Abstract
Anthocyanins are natural water-soluble pigments that are important in plants because they endow a variety of colors to vegetative tissues and reproductive plant organs, mainly ranging from red to purple and blue. The colors regulated by anthocyanins give plants different visual effects through different biosynthetic pathways that provide pigmentation for flowers, fruits and seeds to attract pollinators and seed dispersers. The biosynthesis of anthocyanins is genetically determined by structural and regulatory genes. MYB (v-myb avian myeloblastosis viral oncogene homolog) proteins are important transcriptional regulators that play important roles in the regulation of plant secondary metabolism. MYB transcription factors (TFs) occupy a dominant position in the regulatory network of anthocyanin biosynthesis. The TF conserved binding motifs can be combined with other TFs to regulate the enrichment and sedimentation of anthocyanins. In this study, the regulation of anthocyanin biosynthetic mechanisms of MYB-TFs are discussed. The role of the environment in the control of the anthocyanin biosynthesis network is summarized, the complex formation of anthocyanins and the mechanism of environment-induced anthocyanin synthesis are analyzed. Some prospects for MYB-TF to modulate the comprehensive regulation of anthocyanins are put forward, to provide a more relevant basis for further research in this field, and to guide the directed genetic modification of anthocyanins for the improvement of crops for food quality, nutrition and human health.
1. Introduction
Anthocyanins are abundant natural water-soluble plant pigments in their glycosylated form [1,2]. Anthocyanin biosynthesis in plants is a part of the phenylpropanoid pathway, which produces flavonoids through flavonoid branches. The content and composition of anthocyanins results in a series of orange, red, purple and blue colors in vegetative and reproductive plant organs. Red anthocyanins most likely evolved along with mosses as plants became adapted to the land about 450 million years ago (mya). The blue pigments evolved later, about 300 mya along with the gymnosperms [1]. Anthocyanins or their orthologs are have been found in some mosses [3], ferns [4] and some fungi [5], but not in algae [1]. Dating back to the early 20th century, anthocyanins have been studied by humans [6]. Anthocyanins are now found in 73 taxa and 27 families of plants [6]. The anthocyanins content varies greatly depending on the variety, species, season, climate, 87and stage of plant development. In higher plants, the rich colors of the anthocyanins attract animals for pollination and seed transmission but may also act in the wild as warning coloration [7,8]. Anthocyanins also regulate the negative effects of the external environment as protection against cold or drought [9,10]. Anthocyanin-rich plants are used in landscape gardens and enrich the natural landscape. There are anthocyanins in leaves, stems, roots, flowers and fruits of diverse plant species such as grape (Vitis vinifera), parsley (Petroselinum crispum), eggplant (Solanum melongena), apple (Malus domestica), strawberry (Fragaria vesca), mulberry (Morus alba), and petunia (Petunia hybrida) [11,12,13,14,15,16,17]. Anthocyanins typically accumulate in the vacuoles and their biosynthesis is mediated by many enzymes in the phenylpropanoid metabolic pathways. Anthocyanin synthesis are mainly controlled by two types of genes, one is the anthocyanin biosynthesis structural gene, which can encode the enzyme of anthocyanin biosynthesis pathway, and the other is the regulatory gene, which has three types of transcription factors (TFs): MYB (v-myb avian myeloblastosis viral oncogene homolog) protein, bHLH (basic helix-loop-helix) protein, WD40 (WD-40 has a scaffolding function) protein [18]. In monocotyledon (maize, Zea mays [19], rice, Oryza sativa [20]), MYB-TFs regulate anthocyanin biosynthesis enzymes (such as CHS, F3H and DFR, LDOX, BAN/ANR, UFGT) together with other TFs [21]. In dicotyledon (Arabidopsis thaliana [22], apple, M. domestica [12]), anthocyanin synthases are divided into two classes and the TFs are different [23]. The CHS, CHI, F3H and F3‘H genes are the early biosynthetic genes (EBGs) in the anthocyanin pathway. These genes bind directly and regulate by MYB-TFs [23]. DFR, LDOX, BAN/ANR, and UFGT, are the late biosynthesis genes (LBGs) [24,25], are regulated by the MBW-TF ternary protein complex of MYB-bHLH-WD40 that controls the MBW complex and the downstream accumulation of anthocyanins [26,27,28,29]. The three most common anthocyanins are: delphinidin-3-glycoside (blue/purple) cyanidin-3-glucoside (brick red/magenta) and pelargonidin-3-glycoside (orange/red). Other important anthocyanins are: malvidin, peonidin and leucocyanidin [30,31]. For many anthocyanins, the color depends on the pH. Cyanidin is red/violet at pH 7-8 changing to blue at higher pH. Delphinidin at high pH is a strong blue color that is common in flowers. Peonidin is cherry red at low pH but deep blue at pH8 [32]. Meanwhile, anthocyanins are considered beneficial for human health; presumably as antioxidants that reduce the abundance of free radicals (reactive oxygen species (ROS)), which may de-lay aging, ameliorate cardiovascular and neurogenerative disease, as well as modulating gut microbiota [7,33], however it has been argued that direct evidence of the benefits of dietary supplements are lacking [34].
