Expression and Localization of Thrombospondins, Plastin 3, and STIM1 in Different Cartilage Compartments of the Osteoarthritic Varus Knee
Abstract
1. Introduction
2. Results
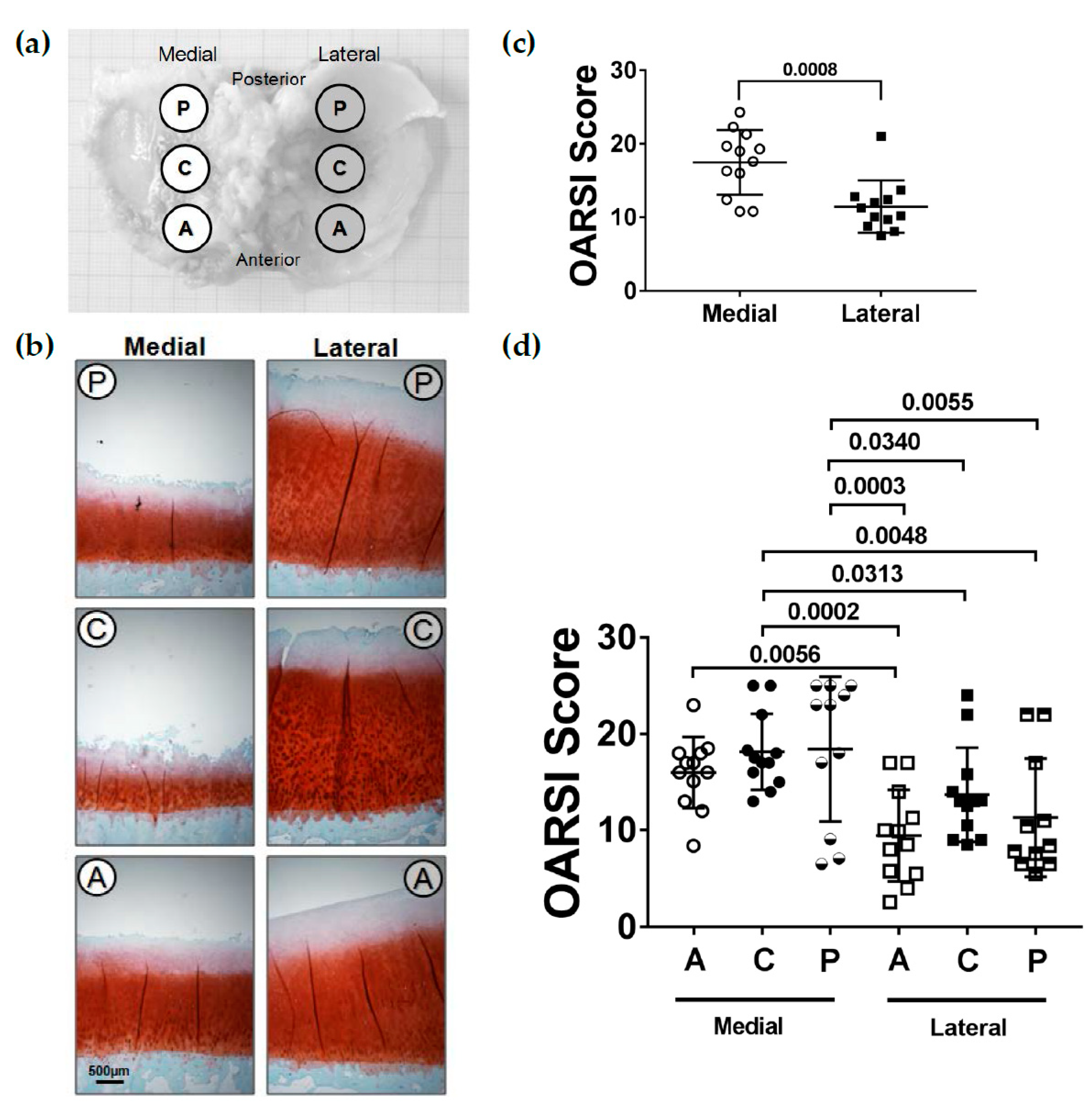
2.1. OARSI-Score
2.2. Localization of COMP
2.3. Localization of TSP-1
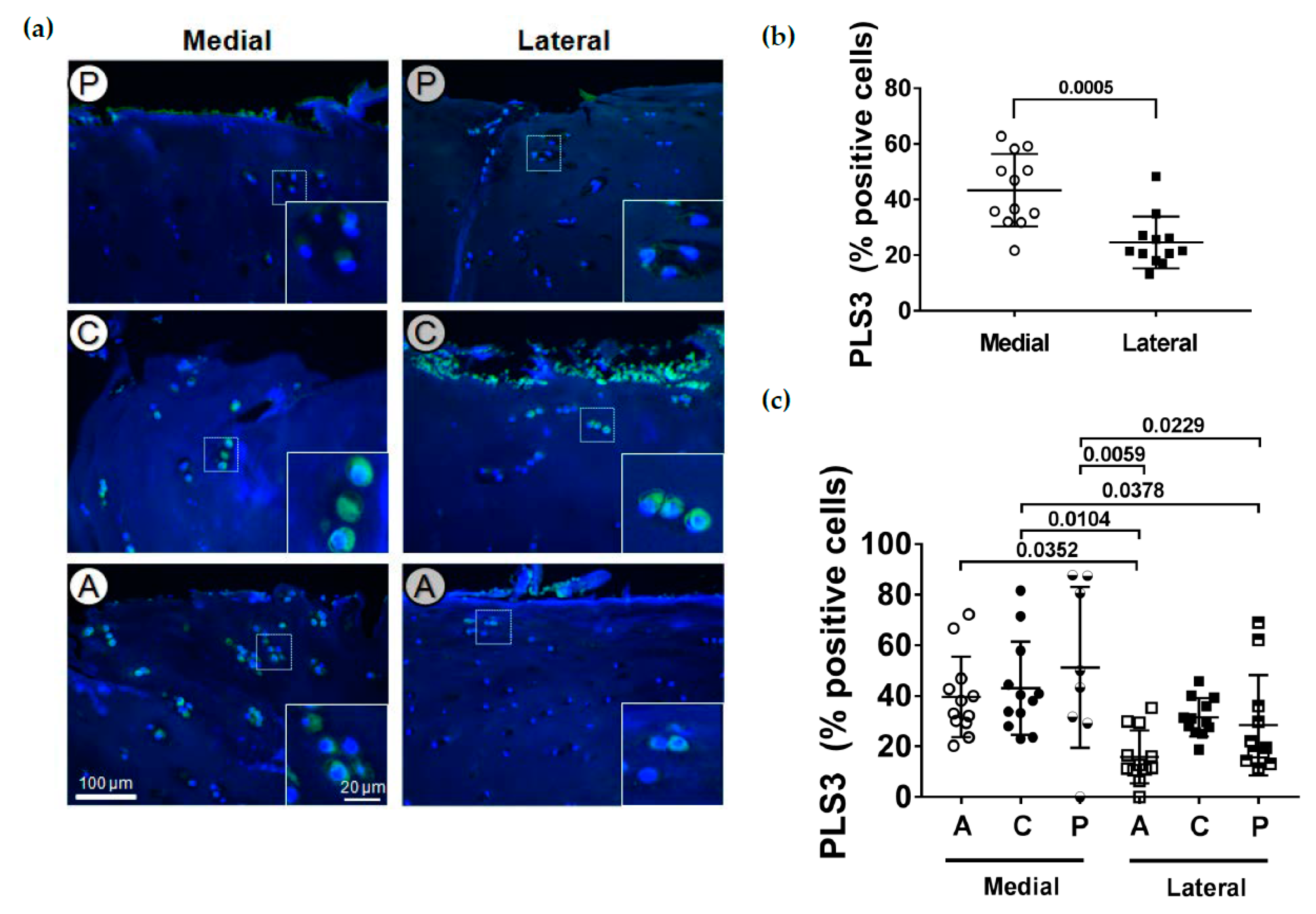
2.4. Localization of PLS3
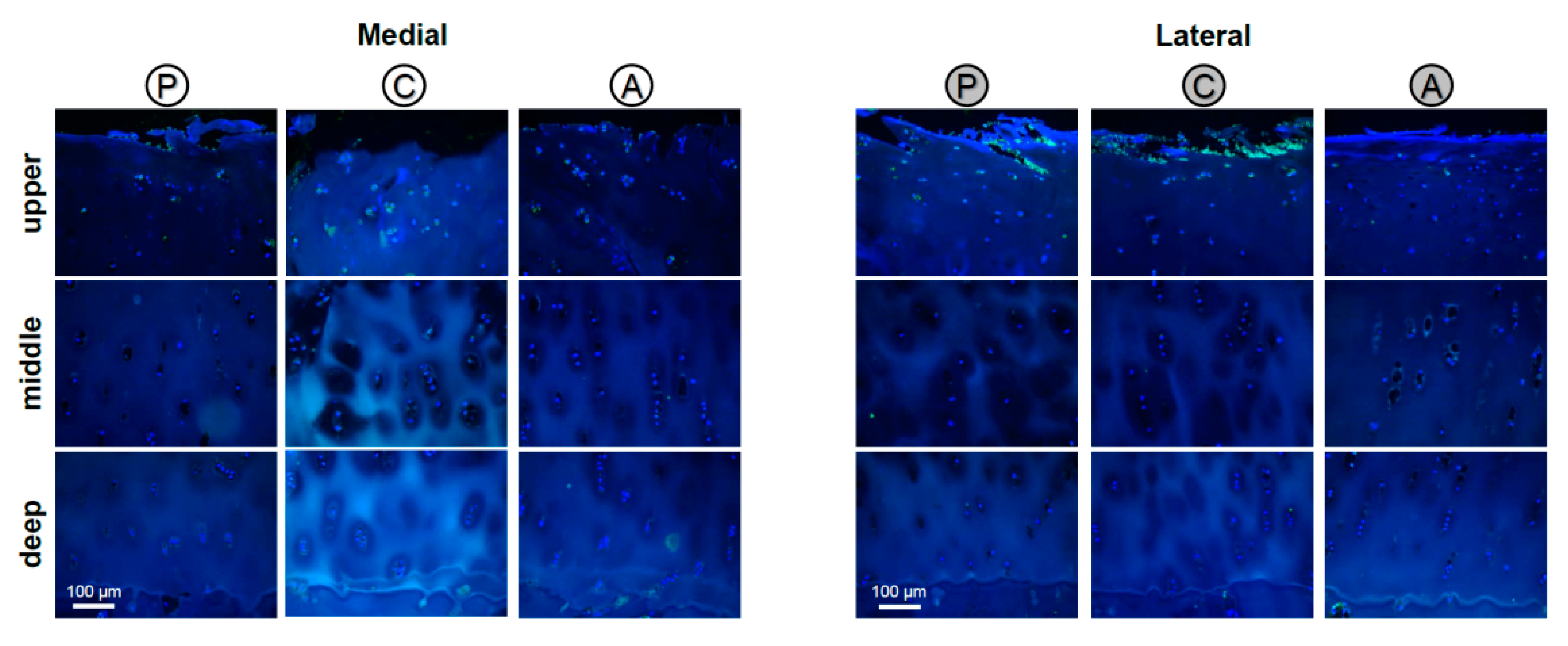
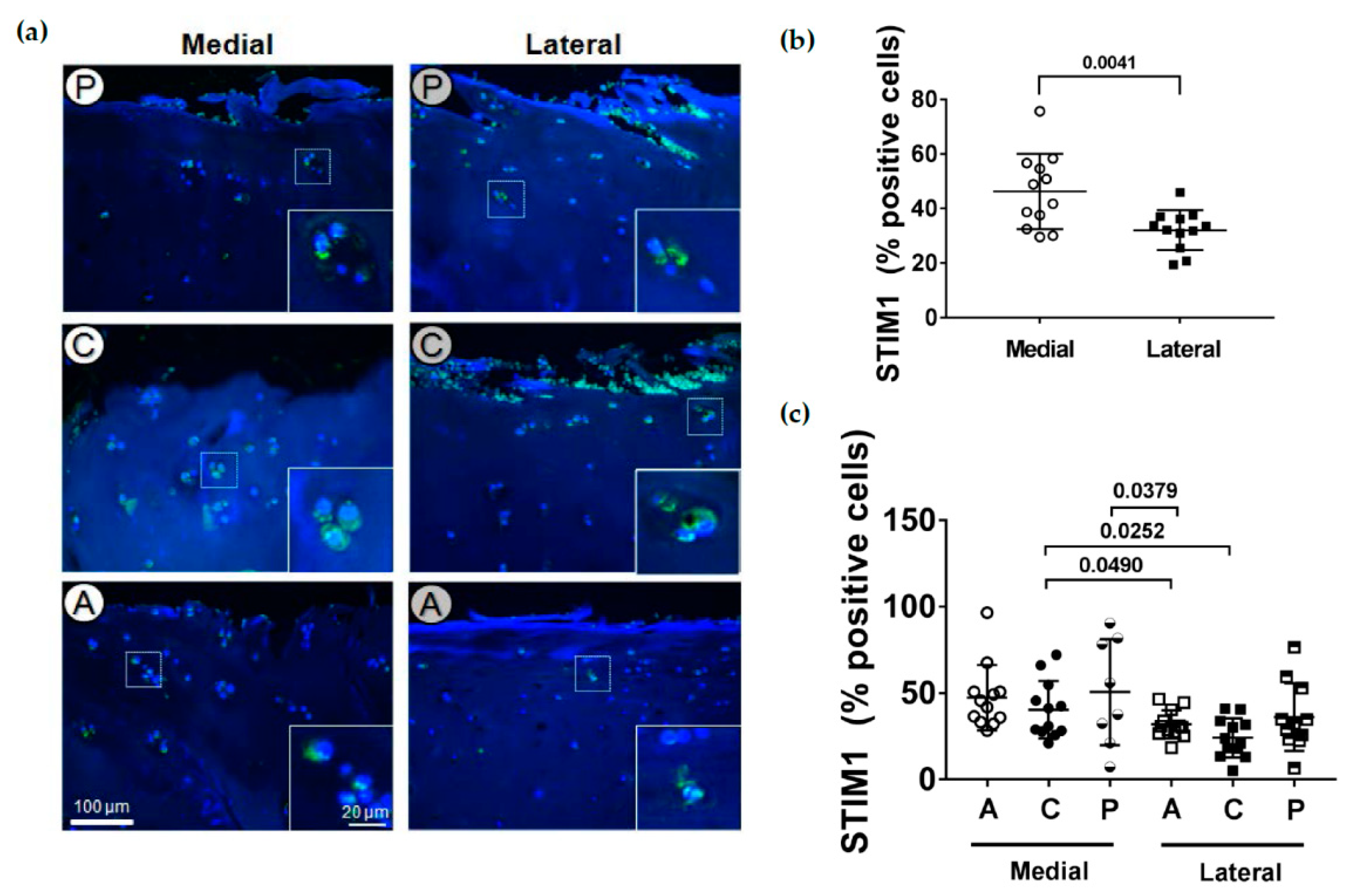
2.5. Localization of STIM1
2.6. PLS3 and STIM1 Staining Correlates with the OARSI Score
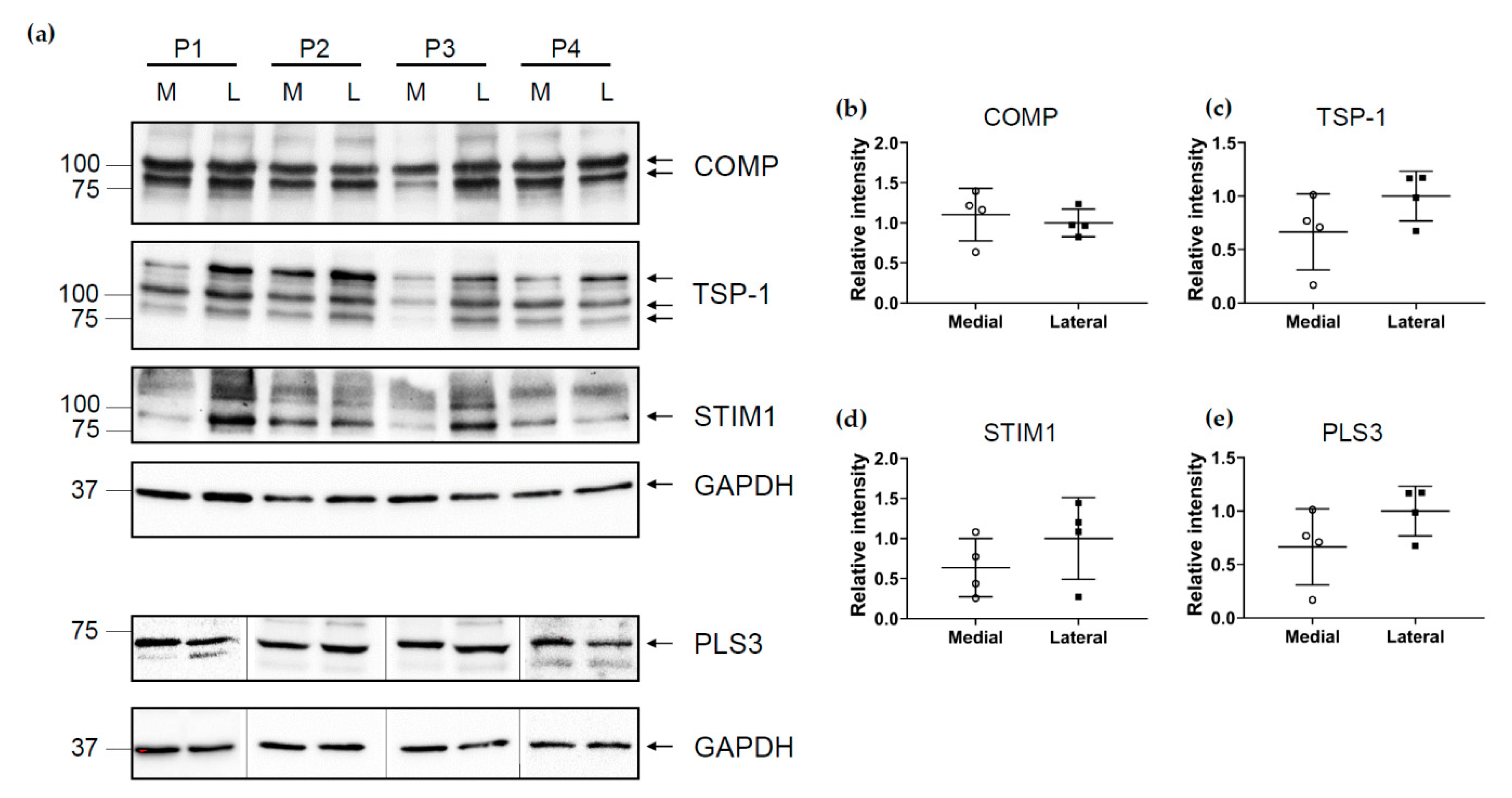
2.7. Expression of TSPs, PLS3 and STIM1 in the Medial and Lateral Compartment
3. Discussion
4. Materials and Methods
4.1. Tissue Samples
4.2. Histological Scoring
4.3. Antibodies
4.4. IHC and Immunofluorescence Analysis
4.5. Immunoblotting
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABD | Actin-binding domain |
| ADAMTS | A disintegrin and metalloproteinase with thrombospondin motif |
| BSA | Bovine serum albumin |
| COMP | Cartilage oligomeric matrix protein |
| DAB | 3,3′-diaminobenzidine |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DPX | Dibutylphthalate polystyrene xylene |
| ECM | Extracellular matrix |
| EDTA | Ethylenediaminetetraacetic acid |
| ER | Endoplasmic reticulum |
| GAPDH | Glycerinaldehyde-3-phosphate-dehydrogenase |
| HCL | Hydrogen chloride |
| HRP | Horseradish peroxidase |
| IgG | Immunoglobulin G |
| IHC | Immunohistochemistry |
| MMP | Matrix metalloproteinase |
| NaCH3COO | Sodium acetate |
| NaH2PO4 | Monosodium phosphate |
| OA | Osteoarthritis |
| OARSI | Osteoarthritis Research Society International |
| PBS | Phosphate-buffered saline |
| PLS | Plastin |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| STIM1 | Stromal interaction molecule 1 |
| TBS | Tris-buffered saline |
| Tris/HCL | Tris(hydroxymethyl)aminomethane hydrochloride |
| TSP | Thrombospondin |
Appendix A
Localization of PLS3 in Human Articular Cartilage

Appendix B
Localization of STIM1 in Human Articular Cartilage
References
- Glyn-Jones, S.; Palmer, A.J.; Agricola, R.; Price, A.J.; Vincent, T.L.; Weinans, H.; Carr, A.J. Osteoarthritis. Lancet 2015, 386, 376–387. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, S.; Huang, J.; Guo, W.; Chen, J.; Zhang, L.; Zhao, B.; Peng, J.; Wang, A.; Wang, Y.; et al. The ECM-cell interaction of cartilage extracellular matrix on chondrocytes. BioMed. Res. Int. 2014, 2014, 648459. [Google Scholar] [CrossRef]
