Abstract
Protein post-translational modifications (PTMs) play key roles in eukaryotes since they finely regulate numerous mechanisms used to diversify the protein functions and to modulate their signaling networks. Besides, these chemical modifications also take part in the viral hijacking of the host, and also contribute to the cellular response to viral infections. All domains of the human immunodeficiency virus type 1 (HIV-1) Gag precursor of 55-kDa (Pr55Gag), which is the central actor for viral RNA specific recruitment and genome packaging, are post-translationally modified. In this review, we summarize the current knowledge about HIV-1 Pr55Gag PTMs such as myristoylation, phosphorylation, ubiquitination, sumoylation, methylation, and ISGylation in order to figure out how these modifications affect the precursor functions and viral replication. Indeed, in HIV-1, PTMs regulate the precursor trafficking between cell compartments and its anchoring at the plasma membrane, where viral assembly occurs. Interestingly, PTMs also allow Pr55Gag to hijack the cell machinery to achieve viral budding as they drive recognition between viral proteins or cellular components such as the ESCRT machinery. Finally, we will describe and compare PTMs of several other retroviral Gag proteins to give a global overview of their role in the retroviral life cycle.
1. Introduction
Post translational modifications (PTMs) introduce a vast diversity in proteome including addition of chemical groups, like phosphorylation, methylation, acetylation, redox-based modifications, or alternatively, addition of polypeptides like ubiquitination, sumoylation or ISGylation. PTMs thus play a key role in functional proteomic by regulating proteins activity, their localization, and the interaction with cellular or viral factors. Even though many proteins are modified shortly after translation, PTMs can also occur at different steps such as after protein folding or protein re-localization to influence their biological activity at those specific sites (for reviews see [1,2]). Besides, depending on the nature of the modification, they can also finely tune reversible processes. Consequently, analysis of PTMs can provide an invaluable insight into cellular functions.
Viruses rely on the protein synthesis machinery of the host to support the production of viral progeny, and several cellular pathways are modulated by viruses to achieve the critical steps in viral replication. Hence, it is not surprising that viruses developed different strategies to either counteract or exploit PTMs of cellular factors, and that many viral proteins carry PTMs. Interestingly, PTMs are strongly involved in the regulation of different steps of the retrovirus viral cycle (for reviews see [3,4]). More specifically, in the HIV-1 (human immunodeficiency virus type 1) context, PTMs within the 55-kDa viral precursor, Pr55Gag (or Gag), were found to be necessary for regulating the last phase of the viral cycle, leading to the assembly of viral particles. Besides, several pieces of evidence have shown that other retroviral Gag carry various PTMs regulate viral replication and pathogenesis. This review will summarize our current knowledge on PTMs observed in HIV-1 Pr55Gag and in other retroviral Gag proteins. Considering the role of the PMTs in the retroviral life cycle, the analysis of PTMs in retroviral Gag precursors could be particularly important for a deeper understanding of the molecular mechanisms driving retroviral replication. In a further step, this knowledge could contribute to the identification of new targets, and the design of new treatments against retroviral replication.
2. HIV-1 Pr55Gag
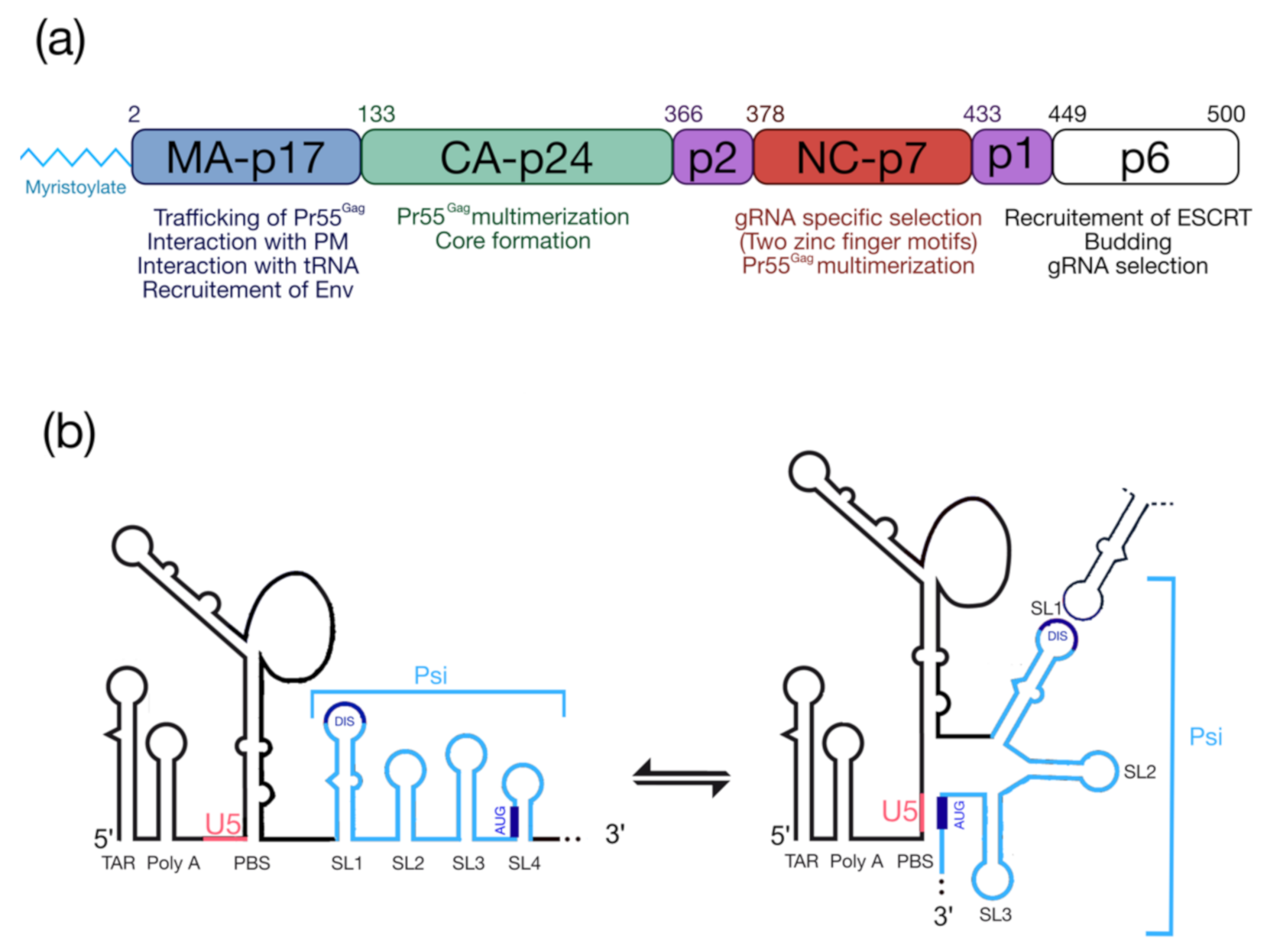
The HIV-1 Pr55Gag precursor (Figure 1a) plays a crucial for genomic RNA (gRNA) packaging, since it specifically selects the full-length gRNA amongst many other RNAs (cellular and spliced viral RNAs) and this process involves specific interactions between Pr55Gag and the highly structured 5′ region of the gRNA [5,6], which contains the packaging signal (Psi) spanning SL (stem-loop)1 to SL4 in the 5′-end region of gRNA [7,8,9] (Figure 1b). In cells, the HIV-1 gRNA dimer in association with low-order Pr55Gag multimers [10,11,12] forms a viral ribonucleoprotein complex that traffics to the plasma membrane (PM) where the assembly of the viral particle occurs (for reviews see [13,14,15]). HIV-1 Pr55Gag is composed of four structural domains named matrix (MA), capsid (CA), nucleocapsid (NC), p6, and two spacer peptides (p2 and p1) (Figure 1a) [16] and each of them carry PTMs.
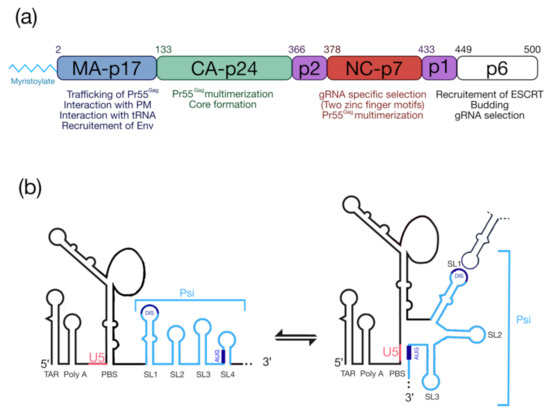
Figure 1.
Pr55Gag and the 5′UTR of HIV-1 genomic RNA. (a) Functional domains of Pr55Gag and a short description of their roles. (b) Schematic representation of the secondary structure model of the 5′UTR (adapted from [29]). TAR: transactivation response element; Poly-A: 5′-copy of the polyadenylation signal; PBS: Primer Biding Site; DIS: Dimerization Initiation Site; Psi: packaging signal spanning SL1 to SL4; U5: unique region at the 5′ end. The structure represents the U5-AUG conformation [5,6].
From the N-terminus, the 17 kDa MA domain that possesses a bipartite signal leads to Pr55Gag interaction with the PM. The first signal corresponds to the N-terminal myristoylated Glycine 2 (G2) (see § “HIV-1 Pr55Gag Myristoylation”), while the second one is constituted by a highly basic region (HBR) at the MA surface (for a review see [17]). MA was also found to interact with nucleic acids such as host tRNAs [18], and recent findings showed that MA-RNA binding ensures the specific interaction between Pr55Gag and the PM, by preventing nonspecific binding of Gag to intracellular membranes [19,20]. The CA is a 24 kDa domain that drives Pr55Gag multimerization and leads to formation of the viral core [21,22,23]. Next, NC is a 7 kDa domain, which is crucial for specific interaction with gRNA and for the incorporation of tRNALys3, which is the primer for reverse transcription. NC displays two zinc finger motifs (CCHC) that specifically interact with the Psi (Figure 1b) [24,25]. This domain also contributes to Pr55Gag multimerization thus promoting viral assembly [26,27,28]. At the C-terminal end of Pr55Gag, the unstructured p6 domain of 6 kDa is required for specific binding to the gRNA [29], and is involved in the recruitment of the ESCRT (Endosomal Sorting Complex Required for Transport) machinery that regulate viral particle budding. Finally, Pr55Gag codes for two spacer peptides, sp1 and sp2 (also named p2 and p1, respectively), regulating the kinetics of Pr55Gag maturation.
The next sections of this revue will describe which PMTs are carried by the different Pr55Gag domains and what are their roles in the viral life cycle.
3. HIV-1 Pr55Gag Myristoylation
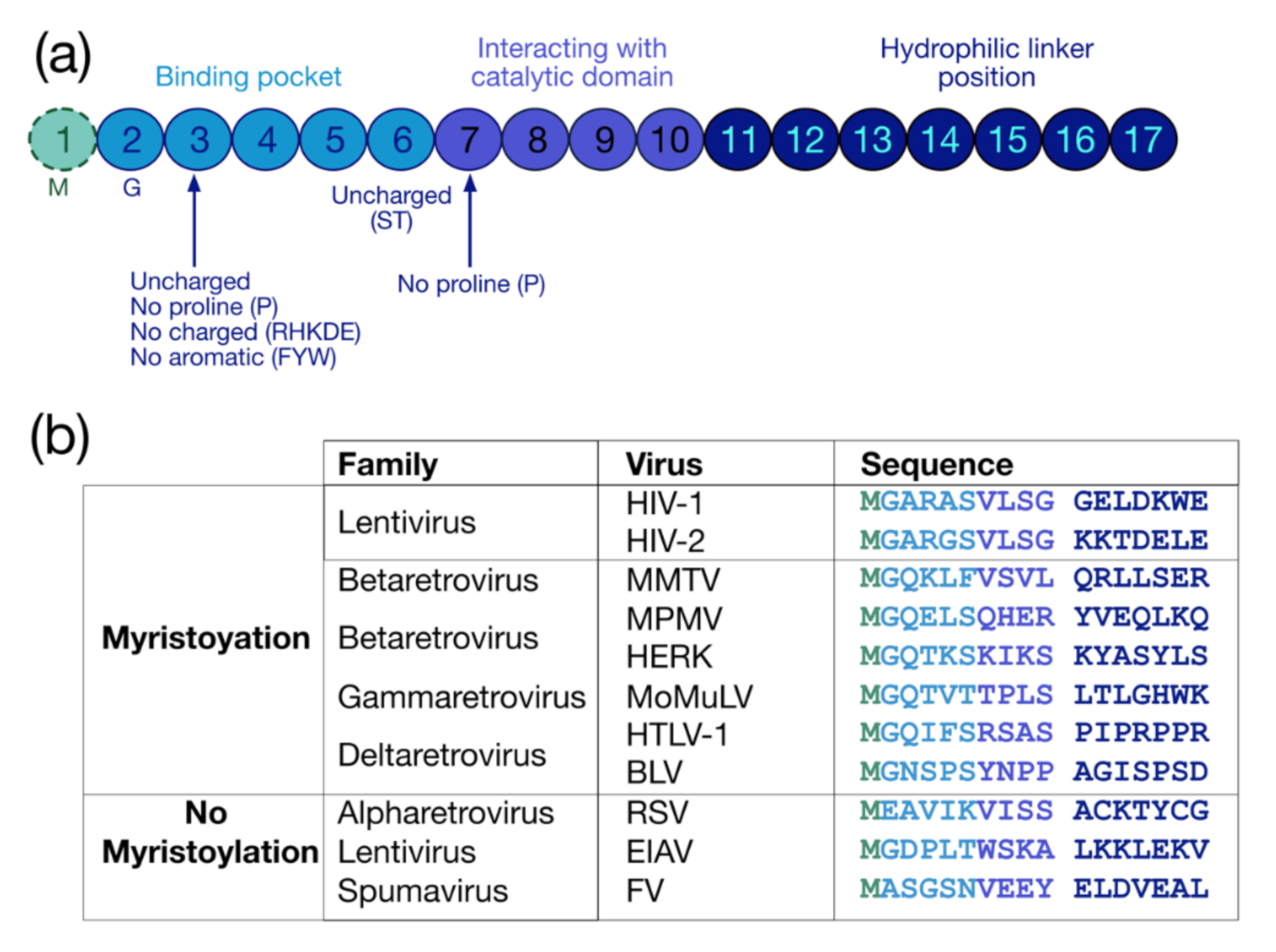
The myristoylation is a co-translational and irreversible modification consisting in the addition of a 14-carbon saturated fatty acid myristate to the protein via an amid bond by the N-myristoyl-transferase (NMT) (for reviews see [30,31,32]). The myristoylation can be achieved on an internal glycine (G) inside a consensus sequence recognized by NMTs, which is G-X2-X3-X4-(S/T/C)-X6 (Figure 2a). The G residue at the first position is necessary for this PMT, while at the second position there is preferentially an uncharged residue (except for proline (P)) or an aromatic amino acid. At the fifth position, uncharged residues are found, preferentially serine (S) and threonine (T) (for a review see [33]), while P is not accepted at the sixth position [34]. In sum, three regions finely regulate myristoylation: the binding pocket (positions from 1 to 6), the catalytic domain (positions from 7 to 10) and the hydrophilic linker (position from 11 to 17) [34,35] (Figure 2a).