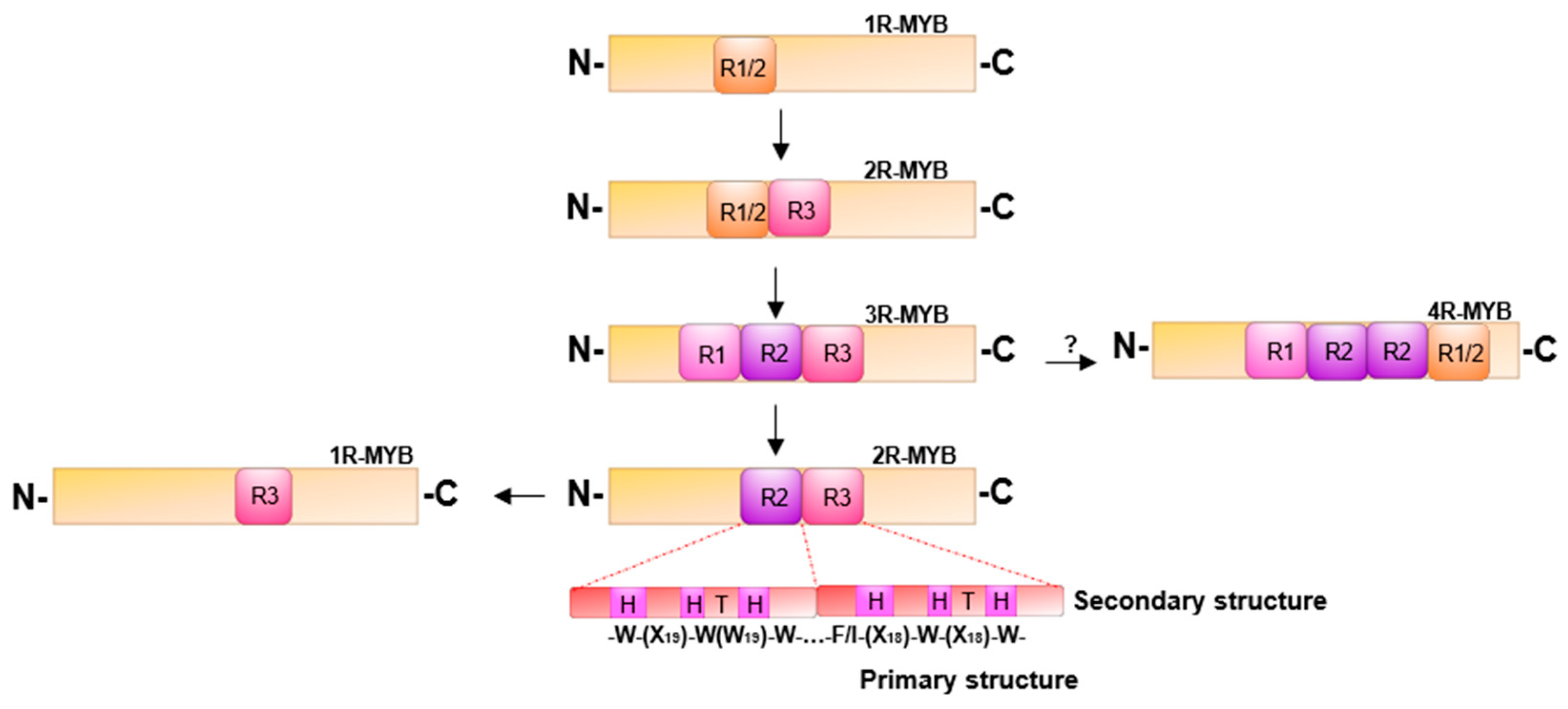
In plants, MYB transcription factors (TFs) are one of the most important classes of transcriptional regulators in the plant metabolic network, controlling secondary metabolism, development, signal transduction and resistance to biotic and abiotic stresses [35]. The MYB-TF family is one of the largest in plants. Anthocyanin are secondary metabolites of flavonoids modify with are a subclass of flavonoids, the anthocyanin biosynthesis as well as that of other phenylpropanoids are regulated by MYB-TFs. For example, MdMYB10a of apple (M. domestica) when overexpressed in Nicotiana tabacum upregulates the biosynthesis of anthocyanins [36], through an increase in the expression levels of structural genes in anthocyanin biosynthesis, thereby activating a hierarchical cascade of the downstream regulatory and structural genes for anthocyanins. Similar results were reported for LdMYB6 from lily (Lilium davidiivar), peach PpMYB7 (Prunus persica), Arabidopsis AtMYB75 and AtMYB90 and eggplant SmMYB113 (Solanium melongena) [37,38,39,40]. In a transient transactivation experiment with strawberry (F. vesca), FvMYB10 co-expression with FvbHLH33 strongly activated the AtDFR, FvDFR and FvUFGT promoters of the structural genes of anthocyanin biosynthesis. Knockouts of FvMYB10 or FVbHLH33 significantly reduced the activity of the AtDFR promoter in tobacco [41]. MYB proteins contain an imperfect repeat sequence ® which has varying forms in the conserved DNA domain. MYB-TFs are classified into four types based on the number of repeats (1R, 2R, 3R, 4R) and the variation in the sequences of the repeats. The MYB that is the major activator of anthocyanin biosynthesis comes from the two-repeat