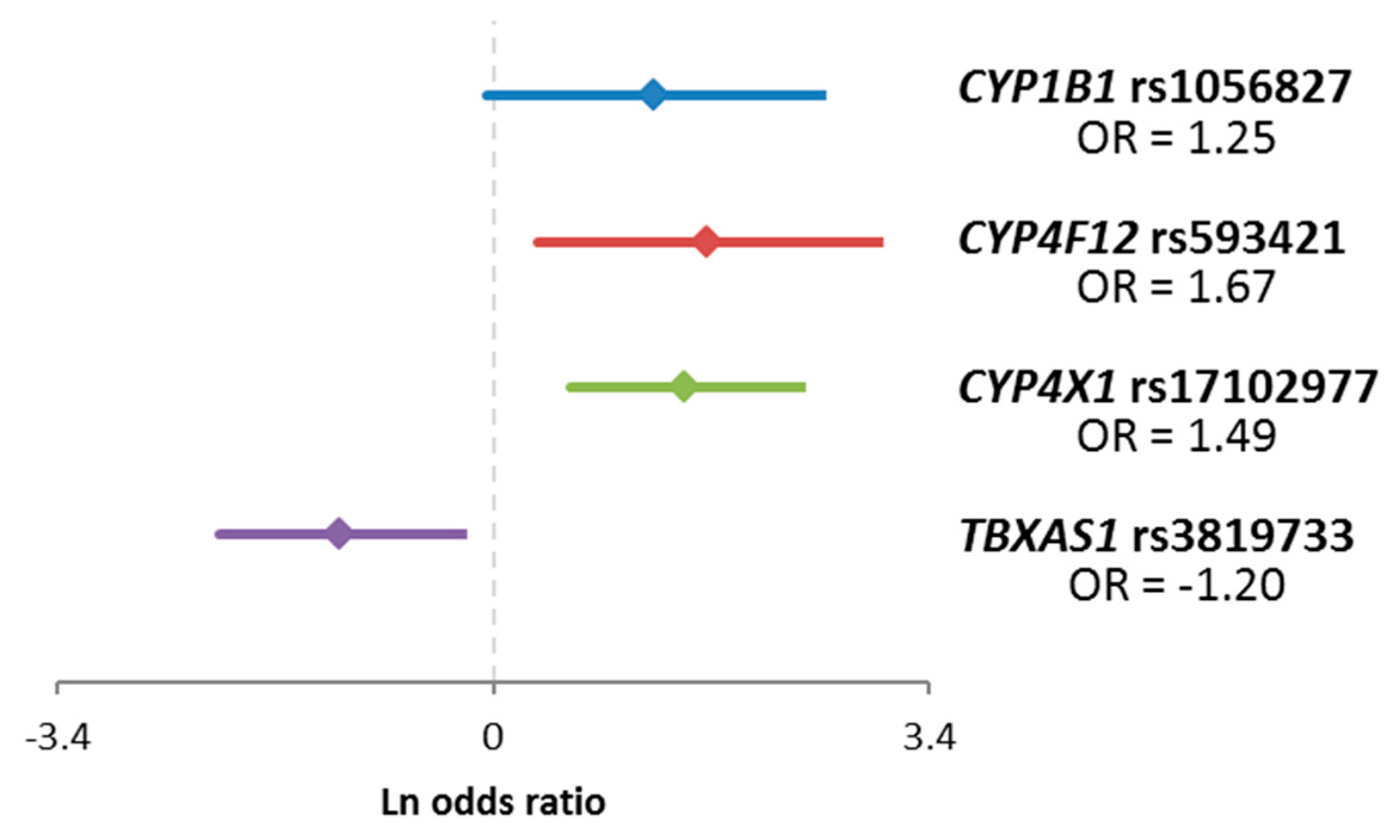
| Gene | SNP ID | Genotype | Good Response 1 | Poor Response 1 | χ−Square | p |
|---|---|---|---|---|---|---|
| CYP1B13 | rs1056827 | C allele | 122 | 35 | 3.96 | 0.047/0.339 2 |
| AA | 5 | 5 | ||||
| CYP4F12 | rs593421 | TT | 63 | 22 | 8.81 | 0.012/0.130 2 |
| TC | 57 | 12 | ||||
| CC | 4 | 6 | ||||
| CYP4X1 | rs17102977 | AA | 111 | 27 | 12.02 | 5.30 × 10−4/0.034 2 |
| G allele | 12 | 13 | ||||
| TBXAS1 | rs3819733 | TT | 81 | 35 | 6.76 | 0.009/0.130 2 |
| C allele | 46 | 6 |
| Gene | SNP ID | Genotypes | Subtypes | |||
|---|---|---|---|---|---|---|
| Luminal A | Luminal B | HER2 | TNBC | |||
| All patients (n = 744) | ||||||
| CYP26B1 | rs62150087 | CC 1 | 174 | 230 | 44 | 73 |
| G allele 1 | 36 | 42 | 12 | 12 | ||
| p2 | 0.754 | 0.086 | 0.010 | 0.178 | ||
| CYP4X1 | rs17102977 | AA 1 | 166 | 223 | 49 | 48 |
| G allele 1 | 44 | 42 | 6 | 16 | ||
| p2 | 0.245 | 0.130 | 0.150 | 0.778 | ||
| Patients treated with cytotoxic therapy (n = 371) | ||||||
| CYP26B1 | rs62150087 | CC 1 | 65 | 128 | 26 | 58 |
| G allele 1 | 9 | 25 | 10 | 8 | ||
| p2 | 0.244 | 0.232 | 0.011 | 0.060 | ||
| CYP24A1 | rs2762934 | GG 1 | 50 | 91 | 27 | 45 |
| A allele 1 | 24 | 60 | 9 | 19 | ||
| p2 | 0.181 | 0.172 | 0.400 | 0.001 | ||
| Patients treated only with hormonal therapy (n = 311) | ||||||
| CYP4X1 | rs17102977 | AA 1 | 102 | 81 | 3 | 1 |
| G allele 1 | 22 | 19 | 0 | 1 | ||