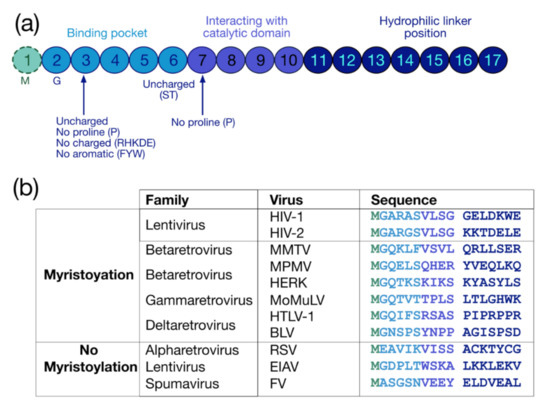
Figure 2.
Protein sequence required for myristoylation and sequences of retroviral myristoylated MA domains. (a) Pro-myristoylated consensus sequence underlying the three regions regulating myristoylation: the binding pocket (positions 1–6), the catalytic domain (positions 7–10) and the hydrophilic linker (positions 11–17) [34,35]. (b) Comparison of the first 17 residues of myristoylated MA domains in different retroviruses. Myristoylation is generally conserved in retroviruses such as lentivirus (HIV-1), betaretrovirus (Mason-Pfizer monkey virus (MPMV), mouse mammary tumor virus (MMTV), and human endogenous retrovirus type K (HERK)), gammaretrovirus (Moloney murine leukemia virus (MoMuLV) and murine leukemia virus (MLV)), and deltaretrovirus (human T-lymphotropic viruses (HTLV-1) and bovine leukemia virus (BLV)), but not in alpharetrovirus (Rous sarcoma virus (RSV)), some other lentivirus (equine infectious anemia virus (EIAV)), and in spumavirus (foamy virus (FV)).
Myristoylation is rather conserved in retroviruses (Figure 2b) (For reviews see [17,36] and [37,38]), and this PTM globally regulates the interaction of retroviral precursors with membranes and sub-membrane domains, such as lipid rafts. However, this modification is not sufficient by itself for membrane binding, and a distant polybasic domain is thus required to complete the optimal attachment of myristoylated proteins to the PM (for reviews see [39,40]). In HIV-1, this task is reached by the HBR spanning residues 17 to 31 of the MA domain, which contributes to a strengthening of the interaction with the PM thanks to electrostatic interactions with the negatively charged PI(4,5)P2 [41,42,43].
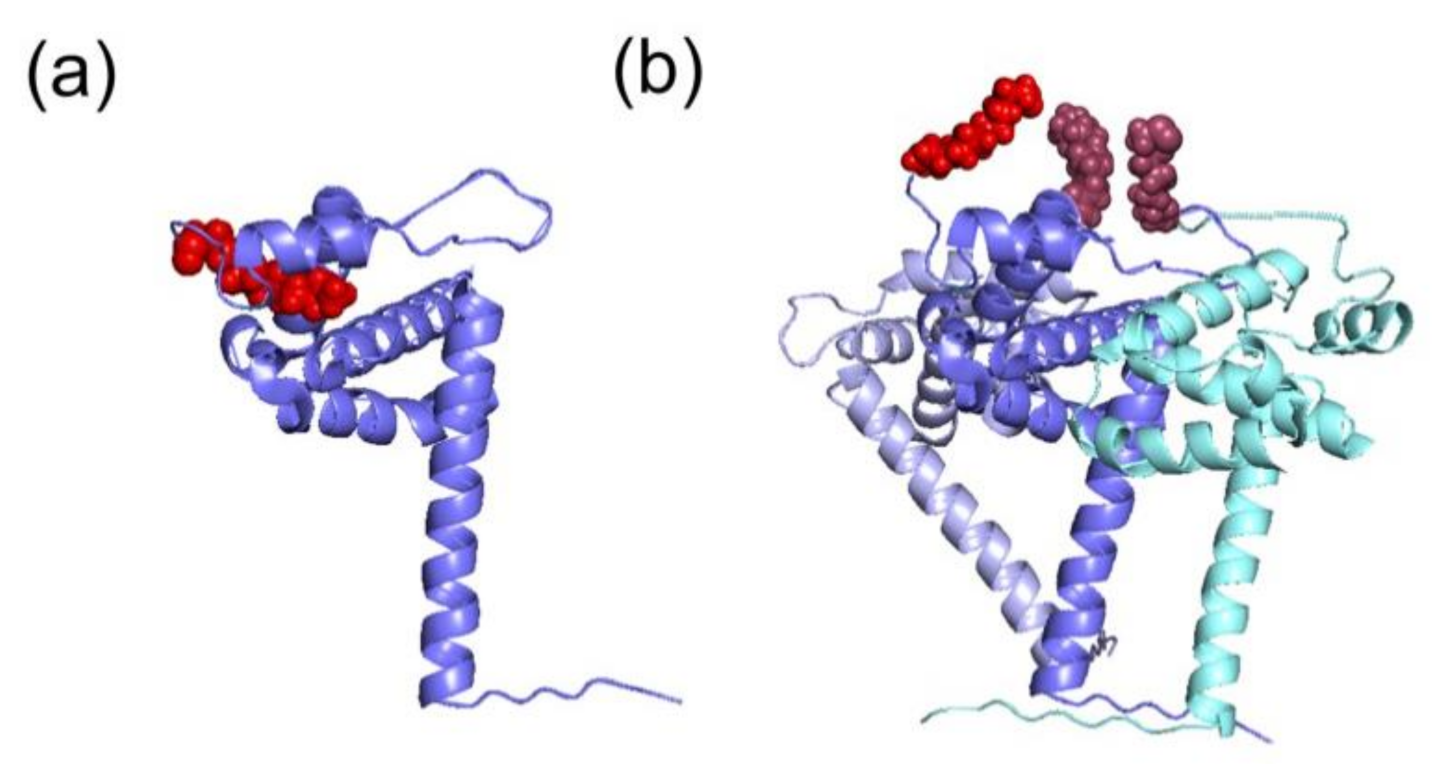
The myristoyl moiety can be exposed or sequestered in the hydrophobic pocket of the mature MA (Figure 3a,b) by the so-called myristoyl-conformational switch [37,39,42,44,45], which controls the exposure of myristoyl group for insertion into the PM, thus contributing to the prevention of aberrant interactions with intracellular membranes. The myristate exposure was found to be triggered by the interactions occurring between Pr55Gag and PI(4,5)P2 [41]. Besides, NMR studies demonstrated that myristate exposure is also regulated by the trimerization of the protein [37,42], and this would explain why the mature MA displays a lower affinity for membranes in comparison with the full-length precursor [37,41,42]. Indeed, several Pr55Gag domains, such as CA, p2 and NC, contribute to the self-association of the precursor and, as a consequence, to the myristate exposure (Figure 3b) [37]. Accordingly, mutational experiments on these domains inhibiting Pr55Gag multimerization, impair Pr55Gag binding to the membrane [37].
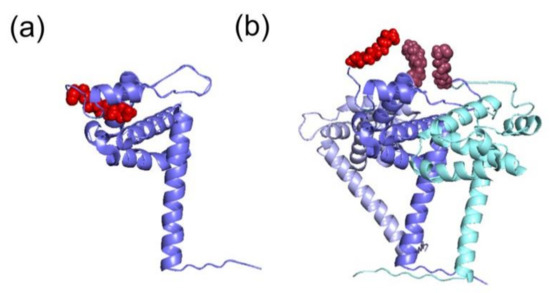
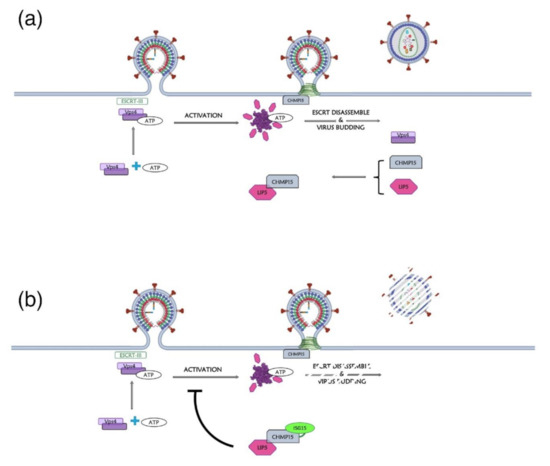
Figure 3.
Different structural conformations of HIV-1 MA monomer or trimer. The tertiary structures of the MA domain in the different conformations of the switch look similar. (a) The MA domain in its monomeric conformation (in blue) displays a sequestered myristoyl group (in red) (PDB: 1UPH [36]). (b) Representation of the trimer of MA (in blue, light blue and cyan) and the corresponding exposed myristoyl groups (in red). This model was proposed according to which the myristoyl group is exposed in the multimeric form, thus allowing its interaction with PM (adapted from [37]).
4. Gag Myristoylation in Other Retroviruses
The MA domains of retroviral Gag polyproteins display two main roles: they participate in genome incorporation, as several analyses recently pointed out, and they are implicated in membrane association. Interestingly, the majority of the retrovirus family displays a myristoylated MA domain (for reviews see [46,47,48,49]).
Among the different genera in which MA is myristoylated, the genus gammaretrovirus is composed by simple and oncogenic retroviruses. One representative virus of this family is MLV inducing leukemias or lymphomas in mice [50]. The MA domain of the primary form of MLV Gag, Pr65Gag, is myristoylated and contains a polybasic region in its globular domain that interacts electrostatically with PI(4,5)P2 at the PM, similarly to HIV-1 [47]. Besides, MLV has the particularity of encoding an additional form of Gag, gPr80Gag, which is glycosylated, but not myristoylated, and this last one is involved in the Pr65Gag trafficking to the PM [51]. However, beside MLV budding at PM, intracellular budding events can also occur into multivesicular bodies (MVBs) or in intracellular compartments as late endosomes in which virus-like particles (VLPs) accumulated [52]. Then, the deltaretrovirus genus contains complex and oncogenic retroviruses, and consists of two different groups, the primate T-lymphotropic viruses (PTLVs) including HTLV-1 and non-primate species, such as BLV [53,54,55]. Similarly, to MLV, the assembly of those retroviruses can occur at the PM, as well as in intracellular compartments such as late endosomes, MVBs or similar compartments [56]. The myristoylation of MA and the presence of basic amino acids leads to membrane binding and is, in this case, a PI(4,5)P2-independent process [57,58,59]. Indeed, the HTLV-I viral precursor Pr53Gag is able to bind membranes by electrostatic interactions involving the zwitterionic phosphatidylcholines (PC) and the negatively charged phosphatidylserines (PS) contained in endocytic membranes [57,60]. Moreover, a model was proposed in which the HBR in the HIV-1 MA domain would bind RNA to prevent premature or non-specific binding to cellular membranes [19,20,61]. Interestingly, a similar regulation between MA and RNA was proposed for BLV [62]. Conversely, the lack of this RNA regulation in HTLV-1 could explain the binding of myristoylated MA to the cellular membranes of intracellular compartments [57]. Finally, betaretroviruses show many similarities with lentiviruses, including a myristoylated MA domain [63]. This genus is composed of two groups: the first one is represented by MMTV [64] and by MPMV; and the second one is represented by HERK [65]. Myristoylated-deficient HERK Gag was observed to localize in the nucleus [66]. Contrary to other lentiviruses, NMR structures of MA domains of MPMV [63] and MMTV [67] show that the myristate group is hidden inside the MA in its oligomeric form. These differences suggest that betaretroviruses have developed different strategies to sequester the myristoyl group until the VLP is bound to the PM. At this site, a conformational change, leading to exposure of the myristate group would occur, similarly to other retroviral Mas that bind PM [67].
In contrast, some retroviral Gag precursors are not myristoylated. Indeed, alpharetroviruses represents simple and oncogenic retroviruses like RSV [68]. At the PM, RSV Pr76Gag interacts with charged lipids PI(4,5)P2 [38,69], and to ensure proper Pr76Gag-PM association, the lack of myristoylation is then counterbalanced by electrostatic interactions occurring between the inositol phosphates and a membrane binding domain (MBD), which is composed of basic residues forming a patch of clustered lysines (K) and arginines (R) on the MA surface ([49,68,70], for a review see [71]). Similarly, the MA domain of the lentivirus EIAV [72,73] is not myristoylated, but binds preferentially with phosphatidylinositol 3-phosphate (PI(3)P) with a higher affinity compared to PI(4,5)P2 [73,74]. Finally, foamy viruses (FV) as the PFV presents interesting differences compared to HIV-1 ([75], for a review see [76]). In particular, the FV Pr74Gag displays a limited number of PTMs compared to the other retroviruses, and strikingly, the FV MA domain contains neither the HBR nor a myristoylation modification. All those elements emphasize a different evolutionary history among retroviruses [75]. Indeed, in this case, viral Env proteins play a major role for viral budding, and the co-expression of Pr74Gag with Env is necessary for VLP production [77].
In sum, there are three main distinct strategies used by retroviruses to target membranes for budding. The first one requires the myristate exposure and a highly basic region (HBR) in the MA domain of retroviral precursors to interact with PM. The two others display dispensable myristoylation to achieve proper membrane binding since the hydrophobic interactions are in this case substituted by electrostatic ones produced by a basic domain in the MA, or alternatively by interactions between the precursor and viral elements such as Env proteins.