- Acharya, C.; Yik, J.H.; Kishore, A.; Van Dinh, V.; Di Cesare, P.E.; Haudenschild, D.R. Cartilage oligomeric matrix protein and its binding partners in the cartilage extracellular matrix: Interaction, regulation and role in chondrogenesis. Matrix Biol. 2014, 37, 102–111. [Google Scholar] [CrossRef]
- Adams, J.C.; Lawler, J. The thrombospondins. Cold Spring Harb. Perspect. Biol. 2011, 3, a009712. [Google Scholar] [CrossRef]
- Schulz, J.N.; Nuchel, J.; Niehoff, A.; Bloch, W.; Schonborn, K.; Hayashi, S.; Kamper, M.; Brinckmann, J.; Plomann, M.; Paulsson, M.; et al. COMP-assisted collagen secretion—A novel intracellular function required for fibrosis. J. Cell Sci. 2016, 129, 706–716. [Google Scholar] [CrossRef]
- Koelling, S.; Clauditz, T.S.; Kaste, M.; Miosge, N. Cartilage oligomeric matrix protein is involved in human limb development and in the pathogenesis of osteoarthritis. Arthritis Res. Ther. 2006, 8, R56. [Google Scholar] [CrossRef]
- Posey, K.L.; Coustry, F.; Hecht, J.T. Cartilage oligomeric matrix protein: COMPopathies and beyond. Matrix Biol. 2018, 71–72, 161–173. [Google Scholar] [CrossRef]
- Petersson, I.F.; Boegard, T.; Svensson, B.; Heinegard, D.; Saxne, T. Changes in cartilage and bone metabolism identified by serum markers in early osteoarthritis of the knee joint. Br. J. Rheumatol. 1998, 37, 46–50. [Google Scholar] [CrossRef]
- Saxne, T.; Heinegard, D. Cartilage oligomeric matrix protein: A novel marker of cartilage turnover detectable in synovial fluid and blood. Br. J. Rheumatol. 1992, 31, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Pfander, D.; Cramer, T.; Deuerling, D.; Weseloh, G.; Swoboda, B. Expression of thrombospondin-1 and its receptor CD36 in human osteoarthritic cartilage. Ann. Rheum. Dis. 2000, 59, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.L.; Shen, P.C.; Shiau, A.L.; Jou, I.M.; Lee, C.H.; Wang, C.R.; Teo, M.L.; Wu, C.L. Intraarticular gene transfer of thrombospondin-1 suppresses the disease progression of experimental osteoarthritis. J. Orthop. Res. 2010, 28, 1300–1306. [Google Scholar] [CrossRef]