class (2R) containing an R2 and an R3 repeat (R2R3-MYB) (Figure 1). Variation in relative abundance and specificity of anthocyanins represents one of the most common evolutionary changes in flower color. MYB-TFs may also act as antagonistic repression of anthocyanin enrichment [42]. The first suppressor of anthocyanins identified was the AmMYB308 gene of snapdragon (Antirrhinum majus) [43]. Subsequently, repressors of anthocyanin synthesis were found in strawberry FaMYB1 (Fragaria × ananassa), Arabidopsis AtMYB4, petunia PhMYBx (Petunia hybrida), peach PpMYB17-20 (P. persica), popular PtrMYB182 (Populus tremula × Populus), and apple MdMYB16 (M. domestica) [11,44,45,46,47,48]. The anthocyanins biosynthesis pathway in the plant concerned is inhibited, resulting in a decrease in the level of anthocyanin in vivo when they were expressed. The repressors identified are in the subgroups of R2R3-MYBs and R3-MYBs. When the putative suppressors were expressed in tobacco, the structural genes of the anthocyanin biosynthesis were inhibited, reducing the abundance of anthocyanin pigments. Environmental factors also affect anthocyanin accumulation [27,49]. In A. thaliana, AtPAP1 is regulated by high light and sucrose levels [50,51], which activate anthocyanin synthesis. During storage of kiwifruit (Actinidia chinensis), low temperature results in the upregulation of AcMYBA1-1 and AcMYB5-1 and induces anthocyanin biosynthesis [51].

Figure 1.
A proposed phylogenetic classification of MYB (v-myb avian myeloblastosis viral oncogene homolog) transcription factors (TFs) (arrows indicate the predicted direction of evolution [52,53]. Different plant protein types vary depending on the number of adjacent MYB duplications (R).