5. HIV-1 Pr55Gag Phosphorylation
Phosphorylation consists of the addition of a phosphate group to the side chain of amino acids. This PMT modifies the local electrostatic potential of proteins, induces conformational modifications, and affects the protein subcellular localization (for a review see [78], and [79,80]). Kinases, which are the enzymes that catalyze the transfer of phosphate group, have a role at multiple steps of HIV-1 viral, and the inhibition of cellular kinases interacting with HIV-1 at the nuclear level has been shown to affect the viral replication cycle [81]. Among HIV-1 viral proteins, which are phosphorylated, there is Pr55Gag (Table 1 and Figure 4). The MA domain is a substrate for the protein kinase C (PKC) [82], which catalyzes S and T phosphorylation. Several studies identified S111 in HIV-1 MA as the substrate for PKC [82]. Interestingly, substituting S111 with an alanine (A) led to decreased association of Pr55Gag with PM, even though MA was myristoylated. This suggests that PKC could also be involved in membrane binding by regulating the exposure of the myristoyl group [83,84].

Table 1.
Summary of different roles of phosphorylated residues in HIV-1 Pr55Gag.

Figure 4.
Phosphorylated residues in HIV-1 Pr55Gag. The different colors represent the Pr55Gag domains, MA (blue), CA (green), spacer peptides p1 and p2 (purple), NC (red), and p6 (black). Phosphorylation positions are highlighted in yellow. TP (in p6) and SP (in CA) motifs involved in the ERK2 recruitment and incorporation into viral particle are indicated in bold [91,93,94,95,96].
Alpha-screen assays allowed us to screen for human kinases interaction with the HIV-1 precursor, and the p6 domain resulted to be a target for PKC. In a further step, mass spectrometry indicated the phosphorylation of S488 residue [85,86]. Its substitution with a hydrophobic aromatic residue such as phenylalanine (F), which can occur spontaneously during anti-retroviral treatments, was found to perturb CA-SP1 processing, virus morphogenesis, maturation and virion infectivity [87,88,89]. On the other hand, the substitution of S488 by another non phosphorylable residue, such as asparagine (N), displayed no global impact on infectivity, thus suggesting that the production of non-mature viral particles would not be due to the lack of phosphorylation, but by the substitution itself [89]. Moreover, the phosphorylation of the p6 domain was found to be also important for the recruitment of the viral factor Vpr. As a consequence, the inhibition of PKC activity reduced Vpr level in virions, and this affected HIV-1 infectivity [85]. The p6 domain is the main phosphorylated domain in Pr55Gag and can be phosphorylated at several positions [86,90]. Indeed, phosphoamino acid analysis [90] and mass spectrometry experiments [86] identified several phosphorylated amino acids (Table 1 and Figure 4), that were found to globally promote viral budding [91]. Moreover, electron microscopy analysis revealed that mutation T471A leads to immature viral particles incompletely separated from PM, and immunoblotting analysis showed an incomplete Pr55Gag proteolytic maturation [91]. In contrast, other findings showed no effects on assembly or on viral release when T471 was substituted with isoleucine (I) or N. Since none of these amino acids can be phosphorylated, it is possible that the observed differences were not due to phosphorylation itself. Furthermore, except for T456 located in the PTAP late domain, the other eleven positions that can be phosphorylated in the p6 domain present redundancy. Mutagenesis experiments confirmed that the modifications of those residues seem to be dispensable for viral release and infectivity [86].
Experiments using an inhibitor of cyclin-dependent kinases [92] showed that also a MAP kinase, the extracellular-signal-regulated kinase 2 (ERK2), is involved in p6 phosphorylation, and this factor can be incorporated into viral particles by interacting with the S148-P149 motif in CA and T471-P472 in p6 [91,93,94,95,96] (Table 1, Figure 4). HIV-1 particles without active ERK2 were found to be poorly infectious due to a defect in reverse transcription [93,95]. Interestingly, ERK2 phosphorylates other viral proteins including Rev [97], Nef [98], Vif [99,100], and mature MA [95]. Besides, the substitution of four highly conserved and major phospho-acceptor S residues in the mature MA (Table 1) with A was found to impair viral replication [95,101].
Finally, the tyrosine kinase Src can also be incorporated into HIV-1 particle [102], and it is involved in the phosphorylation of the tyrosine (Y) 132 in a minority of mature MA proteins. This PMT was shown to play a role in the early phases of HIV-1 replication as the proviral DNA nuclear import [103] and its deletion causes the enhancement of MA accumulation in the cytoplasm at the expense of PM. On the contrary, Src overexpression was found to promote the localization of Pr55Gag at the PM [102].
In sum, HIV-1 Pr55Gag is phosphorylated by at least three kinases, PKC, ERK-2 and Src. Interestingly, mutation of phosphorylated residues in the p6 domain revealed that this domain, in addition to MA, can act as membrane targeting domain of Gag [104]. However, phosphorylation positions in p6 mainly display redundancy, thus hindering the evaluation of the impact of each individual phosphorylated residue.
6. Gag Phosphorylation in Other Retroviruses
Phosphorylation is a conserved modification in the retroviral family (Table 2). In alpharetroviruses, within the RSV MA domain, a small proportion of Y residues results in being phosphorylated [105], as well as S68 and S106 residues (Table 2). However, S68 seems to be transitionally phosphorylated, while S106 is the main phosphorylated signal [106]. Besides, MA phosphorylation could be involved in the recruitment of factors promoting NC phosphorylation [106,107]. In turn, phosphorylation of S529 in NC was found to be necessary for the specific interaction with gRNA [108], but no other notable effects on assembly, or on infectivity, were observed [106].

Table 2.
Summary of phosphorylated positions in the different domains of retroviral Gag precursors.
The deltaretroviruses, HTLV-1 MA is also a phosphoprotein, and S105, which is located close to the two late domains involved in viral release [109], PPPY [110] and PTAP [111], is phosphorylated by ERK-2. Similarly, to HIV-1, ERK-2 is incorporated into HTLV-1 particles, and phosphorylation of the MA domain was found to be involved in virus release and budding efficiency [110].
Interestingly, betaretroviruses such as MPMV encode a phosphoprotein pp24 within the Gag precursor, and its C-terminal cleavage produces the protein pp18 which contains proline-rich motifs (PPPY). Deletion assays indicated that the phosphorylated residue Y205 in pp18 is dispensable for capsid assembly, but is necessary for the viral release [112]. Immunoprecipitation experiments identified the presence of phosphoserines in pp18, [113,114] displaying a redundant character. Similarly, for spumaviruses such as FV, mapping of the p4 domain revealed that seven residues can be phosphorylated (Table 2), but a single substitution of those residues displayed no influence on viral replication [115]. In gammaretrovirus, the phosphorylation of the RNA binding phosphoprotein (p12) within the Gag precursor was found to be necessary for early events of viral life cycle and virion production [116,117]. Mutagenesis experiments identified two residues which can be phosphorylated (S192 and S209). In particular, S192 mainly contributes to p12 phosphorylation and its substitution by A impairs viral assembly and infectivity. However residual phosphorylation can also occur at other positions (Table 2) [117], thus suggesting a redundant character of these modified amino acids. Indeed, the single substitution of one of these residues induced different levels of phosphorylation in p12, displaying no overall effect on the viral cycle [117], even though these PTMs were proposed to modulate p12 early and late functions and p12 viral RNA-binding activity [117,118].
In conclusion, similarly to HIV-1, the kinases PKC and ERK-2 are the main drivers of retroviral Gag phosphorylation. Interestingly, ERK-2 can be incorporated into the viral particle of HTLV-1. Globally, these PTMs generally seem to play a role in viral particle release and in virus infectivity, even though the impact of the phosphorylation rate in retroviral proteins is complicated by the redundancy of phosphorylated positions.
7. HIV-1 Pr55Gag Ubiquitination
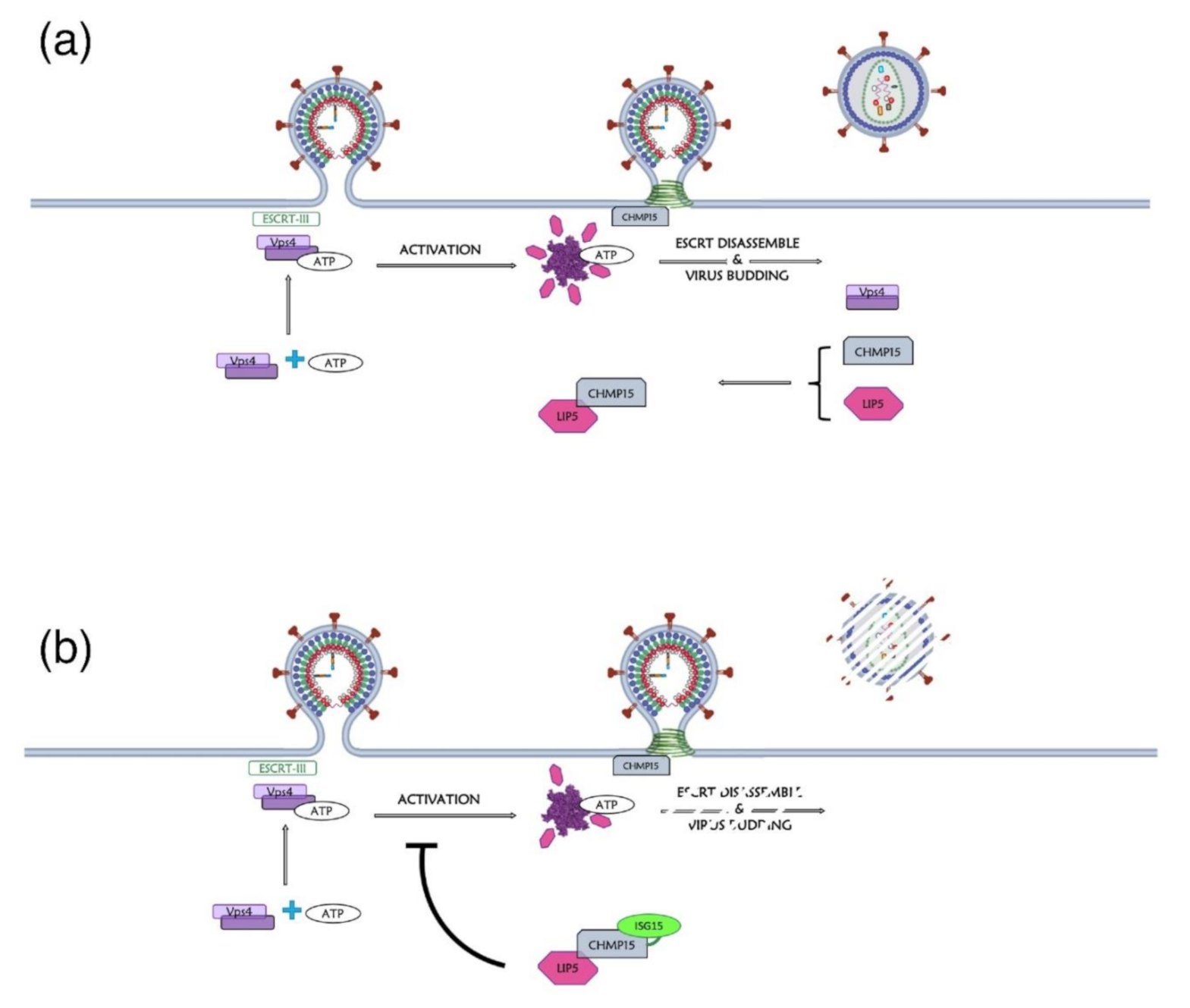
Another crucial PTM for retroviral infectivity is ubiquitination. This PMT consists of intracellular protein modification by adding one or more ubiquitin (Ub) protein(s) (for a review see [119]). Ub is a 76-amino acid polypeptide, which has a conserved structure [120]. The Ub sequence contains seven K residues that can be used for subsequent Ub linkage leading to polyubiquitination (for a review see [121]), even if the two most common polyubiquitination chains consist in the formation of Ub chain connected to residues K48 or K63 of Ub. Monoubiquitylation corresponds to a signal for DNA repair, and vesicle sorting or signal transduction, while polyubiquitinated proteins are often targeted to the 26S proteasome for degradation, or alternatively involved in regulation of the endocytosis of ESCRT-dependent cargo proteins into Multi Vesicular Bodies (MVB) (for a review see [122]) and DNA damage response [123]. Ubiquitination can be reversed by deubiquitinating enzymes (DUB) [124].
HIV-1 Pr55Gag is ubiquitinated in its domains at different levels (Table 3 and Figure 5). Indeed, MA, CA, and NC are monoubiquitinated, while p2 can be mono or bi-ubiquitinated [125]. The cumulative mutations of ubiquitin acceptor sites were observed to cause generally budding defects, even if the substitution of K residues by R in CA (Table 3) revealed very limited effect on viral release, showing that these ubiquitination sites are likely redundant [126]. Besides, it was observed that the level of Pr55Gag ubiquitination increases in cellula when a full-length HIV-1 molecular clone is expressed in comparison to a Pr55Gag expression plasmid, suggesting a role of other viral proteins in Pr55Gag ubiquitination [125]. Globally, the ubiquitination of Pr55Gag was found to be involved in the viral release and, during HIV-1 assembly, viral particles incorporate free Ub proteins corresponding to about 10% of the Pr55Gag level, and around 2–5% of ubiquitinated Pr55Gag are mono-ubiquitinated [125,127,128,129,130]. When the level of free Ub in cells is reduced by proteasomal inhibition, the number of free Ub in viral particles and the number of mono-ubiquitinated residues in the p6 domain of Pr55Gag also decreased [125,127,131]. However free Ub incorporation into viral particles seems to be independent from the global Pr55Gag ubiquitination state [132], and the ubiquitination level in virions increased upon overexpression of free Ub [133]. Furthermore, ubiquitination seems to take place at the PM, and interestingly the level of Pr55Gag mono-ubiquitination was found to be directly correlated with ability of the precursor to bind the PM [134].

Table 3.
Summary of ubiquitinations in HIV-1 Pr55Gag proteins.

Figure 5.
Ubiquitinylated residues in HIV-1 Pr55Gag. The domains of Pr55Gag are represented by different colors (see Figure 4). Experimentally identified ubiquitinylated positions are highlighted in light green. Potential ubiquitinylated positions are highlighted in light blue.