- Blain, E.J. Involvement of the cytoskeletal elements in articular cartilage homeostasis and pathology. Int. J. Exp. Pathol. 2009, 90, 1–15. [Google Scholar] [CrossRef]
- Delanote, V.; Vandekerckhove, J.; Gettemans, J. Plastins: Versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol. Sin. 2005, 26, 769–779. [Google Scholar] [CrossRef]
- Shinomiya, H. Plastin family of actin-bundling proteins: Its functions in leukocytes, neurons, intestines, and cancer. Int. J. Cell Biol. 2012, 2012, 213492. [Google Scholar] [CrossRef]
- Schwebach, C.L.; Kudryashova, E.; Zheng, W.; Orchard, M.; Smith, H.; Runyan, L.A.; Egelman, E.H.; Kudryashov, D.S. Osteogenesis imperfecta mutations in plastin 3 lead to impaired calcium regulation of actin bundling. Bone Res. 2020, 8, 21. [Google Scholar] [CrossRef]
- Miyakawa, T.; Shinomiya, H.; Yumoto, F.; Miyauchi, Y.; Tanaka, H.; Ojima, T.; Kato, Y.S.; Tanokura, M. Different Ca(2)(+)-sensitivities between the EF-hands of T- and L-plastins. Biochem. Biophys. Res. Commun. 2012, 429, 137–141. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, F.S.; Zillikens, M.C.; Micha, D.; Riessland, M.; Marcelis, C.L.; de Die-Smulders, C.E.; Milbradt, J.; Franken, A.A.; Harsevoort, A.J.; Lichtenbelt, K.D.; et al. PLS3 mutations in X-linked osteoporosis with fractures. N. Engl. J. Med. 2013, 369, 1529–1536. [Google Scholar] [CrossRef]
- Fahiminiya, S.; Majewski, J.; Al-Jallad, H.; Moffatt, P.; Mort, J.; Glorieux, F.H.; Roschger, P.; Klaushofer, K.; Rauch, F. Osteoporosis caused by mutations in PLS3: Clinical and bone tissue characteristics. J. Bone Miner. Res. 2014, 29, 1805–1814. [Google Scholar] [CrossRef]
- Kampe, A.J.; Costantini, A.; Makitie, R.E.; Jantti, N.; Valta, H.; Mayranpaa, M.; Kroger, H.; Pekkinen, M.; Taylan, F.; Jiao, H.; et al. PLS3 sequencing in childhood-onset primary osteoporosis identifies two novel disease-causing variants. Osteoporos. Int. 2017, 28, 3023–3032. [Google Scholar] [CrossRef]
- Kannu, P.; Mahjoub, A.; Babul-Hirji, R.; Carter, M.T.; Harrington, J. PLS3 Mutations in X-Linked Osteoporosis: Clinical and Bone Characteristics of Two Novel Mutations. Horm. Res. Paediatr. 2017, 88, 298–304. [Google Scholar] [CrossRef]