Predictive computational analysis has led to new uses for MYB-TFs by changing the amino acid sequence of the MYB domain to expand its regulatory ability. For example, the anthocyanin produced by the MdMYB10 gene can be used as a visible selection marker for the transformation of apples, strawberries and potatoes to replace antibiotic selection for kanamycin resistance [54] by changing the amino acids of the R3 domain of Populus tomentosa MYB6 to generate a MYB6-R3m mutant [55]. It is of significant value to understand more about the regulation of anthocyanins by MYB-TFs because of the commercial value of plant color in agriculture. Here we summarize the recent studies of the regulation of anthocyanin synthesis by MYB-TFs and analyze the specificity of positive and negative regulation of MYBs to lay a foundation for more comprehensive work in the future on the functional mechanisms and evolution of this important class of transcriptional regulators. With sufficient information on the mechanism of MYB-target DNA interactions, designing new MYB based regulators could become an important tool to design new specific interactions for synthetic metabolism and biology.
2. Structure and Evolution of MYB
A MYB-TF (v-myb) was initially identified as an oncogene in an avian myeloblastosis retrovirus (AMV)) that produced leukemia in chicken embryos [56,57]. Initially, Klempnauer et al. found the sequence differences in the v-myb gene and its homolog c-myb, leading to inferred effects on their protein structures [56]. Paz-Ares et al. (1988) discovered a MYB-TF as the C1 gene of wild-type maize (Z. mays) and described it as the first regulatory transcription factor identified in plants [58]. Since then, 63,028 MYB-TF genes have been identified and officially registered in the National Center for Biological Information (NCBI- GeneBank).
A hydrophobic amino acid core consisting of a series of tryptophan residues specifically binding to the main groove of the DNA double helix, form a conserved MYB DNA binding domain. The MYB domain usually binds to a specific DNA sequence (C/TAACG/TG). The three conserved tryptophan residues are separated by 18 to 19 amino acids form a secondary structure [35]. The MYB domain contains three incomplete repeats (R) with approximately 52 amino acid residues at the N-terminus of the protein (Figure 1). Each repeat encodes three helices. The second and third helix form a helix-turn-helix segment that binds to the major groove of the target DNA [52,59,60]. Once bound, transcription can be initiated to activate a hierarchical regulatory network or the MYB-TF may interact with other TFs to regulate the expression of target genes. Animal MYB proteins diverged from a common ancestor, while the MYB proteins in plants are a polyphyletic group related only to a helix-turn-helix ‘‘MYB-box’’ DNA-binding motif [59]. Based on the number of MYB repeats (R), the (1R, 2R, 3R, 4R) MYBs can be divided into four classes. An evolutionary relationship of the four MYB repeat types has been proposed (Figure 1) [53]. The first category has a repetitive sequence contained in a single protein, often with homologous structural domain proteins in combination with DNA, namely, R1/2-MYB. The R1/R2 repeat is thought to be an antecedent that gave rise to the R1 and R2 repetitive sequences by duplication and divergence. The R3-MYB repeat is found as repressor. StMYB1 (from potato) was the first MYB protein found to contain only one MYB domain structure identified in plants [33,52]. The second class consists of two R repeats (R1/2R3-MYB and R2R3-MYB), and both R2 and R3 are required for sequence-specific binding, the C-terminal helix of each repeat being the recognition helix for DNA binding [53]. It has been proposed that R2R3-MYB evolved from an R1R2R3-MYB after the loss of the R1 repeat sequence [53,60,61]. Therefore, members of the superfamily of MYB proteins should be viewed as related principally by their ability to bind DNA, rather than on the basis of their physiological functions [53]. Due to the distinct specific binding ability of different MYB proteins and the differences in target gene recognition sites, there is great flexibility of R2R3-MYB binding sites, leading to a wide range of regulatory roles and functions throughout the process of plant growth, metabolism, development, behavior, or adaptation. The third type of MYB is the three repeats MYB, which is a member of the R1R2R3-MYB class. R1 is more conserved than R2 and R3 in type 3R-MYBs, so it is suggested that 3R-MYBs evolved into R1/2R3-MYBs [60]. Current reports indicate that its main function is to participate in cell cycle and protein control and cell differentiation [59,61]. The fourth class, 4R MYBs, which members contain four R1/R2-like repeats (R1R2R2R1/2) [52]. The 4R-MYB has the longest peptide chain (1350 amino acids) consisting of 38,711 bases in flax [62]. The functional expression of 4R-MYB is still under investigation, and its regulatory function is unclear. Additional variation that is found within types of repeats provides further classification into subtribes.