The C-terminal p6 domain is the most ubiquitinated domain in Pr55Gag [125], and K475 and K481 are the major targets. Even if these mono-ubiquitinated residues are neither directly involved in virus release, nor in infectivity, they were found to be necessary to promote the overall ubiquitination of Pr55Gag [132]. Besides, the mutation of the highly conserved and phosphorylated S488 residue in p6 domain with F (S488F), which can occur spontaneously during anti-retroviral treatments, has not only an impact on virus morphogenesis, maturation and virion infectivity (Table 1) [87,88,89], but it can also induce conformal changes in p6, resulting in an enhanced interaction of Pr55Gag with the PM. This would lead to the polyubiquitination of the precursor and consequently to its proteasomal degradation [104].
The p6 domain is known to be involved in the recruitment of host factors, such as Tsg101 (Tumor susceptibility gene 101) and ALIX (ALG-2 interacting protein X), and ubiquitination of those factors strongly promote viral budding [135]. In this frame, fusion experiments in which the p6 domain was coupled with Ub showed that the affinity of Tsg101 for p6 in this case results in being strengthened [136], and the ubiquitination of Pr55Gag can increase Tsg101 recruitment [137]. Besides, Tsg101 displays an N-terminal Ub E2 variant (UEV) domain that shows homology with E2 Ub ligases, and that can specifically bind Ub proteins, as well the PTAP late domain in Pr55Gag [136]. During assembly, the interaction of Pr55Gag with the PM promotes the intermolecular interaction between Tsg101 and the PTAP domain in Pr55Gag [137]. In this conformation, the di-ubiquitinylated K63 of Tsg101 was found to interact with p6, with the consequence of impairing the potential polyubiquitination of the precursor at PM [137].
Finally, the ESCRT-III-associated ALIX protein is also ubiquitinated [128] and specifically interacts with the E3 ubiquitin-protein ligase NEDD4 that can bind the proline-rich retroviral domain PPPY. The interaction between NEDD4 and the retroviral precursor leads to the recruitment of the ESCRT-III complex, including the eukaryotic sucrose non-fermenting protein 7 (Snf7), and the vacuolar protein sorting-associated proteins Vps 2, Vps20 and Vps24, and Vps4 in order to promote retroviral release [138,139,140,141]. Since in HIV-1 the PPPY domain is absent, ALIX recruits directly NEDD4 to facilitate this step [128,129].
8. Gag Ubiquitination in Other Retroviruses
The role of ubiquitination in the retroviral cycle is not yet fully elucidated. Some retroviruses display a functional contribution of Ub modifications in virus release such as MLV, MPMV or RSV, and for those viruses, it was shown that, similarly to what observed for HIV-1 [131], the inhibition of proteasome not only induces a reduction of the level of free Ubs in the cytoplasm, but also impairs the release of the viral particles (Table 4) [127,132,133,142]. In addition, fusion experiments between RSV Pr76Gag and Ub, or overexpression of Ub, displayed an increase in viral particle release [142], thus supporting the idea that ubiquitination of retroviral precursors is crucial for viral budding [133]. However, for other retroviruses such as MMTV or HTLV-1, to date it was not possible to identify a precise role of ubiquitination [127] (Table 4). Besides, the inhibition of the proteasome did not impair the budding of EIAV [127,143]. On the other hand, similarly to HIV-1, EIAV particles contain free Ubs corresponding to 10–15% of Gag proteins. Likewise, the C-terminal p9 domain is mono-ubiquitinated and contains a YPDL late domain which is involved in the recruitment of the ESCRT machinery [143,144]. Moreover, p9 also contains an Ub-like motif (NVKEKD) that may contribute to virus release, thus suggesting alternative release pathways for EIAV even if Ub quantity is low [133,144].

Table 4.
Summary of ubiquitinations in the different domains of retroviral Gag proteins.
MMTV Gag is monoubiquitinated in its p8 domain, in CA, and is potentially di-ubiquitinated in NC [127]. In comparison with other retroviruses, MMTV does not contain late domains such as PPPY and PTAP, but an alternative PSAP late domain was found in CA, although its functional role was not yet elucidated. Besides, YXXL motifs, which also represent alternative late domains, were identified in MA and in pp21 viral factor. Importantly, since Gag ubiquitination seem to take place mostly in regions close to the late domains [133], it is possible that the presence of these alternative late domains in EIAV and MMTV precursors promote virus release.
In HLTV-1, more than 40% of MA are ubiquitinated [111,145,146], and MA can be mono- and di-ubiquitinated [146]. Furthermore, mutagenesis experiments identified K74 in Pr53Gag as the main substrate for ubiquitination [146]. Indeed, substitution experiments in which the K74 is replaced by an R resulted in a decreased release of infectious particles [146]. The ubiquitination of K74 could also play a role in the recruitment of NEDD4 [146], which is also involved, through its interaction with the PPPY late domains, in the release of other retroviruses such as MMPV [112], avian sarcoma virus (ASV) [147], and MLV [132,148].
In RSV particles, more than 100 free Ubs were found [142,149]. However, contrary to other retroviruses, RSV displays free Ubs exclusively into mature viruses [149]. Since Pr76Gag mono-ubiquitination was found to be necessary for budding and to recruit the ESCRT machinery [142,149], it is thus possible that the presence of free Ubs could be the result of a host-encoded and encapsidated deubiquitinating enzyme (DUB) [124]. Interestingly, this process was also observed to occur during budding or cells lysis [124]. Finally, Gag precursors from spumavirus encode a very limited number of K residue. This observation suggests that Gag of spumavirus could not be a favorable substrate for the ubiquitination machinery [150].
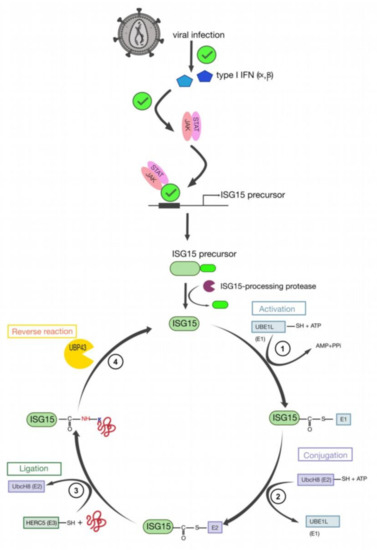
9. HIV-1 Pr55Gag Sumoylation
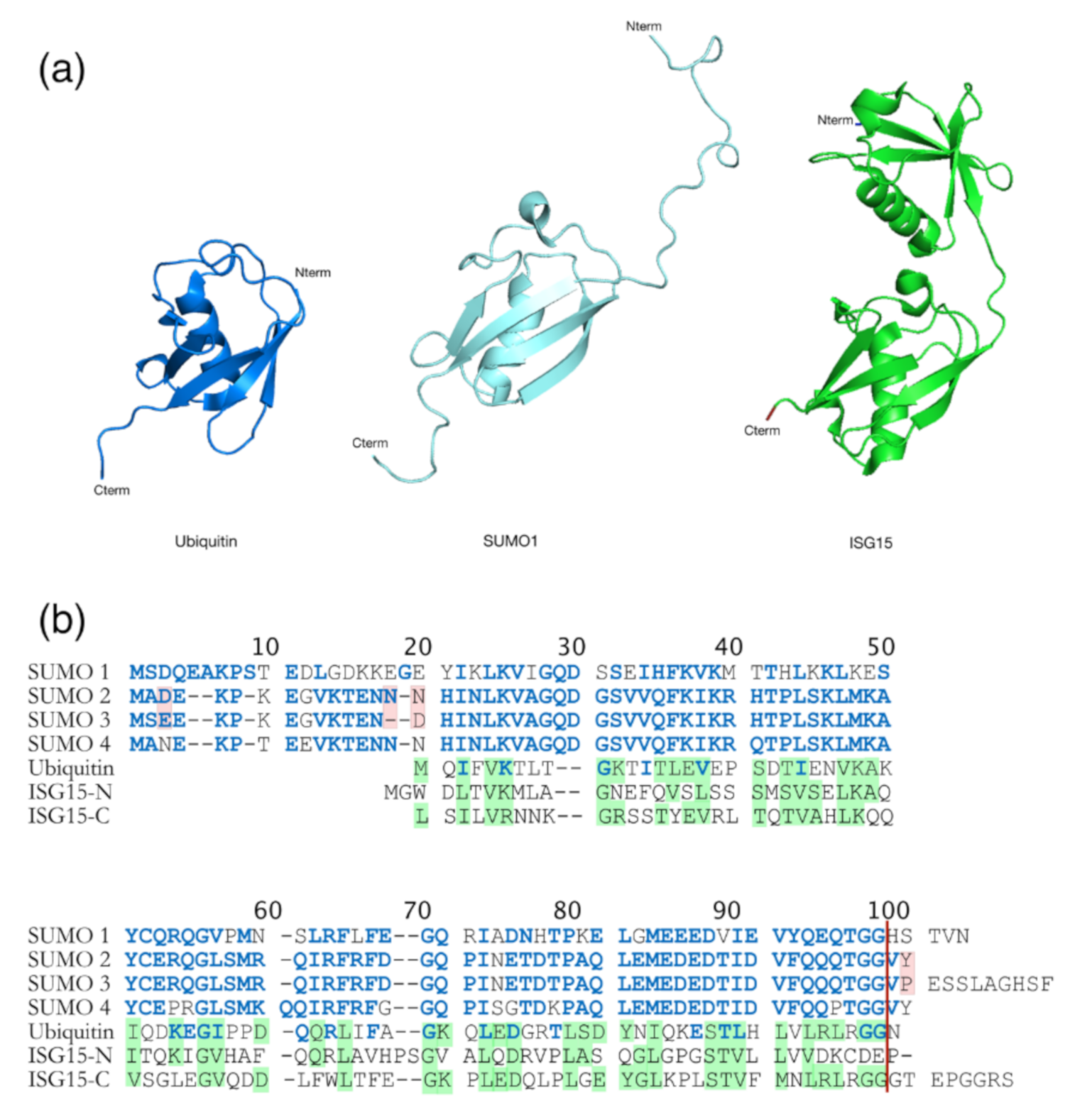
Another modification important for retroviral infectivity is sumoylation, which is a reversible PTM and consists of intracellular protein modification by a covalently attached small Ub related modifier (SUMO) protein to a K substrate (for reviews see [151,152]). Even though SUMO is structurally comparable to Ub (Figure 6a), it presents many differences in amino acids sequence (only 18% of homology) (Figure 6b) [152]. This PTM is usually involved in the maintenance of genomic integrity, with a role in repair of damaged DNA, and in the regulation of transcription and in gene expression. Like ubiquitination, sumoylation is involved in intracellular signal transduction and can regulate biological processes such as apoptosis, immune response, and carcinogenesis. Besides, sumoylation controls protein localization and it can induce protein conformational changes. SUMOs are highly conserved in eukaryotes, and four SUMO isoforms (SUMO-1 to SUMO-4) are present in mammals [152,153,154] (Figure 6b). Similarly, to Ub, the C-terminus region of SUMO-1 is linked to ε-amino groups of K residues in the target protein [155,156]. SUMO-1 was interestingly found to counterbalance the effect of ubiquitination [157]. SUMO-2 and SUMO-3 are mainly involved in the cellular response to environmental stresses [156] and display very similar sequences with more than 95% identity [151,152,156]. For this reason, they are often named SUMO-2/3. Finally, SUMO-4 is less well known, and its mRNA had been found in few organs such as kidney, spleen, and lymph nodes [152].
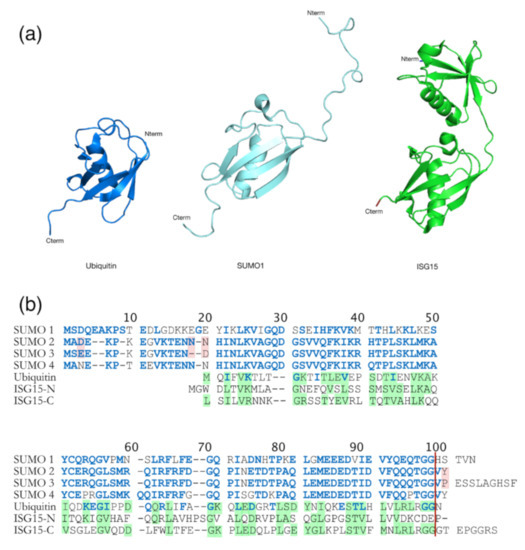
Figure 6.
Comparison between Ub and Ub-like proteins: SUMO and ISG15. (a) Structural comparison between Ub (heavy blue, PDB: 1A5R), SUMO-1 (ligth blue, PDB: 2QHO), and ISG15 (green, PDB: 3PHX). They contain a typical ββαββαβ fold, even if SUMO-1 has long unstructured N-terminal domain which is absent in Ub. ISG15 is composed with two Ub-like domains in N- (TSG15N) and C- (TSG15C) terminus. (b) Amino acid sequence alignments of Ub, the four SUMO homologs and ISG15 from humans. Identities and similarities are indicated between Ub and SUMO (blue residues into Ub sequence) and between Ub and ISG15 (shaded green residues in Ub and ISG15). Differences between SUMO-2 and 3 are highlighted in pink. The red vertical line represents the GG end free after the maturation step required for sumoylation. The amino acid sequence homology between SUMO and Ub is 18% [152], and 30% between Ub and ISG15 [161].