- Laine, C.M.; Wessman, M.; Toiviainen-Salo, S.; Kaunisto, M.A.; Mayranpaa, M.K.; Laine, T.; Pekkinen, M.; Kroger, H.; Valimaki, V.V.; Valimaki, M.J.; et al. A novel splice mutation in PLS3 causes X-linked early onset low-turnover osteoporosis. J. Bone Miner. Res. 2015, 30, 510–518. [Google Scholar] [CrossRef]
- Costantini, A.; Krallis, P.; Kampe, A.; Karavitakis, E.M.; Taylan, F.; Makitie, O.; Doulgeraki, A. A novel frameshift deletion in PLS3 causing severe primary osteoporosis. J. Hum. Genet. 2018, 63, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bian, X.; Cheng, G.; Zhao, P.; Xiang, X.; Tian, W.; Li, T.; Zhai, Q. A novel nonsense variant in PLS3 causes X-linked osteoporosis in a Chinese family. Ann. Hum. Genet. 2020, 84, 92–96. [Google Scholar] [CrossRef]
- Kamioka, H.; Sugawara, Y.; Honjo, T.; Yamashiro, T.; Takano-Yamamoto, T. Terminal differentiation of osteoblasts to osteocytes is accompanied by dramatic changes in the distribution of actin-binding proteins. J. Bone Miner. Res. 2004, 19, 471–478. [Google Scholar] [CrossRef]
- Neugebauer, J.; Heilig, J.; Hosseinibarkooie, S.; Ross, B.C.; Mendoza-Ferreira, N.; Nolte, F.; Peters, M.; Holker, I.; Hupperich, K.; Tschanz, T.; et al. Plastin 3 influences bone homeostasis through regulation of osteoclast activity. Hum. Mol. Genet. 2018, 27, 4249–4262. [Google Scholar]
- Oprea, G.E.; Krober, S.; McWhorter, M.L.; Rossoll, W.; Muller, S.; Krawczak, M.; Bassell, G.J.; Beattie, C.E.; Wirth, B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 2008, 320, 524–527. [Google Scholar] [CrossRef]
- Makitie, R.E.; Niinimaki, T.; Suo-Palosaari, M.; Kampe, A.; Costantini, A.; Toiviainen-Salo, S.; Niinimaki, J.; Makitie, O. PLS3 Mutations Cause Severe Age and Sex-Related Spinal Pathology. Front. Endocrinol. 2020, 11, 393. [Google Scholar] [CrossRef]
- Tsolis, K.C.; Bei, E.S.; Papathanasiou, I.; Kostopoulou, F.; Gkretsi, V.; Kalantzaki, K.; Malizos, K.; Zervakis, M.; Tsezou, A.; Economou, A. Comparative proteomic analysis of hypertrophic chondrocytes in osteoarthritis. Clin. Proteom. 2015, 12, 12. [Google Scholar] [CrossRef]
- Robinson, L.J.; Blair, H.C.; Barnett, J.B.; Soboloff, J. The roles of Orai and Stim in bone health and disease. Cell Calcium 2019, 81, 51–58. [Google Scholar] [CrossRef]