| p2 | 0.123 | 0.202 | N/A | 0.317 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hlaváč, V.; Václavíková, R.; Brynychová, V.; Ostašov, P.; Koževnikovová, R.; Kopečková, K.; Vrána, D.; Gatěk, J.; Souček, P. Role of Genetic Variation in Cytochromes P450 in Breast Cancer Prognosis and Therapy Response. Int. J. Mol. Sci. 2021, 22, 2826. https://doi.org/10.3390/ijms22062826
Hlaváč V, Václavíková R, Brynychová V, Ostašov P, Koževnikovová R, Kopečková K, Vrána D, Gatěk J, Souček P. Role of Genetic Variation in Cytochromes P450 in Breast Cancer Prognosis and Therapy Response. International Journal of Molecular Sciences. 2021; 22(6):2826. https://doi.org/10.3390/ijms22062826
Chicago/Turabian StyleHlaváč, Viktor, Radka Václavíková, Veronika Brynychová, Pavel Ostašov, Renata Koževnikovová, Katerina Kopečková, David Vrána, Jiří Gatěk, and Pavel Souček. 2021. "Role of Genetic Variation in Cytochromes P450 in Breast Cancer Prognosis and Therapy Response" International Journal of Molecular Sciences 22, no. 6: 2826. https://doi.org/10.3390/ijms22062826
APA StyleHlaváč, V., Václavíková, R., Brynychová, V., Ostašov, P., Koževnikovová, R., Kopečková, K., Vrána, D., Gatěk, J., & Souček, P. (2021). Role of Genetic Variation in Cytochromes P450 in Breast Cancer Prognosis and Therapy Response. International Journal of Molecular Sciences, 22(6), 2826. https://doi.org/10.3390/ijms22062826