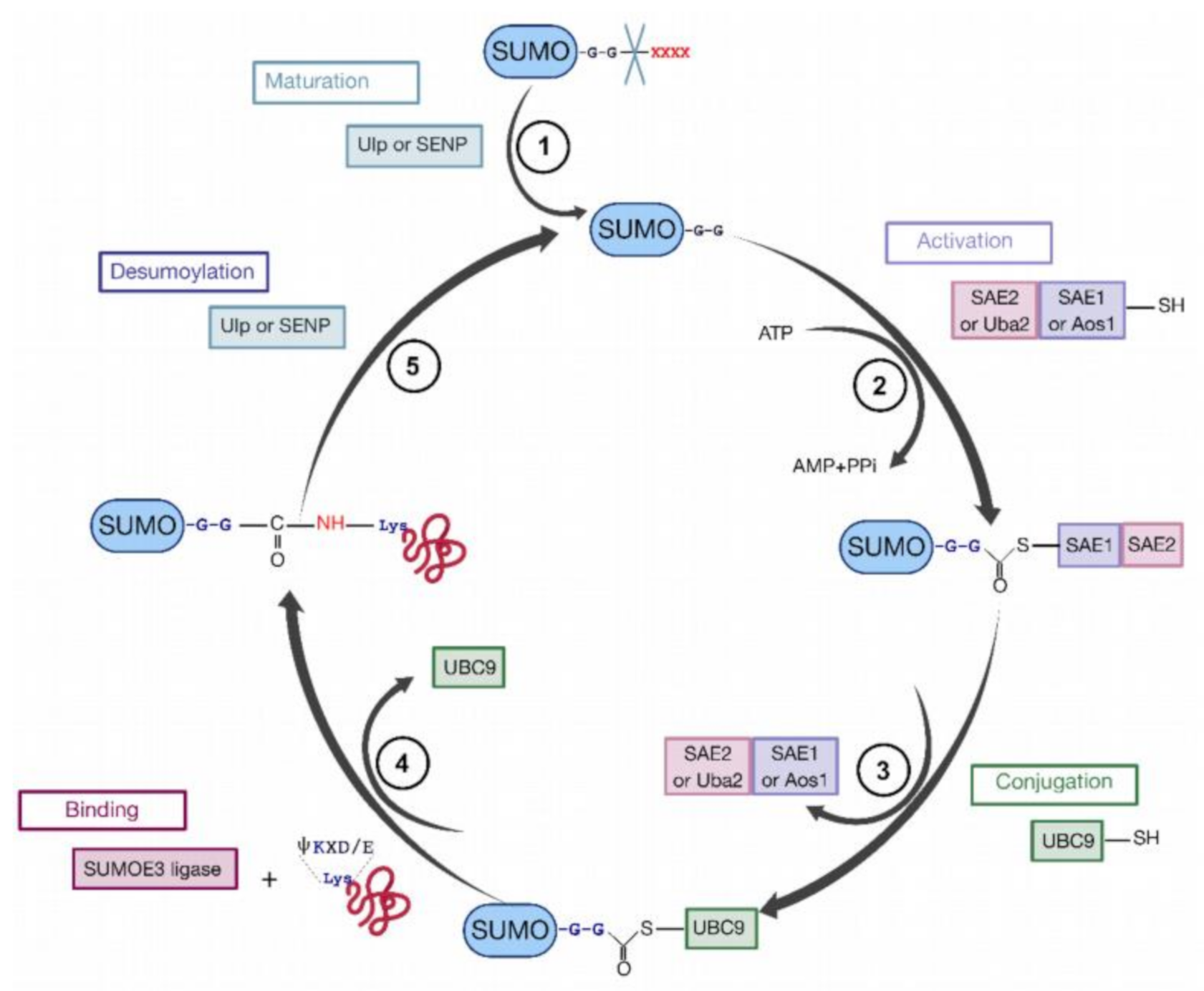
To sumoylate a protein, different successive biochemical reactions are required [152,158,159,160] (Figure 7). Generally, the consensus sequence for K sumoylation is ψKXD/E (ψ stands for a hydrophobic residue). Nevertheless, targets with non-consensus acceptor sites have also been identified [151,152].
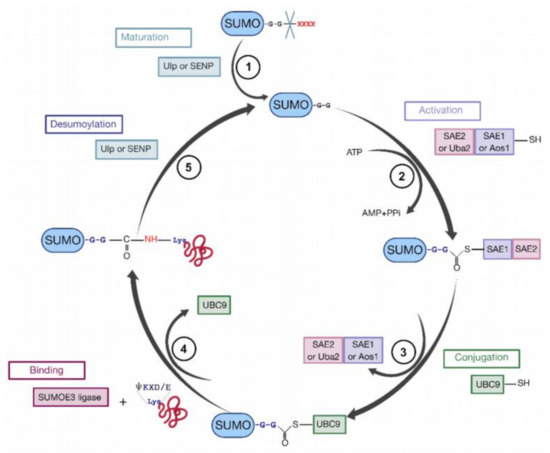
Figure 7.
The cycle of sumoylation. This modification is catalyzed by different enzymes and consists in ligation of SUMO protein to K residues of protein substrates. (1) SUMO is maturated by Ub-like specific protease 1 (Ulp1) or human sentrin-specific protease 1 (SENP1). This proteolytic cleavage exposes the C-terminal GG motif required for the activation step. (2) SUMO is activated by a heterodimer composed with SAE1/SAE2 (Aos1/Uba2) to form the SUMO (E1/E2)-activating enzyme. Heterodimer is bound via a thioester bond between the C-terminal G residue of SUMO and the catalytic C of SAE2. (3) SUMO is transferred to the catalytic C of SUMO-conjugating enzyme E2 (or Ubc9) by a transesterification reaction. (4) SUMO is bound to the target protein by Ubc9 in association with SUMO E3 ligase. Ubc9 forms an amide bond between the SUMO C-terminus and ε-amino groups of the acceptor L residues in the target protein. (5) These reactions are reversible by means of the Ulp or SENP proteases.
The p6 domain of HIV-1 Pr55Gag is sumoylated by SUMO-1, which covalently links K475 in the consensus sequence (ψKXE: QKQE) (Table 5). The K475R substitution partially inhibits binding of the precursor to the SUMO-conjugating enzyme E2 (Ubc9) [162], suggesting that more than one Pr55Gag domain could be involved in the recruitment of Ubc9 [162,163]. It was proposed that SUMO-Ubc9 could be involved in intracellular trafficking of Pr55Gag [164]. Indeed, after translation, the first trafficking complex intermediate observed in the perinuclear region is composed of Pr55Gag, kinesin family motor 4 (KIF4), Ubc9, and SUMO-1 [164]. In contrast, other studies suggested that the recruitment of Ubc9 would be required for the late stages of viral replication, thus participating to Env incorporation into viral particles [163]. Moreover, the overexpression of SUMO-1 was observed to globally decrease viral infectivity, and sumoylation could be then involved in the negative regulation of viral replication [162]. Interestingly, sumoylation and ubiquitination co-regulate each other [165], and sumoylation and mono-ubiquitination of p6 were both found to occur on K475. It is thus possible that SUMO-1 interaction with p6 protects Pr55Gag from proteasomal degradation [162]. Overproduction of SUMO-1 should have no direct effect on viral assembly, but if sumoylation competes with ubiquitination, subsequent decrease of Tsg101 recruitment could produce a negative effect on budding [162].

Table 5.
Summary of sumoylated positions in retroviral Gag proteins.
10. Gag Sumoylation in Other Retroviruses
As for HIV-1, other retroviruses are sumoylated (Table 5); however, the impact of sumoylation is not yet fully elucidated. The Ubc9 factor was found to interact with MLV and MPMV Gag proteins [166,167]. Similarly, to HIV-1, in MPMV this factor was suggested to be involved in the trafficking of Pr78Gag to the PM [167]. Besides, the CA domain of MoMuLV Gag was shown to interact not only with Ubc9 [168], but also with PIASy, a SUMO E3 ligase [168]. These interactions, leading to CA sumoylation during the early stages of the viral life cycle after reverse transcription, might have a role in viral replication [168]. Single K substitutions have generally no effect on the viral cycle, suggesting redundancy between sumoylable positions. On the other hand, the modification of K218 with an R residue was found to reduce the overall viral replication, without affecting the overall SUMO-1 rate on Gag [168].
In the EIAV p9 domain of Gag, K465 is the main target for sumoylation, and mutational experiments showed that this PTM is involved in the regulation of viral replication and infectivity [144,169]. Moreover, sumoylation of K465 seems to regulate the sumoylation of other K residues in different domains of the precursor, such as the MA, the CA and NC (Table 5) [169]. However, a specific role of all those PTMs in viral replication remains to be clarified. Finally, K244 in CA of RSV Pr76Gag was found to be sumoylated, and its substitution with a non-sumoylable R residue (K244R) displayed decreased viral infectivity [142].
Similarly, to ubiquitination, the exact role of sumoylation is still a matter of debate and might be different among retroviruses. Moreover, sumoylation is still very difficult to detect, and thus further technological advances will be required to better identify and characterize this PTM.
13. Conclusions
PTMs create a vast diversity in proteins and thus regulate their functions. Globally, PTMs play a role in many processes such as cell signaling, and protein–protein and protein–RNA interactions. Besides, PTMs are crucial for the life cycle of many viruses and the characterization of viral PTMs would provide a better understanding of the mechanisms of viral processes. HIV-1 Pr55Gag, as many other retroviral Gag precursors, displays several PTMs in its different domains (Figure 10). These PTMs include myristoylation, phosphorylation, ubiquitination, sumoylation, and methylation. All these PTMs can have either antagonistic or cooperative roles, thus allowing fine regulation of the viral cycle. However, up to now, the role of many of these modifications is not fully elucidated and further investigations will be required to better understand their contributions in the viral life cycle. One of the main challenges to study PTMs carried by proteins consists of the development of refined proteomic technologies, allowing the specific detection and characterization of the modifications. The improved knowledge of those regulations would be useful in the future to identify new targets for antiretroviral treatments.

Figure 10.
Summary of post-translational modifications of HIV-1 Pr55Gag residues. The domains of Pr55Gag are represented by different colors (see Figure 4). Experimentally identified modified residues are highlighted: myristoylation (pink), phosphorylation (yellow), ubiquitination (light blue), potential ubiquitinations (light green), and sumoylation (black).
Author Contributions
S.B. supervised the project; C.B. wrote the manuscript with contributions from S.B., J.-C.P. and R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Agence Nationale de Recherche sur le SIDA et les hépatites virales (ANRS), SIDACTION and IdEx (Initiative d’Excellence, Université de Strasbourg, France). CB is supported by a fellowship from the French ministry of Research and Higher Education.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
No applicable.
Data Availability Statement
All figures and discussed literature are available in the main text of this review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Audagnotto, M.; Peraro, M.D. Protein post-translational modifications: In silico prediction tools and molecular modeling. Comput. Struct. Biotechnol. J. 2017, 15, 307–319. [Google Scholar] [CrossRef]
- Spoel, S.H. Orchestrating the proteome with post-translational modifications. J. Exp. Bot. 2018, 69, 4499–4503. [Google Scholar] [CrossRef]
- Biard-Piechaczyk, M.; Borel, S.; Espert, L.; De Bettignies, G.; Coux, O. HIV-1, ubiquitin and ubiquitin-like proteins: The dialectic interactions of a virus with a sophisticated network of post-translational modifications. Biol. Cell 2012, 104, 165–187. [Google Scholar] [CrossRef]
- Chen, L.; Keppler, O.T.; Schölz, C. Post-translational Modification-Based Regulation of HIV Replication. Front. Microbiol. 2018, 9, 2131. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR Detection of Structures in the HIV-15′-Leader RNA That Regulate Genome Packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, A.K.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-Throughput SHAPE Analysis Reveals Structures in HIV-1 Genomic RNA Strongly Conserved across Distinct Biological States. PLoS Biol. 2008, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- El-Wahab, E.W.A.; Smyth, R.P.; Mailler, E.; Bernacchi, S.; Vivet-Boudou, V.; Hijnen, M.; Jossinet, F.; Mak, J.; Paillart, J.-C.; Marquet, R. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat. Commun. 2014, 5, 4304. [Google Scholar] [CrossRef]
- Smyth, R.P.; Despons, L.; Huili, G.; Bernacchi, S.; Hijnen, M.; Mak, J.; Jossinet, F.; Weixi, L.; Paillart, J.-C.; Von Kleist, M.; et al. Mutational interference mapping experiment (MIME) for studying RNA structure and function. Nat. Methods 2015, 12, 866–872. [Google Scholar] [CrossRef]
- Bernacchi, S.; El-Wahab, E.W.A.; Dubois, N.; Hijnen, M.; Smyth, R.P.; Mak, J.; Marquet, R.; Paillart, J.-C. HIV-1 Pr55Gag binds genomic and spliced RNAs with different affinity and stoichiometry. RNA Biol. 2017, 14, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Clerté, C.; Chamontin, C.; Basyuk, E.; Lainé, S.; Hottin, J.; Bertrand, E.; Margeat, E.; Mougel, M. Imaging HIV-1 RNA dimerization in cells by multicolor super-resolution and fluctuation microscopies. Nucleic Acids Res. 2016, 44, 7922–7934. [Google Scholar] [CrossRef]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA 2009, 106, 19114–19119. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Bieniasz, P.D. Analysis of the Initiating Events in HIV-1 Particle Assembly and Genome Packaging. PLoS Pathog. 2010, 6, e1001200. [Google Scholar] [CrossRef]
- Mailler, E.; Bernacchi, S.; Marquet, R.; Paillart, J.-C.; Vivet-Boudou, V.; Smyth, R.P. The Life-Cycle of the HIV-1 Gag–RNA Complex. Viruses 2016, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Bieniasz, P.; Telesnitsky, A. Multiple, Switchable Protein:RNA Interactions Regulate Human Immunodeficiency Virus Type 1 Assembly. Annu. Rev. Virol. 2018, 5, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Comas-Garcia, M.; Davis, S.R.; Rein, A. On the Selective Packaging of Genomic RNA by HIV-1. Viruses 2016, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.M.; Lever, A.M. HIV Gag polyprotein: Processing and early viral particle assembly. Trends Microbiol. 2013, 21, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Olety, B.; Ono, A. Roles played by acidic lipids in HIV-1 Gag membrane binding. Virus Res. 2014, 193, 108–115. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global Changes in the RNA Binding Specificity of HIV-1 Gag Regulate Virion Genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Gaines, C.R.; Tkacik, E.; Rivera-Oven, A.; Somani, P.; Achimovich, A.; Alabi, T.; Zhu, A.; Getachew, N.; Yang, A.L.; McDonough, M.; et al. HIV-1 Matrix Protein Interactions with tRNA: Implications for Membrane Targeting. J. Mol. Biol. 2018, 430, 2113–2127. [Google Scholar] [CrossRef]
- Thornhill, D.; Olety, B.; Ono, A. Relationships between MA-RNA Binding in Cells and Suppression of HIV-1 Gag Mislocalization to Intracellular Membranes. J. Virol. 2019, 93, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nat. Cell Biol. 2013, 497, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.M.; Hagen, W.J.H.; Rumlová, M.; Ruml, T.; Müller, B.; Kräusslich, H.-G.; Briggs, J.A.G. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nat. Cell Biol. 2015, 517, 505–508. [Google Scholar] [CrossRef]
- Pornillos, O.; Ganser-Pornillos, B.K.; Kelly, B.N.; Hua, Y.; Whitby, F.G.; Stout, C.D.; Sundquist, W.I.; Hill, C.P.; Yeager, M. X-Ray Structures of the Hexameric Building Block of the HIV Capsid. Cell 2009, 137, 1282–1292. [Google Scholar] [CrossRef]
- Dannull, J.; Surovoy, A.; Jung, G.; Moelling, K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994, 13, 1525–1533. [Google Scholar] [CrossRef]