- Robinson, L.J.; Blair, H.C.; Barnett, J.B.; Zaidi, M.; Huang, C.L. Regulation of bone turnover by calcium-regulated calcium channels. Ann. N. Y. Acad. Sci. 2010, 1192, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Fodor, J.; Matta, C.; Olah, T.; Juhasz, T.; Takacs, R.; Toth, A.; Dienes, B.; Csernoch, L.; Zakany, R. Store-operated calcium entry and calcium influx via voltage-operated calcium channels regulate intracellular calcium oscillations in chondrogenic cells. Cell Calcium 2013, 54, 1–16. [Google Scholar] [CrossRef]
- Matta, C.; Fodor, J.; Miosge, N.; Takacs, R.; Juhasz, T.; Rybaltovszki, H.; Toth, A.; Csernoch, L.; Zakany, R. Purinergic signalling is required for calcium oscillations in migratory chondrogenic progenitor cells. Pflugers Arch. 2015, 467, 429–442. [Google Scholar] [CrossRef]
- Dziadek, M.A.; Johnstone, L.S. Biochemical properties and cellular localisation of STIM proteins. Cell Calcium 2007, 42, 123–132. [Google Scholar] [CrossRef]
- Soboloff, J.; Rothberg, B.S.; Madesh, M.; Gill, D.L. STIM proteins: Dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 2012, 13, 549–565. [Google Scholar] [CrossRef]
- Duquette, M.; Nadler, M.; Okuhara, D.; Thompson, J.; Shuttleworth, T.; Lawler, J. Members of the thrombospondin gene family bind stromal interaction molecule 1 and regulate calcium channel activity. Matrix Biol. 2014, 37, 15–24. [Google Scholar] [CrossRef]
- Haberkamp, S.; Olah, T.; Orth, P.; Cucchiarini, M.; Madry, H. Analysis of spatial osteochondral heterogeneity in advanced knee osteoarthritis exposes influence of joint alignment. Sci. Transl. Med. 2020, 12, eaba9481. [Google Scholar] [CrossRef]
- Eckstein, F.; Wirth, W.; Hudelmaier, M.; Stein, V.; Lengfelder, V.; Cahue, S.; Marshall, M.; Prasad, P.; Sharma, L. Patterns of femorotibial cartilage loss in knees with neutral, varus, and valgus alignment. Arthritis Rheum. 2008, 59, 1563–1570. [Google Scholar] [CrossRef]
- Pritzker, K.P.; Gay, S.; Jimenez, S.A.; Ostergaard, K.; Pelletier, J.P.; Revell, P.A.; Salter, D.; van den Berg, W.B. Osteoarthritis cartilage histopathology: Grading and staging. Osteoarthr. Cartil. 2006, 14, 13–29. [Google Scholar] [CrossRef]
- Van Rossom, S.; Wesseling, M.; Smith, C.R.; Thelen, D.G.; Vanwanseele, B.; Dieter, V.A.; Jonkers, I. The influence of knee joint geometry and alignment on the tibiofemoral load distribution: A computational study. Knee 2019, 26, 813–823. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Mukai, S.; Yabumoto, H.; Tarumi, E.; Nakamura, T. Cartilage Degeneration and Alignment in Severe Varus Knee Osteoarthritis. Cartilage 2015, 6, 208–215. [Google Scholar] [CrossRef]
- Brouwer, G.M.; van Tol, A.W.; Bergink, A.P.; Belo, J.N.; Bernsen, R.M.; Reijman, M.; Pols, H.A.; Bierma-Zeinstra, S.M. Association between valgus and varus alignment and the development and progression of radiographic osteoarthritis of the knee. Arthritis Rheum. 2007, 56, 1204–1211. [Google Scholar] [CrossRef] [PubMed]
- Vilim, V.; Lenz, M.E.; Vytasek, R.; Masuda, K.; Pavelka, K.; Kuettner, K.E.; Thonar, E.J. Characterization of monoclonal antibodies recognizing different fragments of cartilage oligomeric matrix protein in human body fluids. Arch. Biochem. Biophys. 1997, 341, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Yu, X.P.; Zhang, Y.; Tian, Q.; Song, H.; Mucignat, M.T.; Perris, R.; Samuels, J.; Krasnokutsky, S.; Attur, M.; et al. Enhanced COMP catabolism detected in serum of patients with arthritis and animal disease models through a novel capture ELISA. Osteoarthr. Cartil. 2012, 20, 854–862. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wong, M.; Siegrist, M.; Goodwin, K. Cyclic tensile strain and cyclic hydrostatic pressure differentially regulate expression of hypertrophic markers in primary chondrocytes. Bone 2003, 33, 685–693. [Google Scholar] [CrossRef]
- Lawler, J. Thrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growth. J. Cell Mol. Med. 2002, 6, 1–12. [Google Scholar] [CrossRef]
- Gelse, K.; Klinger, P.; Koch, M.; Surmann-Schmitt, C.; von der Mark, K.; Swoboda, B.; Hennig, F.F.; Gusinde, J. Thrombospondin-1 prevents excessive ossification in cartilage repair tissue induced by osteogenic protein-1. Tissue Eng. Part A 2011, 17, 2101–2112. [Google Scholar] [CrossRef]
- Qian, X.; Tuszynski, G.P. Expression of thrombospondin-1 in cancer: A role in tumor progression. Proc. Soc. Exp. Biol. Med. 1996, 212, 199–207. [Google Scholar] [CrossRef]
- Qian, W.; Zhu, S.; Sobolev, A.Y.; Wek, R.C. Expression of vaccinia virus K3L protein in yeast inhibits eukaryotic initiation factor-2 kinase GCN2 and the general amino acid control pathway. J. Biol. Chem. 1996, 271, 13202–13207. [Google Scholar] [CrossRef]