- Webb, J.A.; Jones, C.P.; Parent, L.J.; Rouzina, I.; Musier-Forsyth, K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: Implications for viral genomic RNA packaging. RNA 2013, 19, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Cimarelli, A.; Sandin, S.; Höglund, S.; Luban, J. Basic Residues in Human Immunodeficiency Virus Type 1 Nucleocapsid Promote Virion Assembly via Interaction with RNA. J. Virol. 2000, 74, 3046–3057. [Google Scholar] [CrossRef] [PubMed]
- Ott, D.E.; Coren, L.V.; Shatzer, T. The Nucleocapsid Region of Human Immunodeficiency Virus Type 1 Gag Assists in the Coordination of Assembly and Gag Processing: Role for RNA-Gag Binding in the Early Stages of Assembly. J. Virol. 2009, 83, 7718–7727. [Google Scholar] [CrossRef]
- El Meshri, S.E.; Dujardin, D.; Godet, J.; Richert, L.; Boudier, C.; Darlix, J.L.; Didier, P.; Mély, Y.; De Rocquigny, H. Role of the Nucleocapsid Domain in HIV-1 Gag Oligomerization and Trafficking to the Plasma Membrane: A Fluorescence Lifetime Imaging Microscopy Investigation. J. Mol. Biol. 2015, 427, 1480–1494. [Google Scholar] [CrossRef]
- Dubois, N.; Khoo, K.K.; Ghossein, S.; Seissler, T.; Wolff, P.; McKinstry, W.J.; Mak, J.; Paillart, J.-C.; Marquet, R.; Bernacchi, S. The C-terminal p6 domain of the HIV-1 Pr55Gag precursor is required for specific binding to the genomic RNA. RNA Biol. 2018, 15, 923–936. [Google Scholar] [CrossRef]
- Farazi, T.A.; Waksman, G.; Gordon, J.I. The Biology and Enzymology of ProteinN-Myristoylation. J. Biol. Chem. 2001, 276, 39501–39504. [Google Scholar] [CrossRef]
- Martin, D.D.; Beauchamp, E.; Berthiaume, L.G. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie 2011, 93, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Dyda, F.; Klein, D.C.; Hickman, A.B. GCN5-Related N-Acetyltransferases: A Structural Overview. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 81–103. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; Bhatnagar, R.S.; Knoll, L.J.; Gordon, I.J. Genetic and Biochemical Studies of Protein N-Myristoylation. Annu. Rev. Biochem. 1994, 63, 869–914. [Google Scholar] [CrossRef]
- Cao, W.; Sumikoshi, K.; Nakamura, S.; Terada, T.; Shimizu, K. Prediction of N-myristoylation modification of proteins by SVM. Bioinformation 2011, 6, 204–206. [Google Scholar] [CrossRef] [PubMed]
- Maurer-Stroh, S.; Eisenhaber, B.; Eisenhaber, F. N-terminal N -myristoylation of proteins: Refinement of the sequence motif and its taxon-specific differences 1 1Edited by J. Thornton. J. Mol. Biol. 2002, 317, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 2003, 101, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Hogue, I.B.; Llewellyn, G.N.; Ono, A. Dynamic Association between HIV-1 Gag and Membrane Domains. Mol. Biol. Int. 2012, 2012, 979765. [Google Scholar] [CrossRef]
- Inlora, J.; Collins, D.R.; Trubin, M.E.; Chung, J.Y.J.; Ono, A. Membrane Binding and Subcellular Localization of Retroviral Gag Proteins Are Differentially Regulated by MA Interactions with Phosphatidylinositol-(4,5)-Bisphosphate and RNA. mBio 2014, 5, e02202-14. [Google Scholar] [CrossRef]
- Resh, M.D. A myristoyl switch regulates membrane binding of HIV-1 Gag. Proc. Natl. Acad. Sci. USA 2004, 101, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Ono, A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol. Cell 2010, 102, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef]
- Saad, J.S.; Loeliger, E.; Luncsford, P.; Liriano, M.; Tai, J.; Kim, A.; Miller, J.; Joshi, A.; Freed, E.O.; Summers, M.F. Point Mutations in the HIV-1 Matrix Protein Turn Off the Myristyl Switch. J. Mol. Biol. 2007, 366, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Dalton, A.K.; Ako-Adjei, D.; Murray, P.S.; Murray, D.; Vogt, V.M. Electrostatic Interactions Drive Membrane Association of the Human Immunodeficiency Virus Type 1 Gag MA Domain. J. Virol. 2007, 81, 6434–6445. [Google Scholar] [CrossRef]
- Paillart, J.-C.; Göttlinger, H.G. Opposing Effects of Human Immunodeficiency Virus Type 1 Matrix Mutations Support a Myristyl Switch Model of Gag Membrane Targeting. J. Virol. 1999, 73, 2604–2612. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Resh, M.D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 1996, 70, 8540–8548. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.S.; Li, Z.; Wang, J.; Tang, C.L.; Honig, B.; Murray, D. Retroviral Matrix Domains Share Electrostatic Homology: Models for Membrane Binding Function throughout the Viral Life Cycle. Structure 2005, 13, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Hamard-Peron, E.; Muriaux, D. Retroviral matrix and lipids, the intimate interaction. Retrovirology 2011, 8, 15. [Google Scholar] [CrossRef]
- Parent, L.J.; Gudleski, N. Beyond Plasma Membrane Targeting: Role of the MA domain of Gag in Retroviral Genome Encapsidation. J. Mol. Biol. 2011, 410, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.; Ben-Tal, N.; Honig, B.; McLaughlin, S. Electrostatic interaction of myristoylated proteins with membranes: Simple physics, complicated biology. Structure 1997, 5, 985–989. [Google Scholar] [CrossRef]
- Kozak, C.A. Origins of the Endogenous and Infectious Laboratory Mouse Gammaretroviruses. Viruses 2014, 7, 1–26. [Google Scholar] [CrossRef]
- Nitta, T.; Kuznetsov, Y.; McPherson, A.; Fan, H. Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proc. Natl. Acad. Sci. USA 2009, 107, 1190–1195. [Google Scholar] [CrossRef]
- Houzet, L.; Gay, B.; Morichaud, Z.; Briant, L.; Mougel, M. Intracellular assembly and budding of the Murine Leukemia Virus in infected cells. Retrovirology 2006, 3, 12. [Google Scholar] [CrossRef]
- Hron, T.; Elleder, D.; Gifford, R.J. Deltaretroviruses have circulated since at least the Paleogene and infected a broad range of mammalian species. Retrovirology 2019, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Egessain, A.; Ecassar, O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front. Microbiol. 2012, 3, 388. [Google Scholar] [CrossRef]
- Barez, P.-Y.; De Brogniez, A.; Carpentier, A.; Gazon, H.; Gillet, N.; Gutiérrez, G.; Hamaidia, M.; Jacques, J.-R.; Perike, S.; Sriramareddy, S.N.; et al. Recent Advances in BLV Research. Viruses 2015, 7, 6080–6088. [Google Scholar] [CrossRef] [PubMed]
- Blot, V.; Perugi, F.; Gay, B.; Prévost, M.-C.; Briant, L.; Tangy, F.; Abriel, H.; Staub, O.; Dokhélar, M.-C.; Pique, C. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 2004, 117, 2357–2367. [Google Scholar] [CrossRef]
- Inlora, J.; Chukkapalli, V.; Derse, D.; Ono, A. Gag Localization and Virus-Like Particle Release Mediated by the Matrix Domain of Human T-Lymphotropic Virus Type 1 Gag Are Less Dependent on Phosphatidylinositol-(4,5)-Bisphosphate than Those Mediated by the Matrix Domain of HIV-1 Gag. J. Virol. 2011, 85, 3802–3810. [Google Scholar] [CrossRef] [PubMed]
- Le Blanc, I.; Rosenberg, A.R.; Dokhélar, M.-C. Multiple Functions for the Basic Amino Acids of the Human T-Cell Leukemia Virus Type 1 Matrix Protein in Viral Transmission. J. Virol. 1999, 73, 1860–1867. [Google Scholar] [CrossRef]
- Fogarty, K.H.; Zhang, W.; Grigsby, I.F.; Johnson, J.L.; Chen, Y.; Mueller, J.D.; Mansky, L.M. New Insights into HTLV-1 Particle Structure, Assembly, and Gag-Gag Interactions in Living Cells. Viruses 2011, 3, 770–793. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.; Gilbert, G.E.; Shi, J.; Silvius, J.; Kapus, A.; Grinstein, S. Membrane Phosphatidylserine Regulates Surface Charge and Protein Localization. Science 2008, 319, 210–213. [Google Scholar] [CrossRef]
- Chukkapalli, V.; Oh, S.J.; Ono, A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA 2010, 107, 1600–1605. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Norris, K.M.; Mansky, L.M. Involvement of the Matrix and Nucleocapsid Domains of the Bovine Leukemia Virus Gag Polyprotein Precursor in Viral RNA Packaging. J. Virol. 2003, 77, 9431–9438. [Google Scholar] [CrossRef]
- Prchal, J.; Srb, P.; Hunter, E.; Ruml, T.; Hrabal, R. The Structure of Myristoylated Mason-Pfizer Monkey Virus Matrix Protein and the Role of Phosphatidylinositol-(4,5)-Bisphosphate in Its Membrane Binding. J. Mol. Biol. 2012, 423, 427–438. [Google Scholar] [CrossRef]
- Ross, S.R. Mouse Mammary Tumor Virus Molecular Biology and Oncogenesis. Viruses 2010, 2, 2000–2012. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.J.; Tachedjian, M.; Cui, J.; Field, H.; Holmes, E.C.; Wang, L.-F.; Tachedjian, G. Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology 2013, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Monde, K.; Contreras-Galindo, R.; Kaplan, M.H.; Markovitz, D.M.; Ono, A. Human Endogenous Retrovirus K Gag Coassembles with HIV-1 Gag and Reduces the Release Efficiency and Infectivity of HIV-1. J. Virol. 2012, 86, 11194–11208. [Google Scholar] [CrossRef]
- Doležal, M.; Zábranský, A.; Dostál, J.; Vaněk, O.; Brynda, J.; Lepšík, M.; Hadravová, R.; Pichová, I. Myristoylation drives dimerization of matrix protein from mouse mammary tumor virus. Retrovirology 2016, 13, 1–15. [Google Scholar] [CrossRef]
- Dalton, A.K.; Murray, P.S.; Murray, D.; Vogt, V.M. Biochemical Characterization of Rous Sarcoma Virus MA Protein Interaction with Membranes. J. Virol. 2005, 79, 6227–6238. [Google Scholar] [CrossRef][Green Version]
- Nadaraia-Hoke, S.; Bann, D.V.; Lochmann, T.L.; Gudleski-O’Regan, N.; Parent, L.J. Alterations in the MA and NC Domains Modulate Phosphoinositide-Dependent Plasma Membrane Localization of the Rous Sarcoma Virus Gag Protein. J. Virol. 2013, 87, 3609–3615. [Google Scholar] [CrossRef] [PubMed]
- Callahan, E.M.; Wills, J.W. Repositioning Basic Residues in the M Domain of the Rous Sarcoma Virus Gag Protein. J. Virol. 2000, 74, 11222–11229. [Google Scholar] [CrossRef]
- Dick, R.A.; Vogt, V.M. Membrane interaction of retroviral Gag proteins. Front. Microbiol. 2014, 5, 187. [Google Scholar] [CrossRef]
- Hatanaka, H.; Iourin, O.; Rao, Z.; Fry, E.; Kingsman, A.; Stuart, D.I. Structure of Equine Infectious Anemia Virus Matrix Protein. J. Virol. 2002, 76, 1876–1883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ma, J.; Zhang, X.; Su, C.; Yao, Q.-C.; Wang, X. Equine Infectious Anemia Virus Gag Assembly and Export Are Directed by Matrix Protein throughtrans-Golgi Networks and Cellular Vesicles. J. Virol. 2015, 90, 1824–1838. [Google Scholar] [CrossRef]
- Fernandes, F.; Chen, K.; Ehrlich, L.S.; Jin, J.; Chen, M.H.; Medina, G.N.; Symons, M.; Montelaro, R.; Donaldson, J.; Tjandra, N.; et al. Phosphoinositides Direct Equine Infectious Anemia Virus Gag Trafficking and Release. Traffic 2010, 12, 438–451. [Google Scholar] [CrossRef]
- Goldstone, D.C.; Flower, T.G.; Ball, N.J.; Sanz-Ramos, M.; Yap, M.W.; Ogrodowicz, R.W.; Stanke, N.; Reh, J.; Lindemann, D.; Stoye, J.P.; et al. A Unique Spumavirus Gag N-terminal Domain with Functional Properties of Orthoretroviral Matrix and Capsid. PLoS Pathog. 2013, 9, e1003376. [Google Scholar] [CrossRef]
- Heneine, W.; Schweizer, M.; Sandstrom, P.; Folks, T. Human Infection with Foamy Viruses. Curr. Top. Microbiol. Immunol. 2003, 277, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.L.; Lindemann, D.; Mulligan, M.J.; Goepfert, P.A. Foamy Virus Envelope Glycoprotein Is Sufficient for Particle Budding and Release. J. Virol. 2003, 77, 2338–2348. [Google Scholar] [CrossRef] [PubMed]
- Nishi, H.; Shaytan, A.; Panchenko, A.R. Physicochemical mechanisms of protein regulation by phosphorylation. Front. Genet. 2014, 5, 270. [Google Scholar] [CrossRef] [PubMed]
- Mandell, D.J.; Chorny, I.; Groban, E.S.; Wong, S.E.; Levine, E.; Rapp, A.C.S.; Jacobson, M.P. Strengths of Hydrogen Bonds Involving Phosphorylated Amino Acid Side Chains. J. Am. Chem. Soc. 2007, 129, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Burnett, G.; Kennedy, E.P. The Enzymatic Phosphorylation of Proteins. J. Biol. Chem. 1954, 211, 969–980. [Google Scholar] [CrossRef]