- Bader, D.L.; Salter, D.M.; Chowdhury, T.T. Biomechanical influence of cartilage homeostasis in health and disease. Arthritis 2011, 2011, 979032. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.J.; Hardin, J.A.; Cobelli, N.J.; Sun, H.B. Mechanotransduction and cartilage integrity. Ann. N. Y. Acad. Sci. 2011, 1240, 32–37. [Google Scholar] [CrossRef]
- Varady, N.H.; Grodzinsky, A.J. Osteoarthritis year in review 2015: Mechanics. Osteoarthr. Cartil. 2016, 24, 27–35. [Google Scholar] [CrossRef]
- Lee, W.; Guilak, F.; Liedtke, W. Role of Piezo Channels in Joint Health and Injury. Curr. Top. Membr. 2017, 79, 263–273. [Google Scholar] [PubMed]
- Mobasheri, A.; Matta, C.; Uzieliene, I.; Budd, E.; Martin-Vasallo, P.; Bernotiene, E. The chondrocyte channelome: A narrative review. Joint Bone Spine 2019, 86, 29–35. [Google Scholar] [CrossRef]
- Guilak, F.; Zell, R.A.; Erickson, G.R.; Grande, D.A.; Rubin, C.T.; McLeod, K.J.; Donahue, H.J. Mechanically induced calcium waves in articular chondrocytes are inhibited by gadolinium and amiloride. J. Orthop. Res. 1999, 17, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Frischauf, I.; Schindl, R.; Derler, I.; Bergsmann, J.; Fahrner, M.; Romanin, C. The STIM/Orai coupling machinery. Channels 2008, 2, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Wang, X.; Qiu, X.; Wu, Z.; Gao, B.; Liu, L.; Liang, G.; Zhou, H.; Yang, X.; Peng, Y.; et al. Collagen type II suppresses articular chondrocyte hypertrophy and osteoarthritis progression by promoting integrin beta1-SMAD1 interaction. Bone Res. 2019, 7, 8. [Google Scholar] [CrossRef]
- Chen, H.; Deere, M.; Hecht, J.T.; Lawler, J. Cartilage oligomeric matrix protein is a calcium-binding protein, and a mutation in its type 3 repeats causes conformational changes. J. Biol. Chem. 2000, 275, 26538–26544. [Google Scholar] [CrossRef] [PubMed]
- Tanaka-Kamioka, K.; Kamioka, H.; Ris, H.; Lim, S.S. Osteocyte shape is dependent on actin filaments and osteocyte processes are unique actin-rich projections. J. Bone Miner. Res. 1998, 13, 1555–1568. [Google Scholar] [CrossRef]
- Makitie, R.E.; Costantini, A.; Kampe, A.; Alm, J.J.; Makitie, O. New Insights into Monogenic Causes of Osteoporosis. Front. Endocrinol. 2019, 10, 70. [Google Scholar] [CrossRef]
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef]
- Martino, F.; Perestrelo, A.R.; Vinarsky, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- Schwebach, C.L.; Kudryashova, E.; Kudryashov, D.S. Plastin 3 in X-Linked Osteoporosis: Imbalance of Ca(2+)-Dependent Regulation Is Equivalent to Protein Loss. Front. Cell Dev. Biol. 2020, 8, 635783. [Google Scholar] [CrossRef] [PubMed]
- DiCesare, P.E.; Morgelin, M.; Carlson, C.S.; Pasumarti, S.; Paulsson, M. Cartilage oligomeric matrix protein: Isolation and characterization from human articular cartilage. J. Orthop. Res. 1995, 13, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mählich, D.; Glasmacher, A.; Müller, I.; Oppermann, J.; Grevenstein, D.; Eysel, P.; Heilig, J.; Wirth, B.; Zaucke, F.; Niehoff, A. Expression and Localization of Thrombospondins, Plastin 3, and STIM1 in Different Cartilage Compartments of the Osteoarthritic Varus Knee. Int. J. Mol. Sci. 2021, 22, 3073. https://doi.org/10.3390/ijms22063073
Mählich D, Glasmacher A, Müller I, Oppermann J, Grevenstein D, Eysel P, Heilig J, Wirth B, Zaucke F, Niehoff A. Expression and Localization of Thrombospondins, Plastin 3, and STIM1 in Different Cartilage Compartments of the Osteoarthritic Varus Knee. International Journal of Molecular Sciences. 2021; 22(6):3073. https://doi.org/10.3390/ijms22063073
Chicago/Turabian StyleMählich, Daniela, Anne Glasmacher, Ilka Müller, Johannes Oppermann, David Grevenstein, Peer Eysel, Juliane Heilig, Brunhilde Wirth, Frank Zaucke, and Anja Niehoff. 2021. "Expression and Localization of Thrombospondins, Plastin 3, and STIM1 in Different Cartilage Compartments of the Osteoarthritic Varus Knee" International Journal of Molecular Sciences 22, no. 6: 3073. https://doi.org/10.3390/ijms22063073
APA StyleMählich, D., Glasmacher, A., Müller, I., Oppermann, J., Grevenstein, D., Eysel, P., Heilig, J., Wirth, B., Zaucke, F., & Niehoff, A. (2021). Expression and Localization of Thrombospondins, Plastin 3, and STIM1 in Different Cartilage Compartments of the Osteoarthritic Varus Knee. International Journal of Molecular Sciences, 22(6), 3073. https://doi.org/10.3390/ijms22063073