- Francis, A.C.; Di Primio, C.; Allouch, A.; Cereseto, A. Role of phosphorylation in the nuclear biology of HIV-1. Curr. Med. Chem. 2011, 18, 2904–2912. [Google Scholar] [CrossRef] [PubMed]
- Burnette, B.; Yu, G.; Felsted, R. Phosphorylation of HIV-1 gag proteins by protein kinase C. J. Biol. Chem. 1993, 268, 8698–8703. [Google Scholar] [CrossRef]
- Yu, G.; Shen, F.S.; Sturch, S.; Aquino, A.; Glazer, R.I.; Felsted, R.L. Regulation of HIV-1 gag Protein Subcellular Targeting by Protein Kinase C. J. Biol. Chem. 1995, 270, 4792–4796. [Google Scholar] [CrossRef] [PubMed]
- Kräusslich, H.G. Morphogenesis and Maturation of Retroviruses—Google Livres, 1st ed.; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1996. [Google Scholar]
- Kudoh, A.; Takahama, S.; Sawasaki, T.; Ode, H.; Yokoyama, M.; Okayama, A.; Ishikawa, A.; Miyakawa, K.; Matsunaga, S.; Kimura, H.; et al. The phosphorylation of HIV-1 Gag by atypical protein kinase C facilitates viral infectivity by promoting Vpr incorporation into virions. Retrovirology 2014, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Radestock, B.; Morales, I.; Rahman, S.A.; Radau, S.; Glass, B.; Zahedi, R.P.; Müller, B.; Kräusslich, H.-G. Comprehensive Mutational Analysis Reveals p6Gag Phosphorylation to Be Dispensable for HIV-1 Morphogenesis and Replication. J. Virol. 2012, 87, 724–734. [Google Scholar] [CrossRef]
- Votteler, J.; Neumann, L.; Hahn, S.; Hahn, F.; Rauch, P.; Schmidt, K.; Studtrucker, N.; Solbak, S.M.; Fossen, T.; Henklein, P.; et al. Highly conserved serine residue 40 in HIV-1 p6 regulates capsid processing and virus core assembly. Retrovirology 2011, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.M.; Chen, M.-H.; Khan, M.; Ehrlich, L.; Kemal, K.S.; Weiser, B.; Shi, B.; Chen, C.; Powell, M.; Anastos, K.; et al. The S40 residue in HIV-1 Gag p6 impacts local and distal budding determinants, revealing additional late domain activities. Retrovirology 2013, 10, 143. [Google Scholar] [CrossRef]
- Radestock, B.; Burk, R.; Müller, B.; Kräusslich, H.-G. Re-visiting the functional Relevance of the highly conserved Serine 40 Residue within HIV-1 p6Gag. Retrovirology 2014, 11, 1–5. [Google Scholar] [CrossRef]
- Müller, B.; Patschinsky, T.; Kräusslich, H.-G. The Late-Domain-Containing Protein p6 Is the Predominant Phosphoprotein of Human Immunodeficiency Virus Type 1 Particles. J. Virol. 2002, 76, 1015–1024. [Google Scholar] [CrossRef]
- Hemonnot, B.; Cartier, C.; Gay, B.; Rebuffat, S.; Bardy, M.; Devaux, C.; Boyer, V.; Briant, L. The Host Cell MAP Kinase ERK-2 Regulates Viral Assembly and Release by Phosphorylating the p6 Protein of HIV-1. J. Biol. Chem. 2004, 279, 32426–32434. [Google Scholar] [CrossRef]
- Wang, Z.; Canagarajah, B.J.; Boehm, J.C.; Kassisà, S.; Cobb, M.H.; Young, P.R.; Abdel-Meguid, S.; Adams, J.L.; Goldsmith, E.J. Structural basis of inhibitor selectivity in MAP kinases. Structure 1998, 6, 1117–1128. [Google Scholar] [CrossRef]
- Cartier, C.; Deckert, M.; Grangeasse, C.; Trauger, R.; Jensen, F.; Bernard, A.; Cozzone, A.; Desgranges, C.; Boyer, V. Association of ERK2 mitogen-activated protein kinase with human immunodeficiency virus particles. J. Virol. 1997, 71, 4832–4837. [Google Scholar] [CrossRef]
- Cartier, C.; Hemonnot, B.; Gay, B.; Bardy, M.; Sanchiz, C.; Devaux, C.; Briant, L. Active cAMP-dependent Protein Kinase Incorporated within Highly Purified HIV-1 Particles Is Required for Viral Infectivity and Interacts with Viral Capsid Protein. J. Biol. Chem. 2003, 278, 35211–35219. [Google Scholar] [CrossRef]
- Jacqué, J.; Mann, A.; Enslen, H.; Sharova, N.; Brichacek, B.; Davis, R.J.; Stevenson, M. Modulation of HIV-1 infectivity by MAPK, a virion-associated kinase. EMBO J. 1998, 17, 2607–2618. [Google Scholar] [CrossRef]
- Dochi, T.; Nakano, T.; Inoue, M.; Takamune, N.; Shoji, S.; Sano, K.; Misumi, S. Phosphorylation of human immunodeficiency virus type 1 capsid protein at serine 16, required for peptidyl-prolyl isomerase-dependent uncoating, is mediated by virion-incorporated extracellular signal-regulated kinase 2. J. Gen. Virol. 2014, 95, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gabuzda, D. Regulation of Human Immunodeficiency Virus Type 1 Infectivity by the ERK Mitogen-Activated Protein Kinase Signaling Pathway. J. Virol. 1999, 73, 3460–3466. [Google Scholar] [CrossRef]
- Greenway, A.; Azad, A.; Mills, J.; McPhee, D. Human immunodeficiency virus type 1 Nef binds directly to Lck and mitogen-activated protein kinase, inhibiting kinase activity. J. Virol. 1996, 70, 6701–6708. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Goncalves, J.; Gabuzda, D. Phosphorylation of Vif and Its Role in HIV-1 Replication. J. Biol. Chem. 1996, 271, 10121–10129. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gabuzda, D. Mitogen-activated Protein Kinase Phosphorylates and Regulates the HIV-1 Vif Protein. J. Biol. Chem. 1998, 273, 29879–29887. [Google Scholar] [CrossRef]
- Kaushik, R.; Ratner, L. Role of Human Immunodeficiency Virus Type 1 Matrix Phosphorylation in an Early Postentry Step of Virus Replication. J. Virol. 2004, 78, 2319–2326. [Google Scholar] [CrossRef] [PubMed]
- Strasner, A.B.; Natarajan, M.; Doman, T.; Key, D.; August, A.; Henderson, A.J. The Src Kinase Lck Facilitates Assembly of HIV-1 at the Plasma Membrane. J. Immunol. 2008, 181, 3706–3713. [Google Scholar] [CrossRef]
- Gallay, P.; Swingler, S.; Song, J.; Bushman, F.; Trono, D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 1995, 83, 569–576. [Google Scholar] [CrossRef]
- Hahn, F.; Setz, C.; Friedrich, M.; Rauch, P.; Solbak, S.M.; Frøystein, N.Å.; Henklein, P.; Votteler, J.; Fossen, T.; Schubert, U. Mutation of the Highly Conserved Ser-40 of the HIV-1 p6 Gag Protein to Phe Causes the Formation of a Hydrophobic Patch, Enhances Membrane Association, and Polyubiquitination of Gag. Viruses 2014, 6, 3738–3765. [Google Scholar] [CrossRef] [PubMed]
- Leis, J.; Phillips, N.; Fu, X.; Tuazon, P.T.; Traugh, J.A. Phosphorylation of avian retrovirus matrix protein by Ca2+/phospholipid-dependent protein kinase. JBIC J. Biol. Inorg. Chem. 1989, 179, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Nelle, T.D.; Verderame, M.F.; Leis, J.; Wills, J.W. The Major Site of Phosphorylation within the Rous Sarcoma Virus MA Protein Is Not Required for Replication. J. Virol. 1998, 72, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- Pepinsky, R.B.; Papayannopoulos, A.I.; Campbell, S.; Vogt, V.M. Analysis of Rous sarcoma virus Gag protein by mass spectrometry indicates trimming by host exopeptidase. J. Virol. 1996, 70, 3313–3318. [Google Scholar] [CrossRef]
- Fu, X.; Tuazon, P.T.; Traugh, J.A.; Leis, J. Site-Directed Mutagenesis of the Avian Retrovirus Nucleocapsid Protein, Pp12, at Serine 40, the Primary Site of Phosphorylation in Vivo. J. Biol. Chem. 1988, 268, 2134–2139. [Google Scholar] [CrossRef]
- Wang, H.; Machesky, N.J.; Mansky, L.M. Both the PPPY and PTAP Motifs Are Involved in Human T-Cell Leukemia Virus Type 1 Particle Release. J. Virol. 2004, 78, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Hemonnot, B.; Molle, D.; Bardy, M.; Gay, B.; Laune, D.; Devaux, C.; Briant, L. Phosphorylation of the HTLV-1 matrix L-domain-containing protein by virus-associated ERK-2 kinase. Virology 2006, 349, 430–439. [Google Scholar] [CrossRef]
- Bouamr, F.; Melillo, J.A.; Wang, M.Q.; Nagashima, K.; Santos, M.D.L.; Rein, A.; Goff, S.P. PPPYEPTAP Motif Is the Late Domain of Human T-Cell Leukemia Virus Type 1 Gag and Mediates Its Functional Interaction with Cellular Proteins Nedd4 and Tsg101. J. Virol. 2003, 77, 11882–11895. [Google Scholar] [CrossRef]
- Yasuda, J.; Hunter, E. A Proline-Rich Motif (PPPY) in the Gag Polyprotein of Mason-Pfizer Monkey Virus Plays a Maturation-Independent Role in Virion Release. J. Virol. 1998, 72, 4095–4103. [Google Scholar] [CrossRef]
- Henderson, E.L.; Sowder, R.; Smythers, G.; Benveniste, R.E.; Oroszlan, S. Purification and N-terminal amino acid sequence comparisons of structural proteins from retrovirus-D/Washington and Mason-Pfizer monkey virus. J. Virol. 1985, 55, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Uckert, W.; Wunderlich, V.; Fiebach, H.; Hertling, I.; Stein, U.; Kraft, R.; Desrosiers, R. Biochemical and immunological characterization of structural proteins from retrovirus-D/New England and comparison to Mason-Pfizer monkey virus and permanent human fibroblast virus. Arch. Virol. 1987, 94, 267–282. [Google Scholar] [CrossRef]
- Enssle, J.; Fischer, N.; Moebes, A.; Mauer, B.; Smola, U.; Rethwilm, A. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J. Virol. 1997, 71, 7312–7317. [Google Scholar] [CrossRef]
- Yuan, B.; Li, X.; Goff, S.P. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999, 18, 4700–4710. [Google Scholar] [CrossRef] [PubMed]
- Yueh, A.; Goff, S.P. Phosphorylated Serine Residues and an Arginine-Rich Domain of the Moloney Murine Leukemia Virus p12 Protein Are Required for Early Events of Viral Infection. J. Virol. 2003, 77, 1820–1829. [Google Scholar] [CrossRef][Green Version]
- Brzezinski, J.D.; Felkner, R.; Modi, A.; Liu, M.; Roth, M.J. Phosphorylation Requirement of Murine Leukemia Virus p12. J. Virol. 2016, 90, 11208–11219. [Google Scholar] [CrossRef] [PubMed]
- Callis, J. The Ubiquitination Machinery of the Ubiquitin System. Arab. Book 2014, 12, e0174. [Google Scholar] [CrossRef]
- Vijay-Kumar, S.; Bugg, C.; Wilkinson, K.; Vierstra, R.; Hatfield, P.; Cook, W. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J. Biol. Chem. 1987, 262, 6396–6399. [Google Scholar] [CrossRef]
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953. [Google Scholar] [CrossRef]
- Shields, S.B.; Piper, R.C. How Ubiquitin Functions with ESCRTs. Traffic 2011, 12, 1306–1317. [Google Scholar] [CrossRef]
- Ohtake, F.; Saeki, Y.; Ishido, S.; Kanno, J.; Tanaka, K. The K48-K63 Branched Ubiquitin Chain Regulates NF-κB Signaling. Mol. Cell 2016, 64, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Suresh, B.; Lee, J.; Kim, K.-S.; Ramakrishna, S. The Importance of Ubiquitination and Deubiquitination in Cellular Reprogramming. Stem Cells Int. 2016, 2016, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Kräusslich, H.-G. Analysis of Human Immunodeficiency Virus Type 1 Gag Ubiquitination. J. Virol. 2005, 79, 9134–9144. [Google Scholar] [CrossRef]
- Gottwein, E.; Jäger, S.; Habermann, A.; Kräusslich, H.-G. Cumulative Mutations of Ubiquitin Acceptor Sites in Human Immunodeficiency Virus Type 1 Gag Cause a Late Budding Defect. J. Virol. 2006, 80, 6267–6275. [Google Scholar] [CrossRef]
- Ott, D.E.; Coren, L.V.; Ii, R.C.S.; Adams, J.; Schubert, U. Retroviruses Have Differing Requirements for Proteasome Function in the Budding Process. J. Virol. 2003, 77, 3384–3393. [Google Scholar] [CrossRef] [PubMed]
- Sette, P.; Jadwin, J.A.; Dussupt, V.; Bello, N.F.; Bouamr, F. The ESCRT-Associated Protein Alix Recruits the Ubiquitin Ligase Nedd4-1 To Facilitate HIV-1 Release through the LYPXnL L Domain Motif. J. Virol. 2010, 84, 8181–8192. [Google Scholar] [CrossRef]
- Sette, P.; Nagashima, K.; Piper, R.C.; Bouamr, F. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 2013, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Ott, D.E.; Coren, L.V.; Copeland, T.D.; Kane, B.P.; Johnson, D.G.; Sowder, R.C.; Yoshinaka, Y.; Oroszlan, S.; Arthur, L.O.; Henderson, L.E. Ubiquitin Is Covalently Attached to the p6GagProteins of Human Immunodeficiency Virus Type 1 and Simian Immunodeficiency Virus and to the p12Gag Protein of Moloney Murine Leukemia Virus. J. Virol. 1998, 72, 2962–2968. [Google Scholar] [CrossRef]
- Schubert, U.; Ott, D.E.; Chertova, E.N.; Welker, R.; Tessmer, U.; Princiotta, M.F.; Bennink, J.R.; Kräusslich, H.-G.; Yewdell, J.W. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 2000, 97, 13057–13062. [Google Scholar] [CrossRef] [PubMed]
- Ott, D.E.; Coren, L.V.; Chertova, E.N.; Gagliardi, T.D.; Schubert, U. Ubiquitination of HIV-1 and MuLV Gag. Virology 2000, 278, 111–121. [Google Scholar] [CrossRef]
- Patnaik, A.; Chau, V.; Wills, J.W. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 2000, 97, 13069–13074. [Google Scholar] [CrossRef]
- Jäger, S.; Gottwein, E.; Kräusslich, H.-G. Ubiquitination of Human Immunodeficiency Virus Type 1 Gag Is Highly Dependent on Gag Membrane Association. J. Virol. 2007, 81, 9193–9201. [Google Scholar] [CrossRef] [PubMed]
- Von Schwedler, U.K.; Stuchell, M.; Müller, B.; Ward, D.M.; Chung, H.Y.; Morita, E.; Wang, H.E.; Davis, T.; He, G.P.; Cimbora, D.M.; et al. The protein network of HIV budding. Cell 2003, 114, 701–713. [Google Scholar] [CrossRef]
- Pornillos, O.; Alam, S.L.; Rich, R.L.; Myszka, D.G.; Davis, D.R.; Sundquist, W.I. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002, 21, 2397–2406. [Google Scholar] [CrossRef]
- Watanabe, S.M.; Strickland, M.; Tjandra, N.; Carter, C.A. RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane. Viruses 2020, 12, 447. [Google Scholar] [CrossRef]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Côté, M.; Rich, R.L.; et al. Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef]
- Staub, O.; Dho, S.; Henry, P.; Correa, J.; Ishikawa, T.; McGlade, J.; Rotin, D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996, 15, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Babst, M.; Katzmann, D.J.; Estepa-Sabal, E.J.; Meerloo, T.; Emr, S.D. Escrt-III. Dev. Cell 2002, 3, 271–282. [Google Scholar] [CrossRef]
- Dunn, R.; Klos, D.A.; Adler, A.S.; Hicke, L. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 2004, 165, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Spidel, J.L.; Craven, R.C.; Wilson, C.B.; Patnaik, A.; Wang, H.; Mansky, L.M.; Wills, J.W. Lysines Close to the Rous Sarcoma Virus Late Domain Critical for Budding. J. Virol. 2004, 78, 10606–10616. [Google Scholar] [CrossRef] [PubMed]
- Ott, D.E.; Coren, L.V.; Sowder, R.C.; Adams, J.; Nagashima, K.; Schubert, U. Equine Infectious Anemia Virus and the Ubiquitin-Proteasome System. J. Virol. 2002, 76, 3038–3044. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, F.; Montelaro, R.C. Functional Roles of Equine Infectious Anemia Virus Gag p9 in Viral Budding and Infection. J. Virol. 2001, 75, 9762–9770. [Google Scholar] [CrossRef][Green Version]
- Heidecker, G.; Lloyd, P.A.; Fox, K.; Nagashima, K.; Derse, D. Late Assembly Motifs of Human T-Cell Leukemia Virus Type 1 and Their Relative Roles in Particle Release. J. Virol. 2004, 78, 6636–6648. [Google Scholar] [CrossRef]
- Heidecker, G.; Lloyd, P.A.; Soheilian, F.; Nagashima, K.; Derse, D. The Role of WWP1-Gag Interaction and Gag Ubiquitination in Assembly and Release of Human T-Cell Leukemia Virus Type 1. J. Virol. 2007, 81, 9769–9777. [Google Scholar] [CrossRef] [PubMed]
- Medina, G.; Pincetic, A.; Ehrlich, L.S.; Zhang, Y.; Tang, Y.; Leis, J.; Carter, C.A. Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting. Virology 2008, 377, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Bartusch, C.; Prange, R. ESCRT Requirements for Murine Leukemia Virus Release. Viruses 2016, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Vana, M.L.; Tang, Y.; Chen, A.; Medina, G.; Carter, C.; Leis, J. Role of Nedd4 and Ubiquitination of Rous Sarcoma Virus Gag in Budding of Virus-Like Particles from Cells. J. Virol. 2004, 78, 13943–13953. [Google Scholar] [CrossRef]
- Stanke, N.; Stange, A.; Lüftenegger, D.; Zentgraf, H.; Lindemann, D. Ubiquitination of the Prototype Foamy Virus Envelope Glycoprotein Leader Peptide Regulates Subviral Particle Release. J. Virol. 2005, 79, 15074–15083. [Google Scholar] [CrossRef]
- Yang, Y.; He, Y.; Wang, X.; Liang, Z.; He, G.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Protein SUMOylation modification and its associations with disease. Open Biol. 2017, 7, 170167. [Google Scholar] [CrossRef]
- Han, Z.-J.; Feng, Y.-H.; Gu, B.-H.; Li, Y.-M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef]
- Matunis, M.J.; Coutavas, E.; Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996, 135, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Owerbach, D.; McKay, E.M.; Yeh, E.T.; Gabbay, K.H.; Bohren, K.M. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 2005, 337, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, R.; Gerace, L.; Melchior, F. Molecular Characterization of the SUMO-1 Modification of RanGAP1 and Its Role in Nuclear Envelope Association. J. Cell Biol. 1998, 140, 259–270. [Google Scholar] [CrossRef]
- Saitoh, H.; Hinchey, J. Functional Heterogeneity of Small Ubiquitin-related Protein Modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef]
- Desterro, J.M.; Rodriguez, M.S.; Hay, R.T. SUMO-1 Modification of IκBα Inhibits NF-κB Activation. Mol. Cell 1998, 2, 233–239. [Google Scholar] [CrossRef]
- Nayak, A.; Müller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014, 15, 422. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Kim, S.; Jaffray, E.; Song, J.; Chen, Y.; Hay, R.T. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 2004, 12, 67–74. [Google Scholar] [CrossRef]
- Eisenhardt, N.; Ilic, D.; Nagamalleswari, E.; Pichler, A. Biochemical characterization of SUMO-conjugating enzymes by in vitro sumoylation assays. Methods Enzym. 2019, 618, 167–185. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Yoo, H.M.; Chung, C.H. ISG15 and immune diseases. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2010, 1802, 485–496. [Google Scholar] [CrossRef]
- Gurer, C.; Berthoux, L.; Luban, J. Covalent Modification of Human Immunodeficiency Virus Type 1 p6 by SUMO-1. J. Virol. 2005, 79, 910–917. [Google Scholar] [CrossRef]
- Jaber, T.; Bohl, C.R.; Lewis, G.L.; Wood, C.; West, J.T.; Weldon, R.A. Human Ubc9 Contributes to Production of Fully Infectious Human Immunodeficiency Virus Type 1 Virions. J. Virol. 2009, 83, 10448–10459. [Google Scholar] [CrossRef]
- Martinez, N.W.; Xue, X.; Berro, R.G.; Kreitzer, G.; Resh, M.D. Kinesin KIF4 Regulates Intracellular Trafficking and Stability of the Human Immunodeficiency Virus Type 1 Gag Polyprotein. J. Virol. 2008, 82, 9937–9950. [Google Scholar] [CrossRef] [PubMed]
- Lamoliatte, F.; McManus, F.P.; Maarifi, G.; Chelbi-Alix, M.K.; Thibault, P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 2017, 8, 14109. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Tang, Y.; Okada, Y.; Torrey, T.A.; Chattopadhyay, S.K.; Pfleiderer, M.; Falkner, F.G.; Dorner, F.; Choi, W.; Hirokawa, N.; et al. Binding of Murine Leukemia Virus Gag Polyproteins to KIF4, a Microtubule-Based Motor Protein. J. Virol. 1998, 72, 6898–6901. [Google Scholar] [CrossRef]
- Weldon, A.R.; Sarkar, P.; Brown, S.M.; Weldon, S.K. Mason–Pfizer monkey virus Gag proteins interact with the human sumo conjugating enzyme, hUbc9. Virology 2003, 314, 62–73. [Google Scholar] [CrossRef]
- Yueh, A.; Leung, J.; Bhattacharyya, S.; Perrone, L.A.; Santos, K.D.L.; Pu, S.-Y.; Goff, S.P. Interaction of Moloney Murine Leukemia Virus Capsid with Ubc9 and PIASy Mediates SUMO-1 Addition Required Early in Infection. J. Virol. 2006, 80, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wen, S.; Zhao, R.; Qi, J.; Liu, Z.; Li, W.; An, J.; Wood, C.; Wang, Y. Covalent conjugation of the equine infectious anemia virus Gag with SUMO. Biochem. Biophys. Res. Commun. 2017, 486, 712–719. [Google Scholar] [CrossRef]
- Perng, Y.-C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Genet. 2018, 16, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Potter, J.L.; Narasimhan, J.; Mende-Mueller, L.; Haas, A.L. Precursor Processing of Pro-ISG15/UCRP, an Interferon-β-induced Ubiquitin-like Protein. J. Biol. Chem. 1999, 274, 25061–25068. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation—A key to lock the cell gates for preventing the spread of threats. J. Cell Sci. 2017, 130, 2961–2969. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Collins, M.N.; Hsiang, T.-Y.; Krug, R.M. Interferon-induced ISG15 pathway: An ongoing virus–host battle. Trends Microbiol. 2013, 21, 181–186. [Google Scholar] [CrossRef]
- Hou, S.X.; Zheng, Z.; Chen, X.; Perrimon, N. The JAK/STAT Pathway in Model Organisms. Dev. Cell 2002, 3, 765–778. [Google Scholar] [CrossRef]
- Malakhov, M.P.; Malakhova, O.A.; Kim, K.I.; Ritchie, K.J.; Zhang, D.-E. UBP43 (USP18) Specifically Removes ISG15 from Conjugated Proteins. J. Biol. Chem. 2002, 277, 9976–9981. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.J.; Scott, I.; Mackewicz, C. Protection from HIV/AIDS: The importance of innate immunity. Clin. Immunol. 2003, 108, 167–174. [Google Scholar] [CrossRef]
- Shirazi, Y.; Pitha, P.M. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 1992, 66, 1321–1328. [Google Scholar] [CrossRef]
- Cheney, K.M.; Áine, M. Interferon-Alpha Mediates Restriction of Human Immunodeficiency Virus Type-1 Replication in Primary Human Macrophages at an Early Stage of Replication. PLoS ONE 2010, 5, e13521. [Google Scholar] [CrossRef] [PubMed]
- Pincetic, A.; Kuang, Z.; Seo, E.J.; Leis, J. The Interferon-Induced Gene ISG15 Blocks Retrovirus Release from Cells Late in the Budding Process. J. Virol. 2010, 84, 4725–4736. [Google Scholar] [CrossRef]
- Woods, M.W.; Kelly, J.N.; Hattlmann, C.J.; Tong, J.G.K.; Xu, L.S.; Coleman, M.D.; Quest, G.R.; Smiley, J.R.; Barr, S.D. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology 2011, 8, 95. [Google Scholar] [CrossRef]
- Okumura, A.; Lu, G.; Pitha-Rowe, I.; Pitha, P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. USA 2006, 103, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.J.; Lenschow, D.J. The antiviral activities of ISG15. J. Mol. Biol. 2013, 425, 4995–5008. [Google Scholar] [CrossRef]
- Zou, W.; Papov, V.; Malakhova, O.; Kim, K.I.; Dao, C.; Li, J.; Zhang, D.-E. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem. Biophys. Res. Commun. 2005, 336, 61–68. [Google Scholar] [CrossRef]
- Ambler, R.P.; Rees, M.W. ɛ-N-Methyl-lysine in Bacterial Flagellar Protein. Nat. Cell Biol. 1959, 184, 56–57. [Google Scholar] [CrossRef]
- Levy, D. Lysine methylation signaling of non-histone proteins in the nucleus. Cell. Mol. Life Sci. 2019, 76, 2873–2883. [Google Scholar] [CrossRef]
- Lanouette, S.; Mongeon, V.; Figeys, D.; Couture, J. The functional diversity of protein lysine methylation. Mol. Syst. Biol. 2014, 10, 724. [Google Scholar] [CrossRef]
- Blanchet, F.; Cardona, A.; Letimier, F.A.; Hershfield, M.S.; Acuto, O. CD28 costimulatory signal induces protein arginine methylation in T cells. J. Exp. Med. 2005, 202, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.A.; Schurter, B.T.; Wong-Staal, F.; David, M. Arginine Methylation of RNA Helicase A Determines Its Subcellular Localization. J. Biol. Chem. 2004, 279, 22795–22798. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, N.M.; Hitchen, E.M.; Bodetti, T.J.; Apolloni, A.; Warrilow, D.; Piller, S.C.; Harrich, D. Protein methylation is required to maintain optimal HIV-1 infectivity. Retrovirology 2006, 3, 92. [Google Scholar] [CrossRef]
- Lochmann, T.L.; Bann, D.V.; Ryan, E.P.; Beyer, A.R.; Mao, A.; Cochrane, A.; Parent, L.J. NC-mediated nucleolar localization of retroviral gag proteins. Virus Res. 2013, 171, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Invernizzi, C.D.F.; Xie, B.; Frankel, F.A.; Feldhammer, M.; Roy, B.B.; Richard, S.; Wainberg, M.A. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS 2007, 21, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.; Tobaly-Tapiero, J.; Giron, M.-L.; Burlaud-Gaillard, J.; Buseyne, F.; Roingeard, P.; Lesage, P.; Zamborlini, A.; Saïb, A. The invariant arginine within the chromatin-binding motif regulates both nucleolar localization and chromatin binding of Foamy virus Gag. Retrovirology 2018, 15, 48. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).