3. Anthocyanin Biosynthesis Pathway
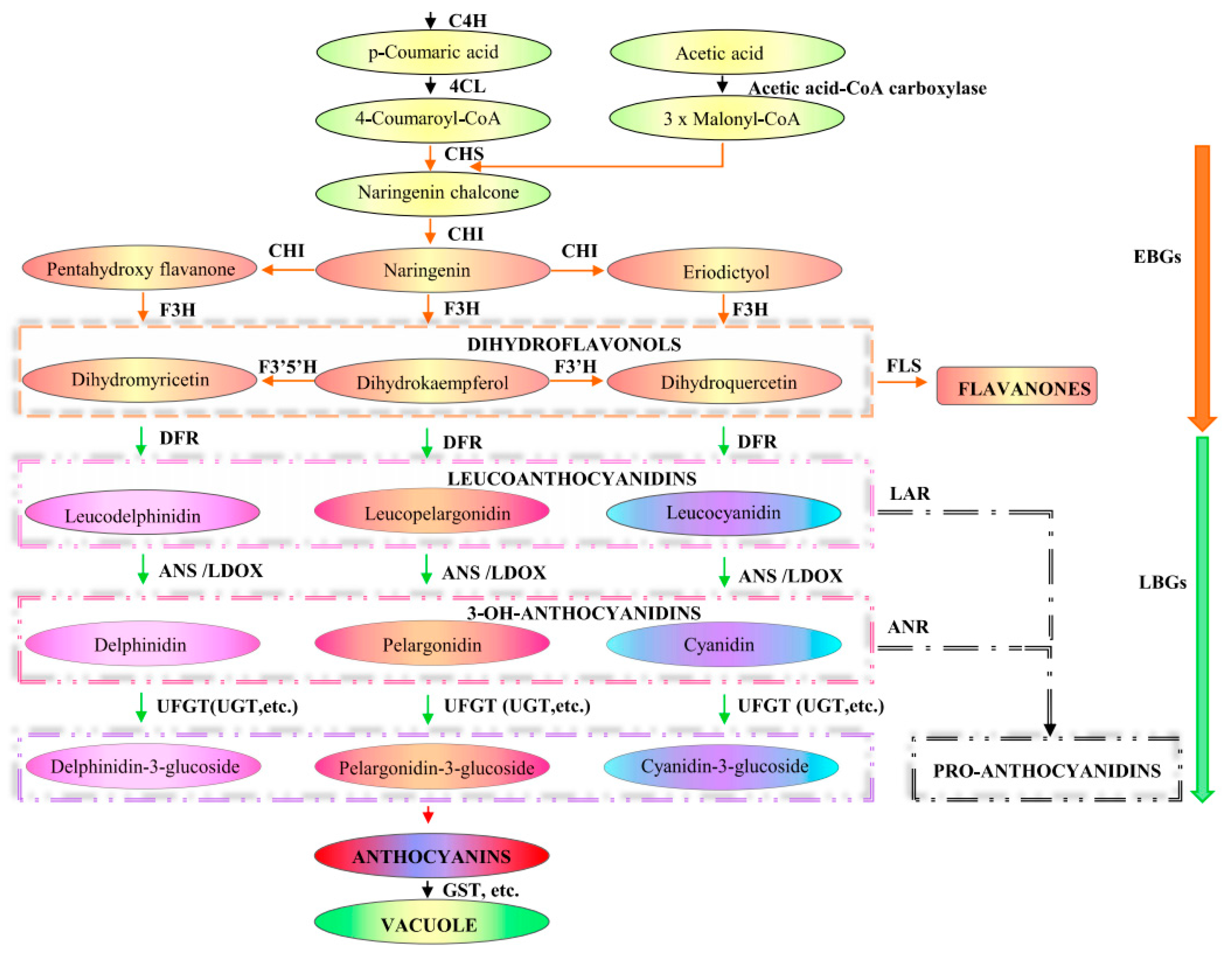
Anthocyanins are highly abundant and exceptionally diverse in structure and function among plant phenolic metabolites [63]. Walheldale (1911) explored the process of anthocyanin formation through the phenylpropionic acid and flavonoid pathways [6]. The basic anthocyanin structure consists of two benzene rings and three connecting carbon atoms (C6-C3-C6) [64]. The main metabolic pathways of anthocyanins are mapped in Figure 2 [23,26,65]. Anthocyanin biosynthesis is mediated and regulated by a variety of enzymes to regulate synthesis. The EBGs (CHS, CHI, F3H and F3‘H) and LBGs (DFR, LDOX, BAN/ANR, and UFGT) were initially categorized accordingly to their coordinate expression in response to environmental cues, such as light, at distinct developmental stages, in a species-dependent manner [23,26,27,28,29]. The biosynthesis does not follow a strictly linear pathway but is better described as a metabolic grid, with many branches and alternative metabolic routes. The biosynthesis begins with the deamination of phenylalanine by phenylalanine ammonia lyase (PAL) to cinnamic acid, followed by the hydroxylation of cinnamic acid by cinnamate 4-hydroxylase (C4H) to 4-coumaric acid. 4-coumaric acids activated by 4-cinnamate-CoA ligase (4CL) to 4-coumaryl CoA. Chalcone synthase (CHS) is the key reaction involved in flavonoid biosynthesis [66]. In plants 4-coumaryl CoA is condensed with 3 molecules of malonyl-CoA to form a new aromatic ring system of polytetraketone [67]. As one of the important branches of flavonoid and anthocyanin synthesis, flavonone 3-hydroxylase (F3H), with a 4,5,7-trihydroxyflavanone-catalyzed reaction produces dihydroflavonol [68]. Then, dihydroflavonol is catalyzed by dihydrofavonol 4-reductase (DFR) to synthesize colorless anthocyanins, which are the basic skeletons of flavonoids and anthocyinodins, which are the basic skeletons of flavonoids and anthocyanins [69]. Finally, the colorless dihydroflavonols are converted into colored anthocyanins by anthocyanidin synthase (ANS) [67] and flavonoid 3-O-glucosyltransferase (UFGT), which transforms the colorless dihydrogen flavonols into colored anthocyanins [70]. Subsequently, after UDP -glucosyltransferase (UGT) modification, such as methylation, different anthocyanins are generated and collected into vacuoles for storage under the action of glutathione S-transferase (GST) transporters [71]. Finally, colorless dihydroflavanols are converted into colored anthocyanins by anthocyanin synthase ANS [67]. After modification, such as glycation (Flavonoid 3-glucosyltransferase [UFGT]) and other UGT [70] sequestered into vacuoles for storage under the action of Glutathione S-transferase (GST) transporters [71].

Figure 2.
Model of the anthocyanin biosynthetic pathway. Enzymes are indicated in bold uppercase letters [23,26,65]. PAL, Phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, Cinnamic acid-4-hydroxylase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, Flavanone 3-hydroxylase; F3′H, Flavonoid 3′-hydroxylase; F3′5′H, Flavonoid 3′,5′-hydroxylase; ANS, Anthocyanidin synthase; LDOX, Leucoanthocyanidin dioxygenase; UFGT, UDP flavonoid 3-O-glucosyltransferase; UGT, UDP -glucosyltransferase; GST, Glutathione S-transferase; FLS, Flavonol synthase; ANR, Anthocyanidin reductase; EBGs, early biosynthetic gene; LBGs, late biosynthetic gene.
4. Transcriptional Regulation Mechanism of Anthocyanins by MYB-TFs
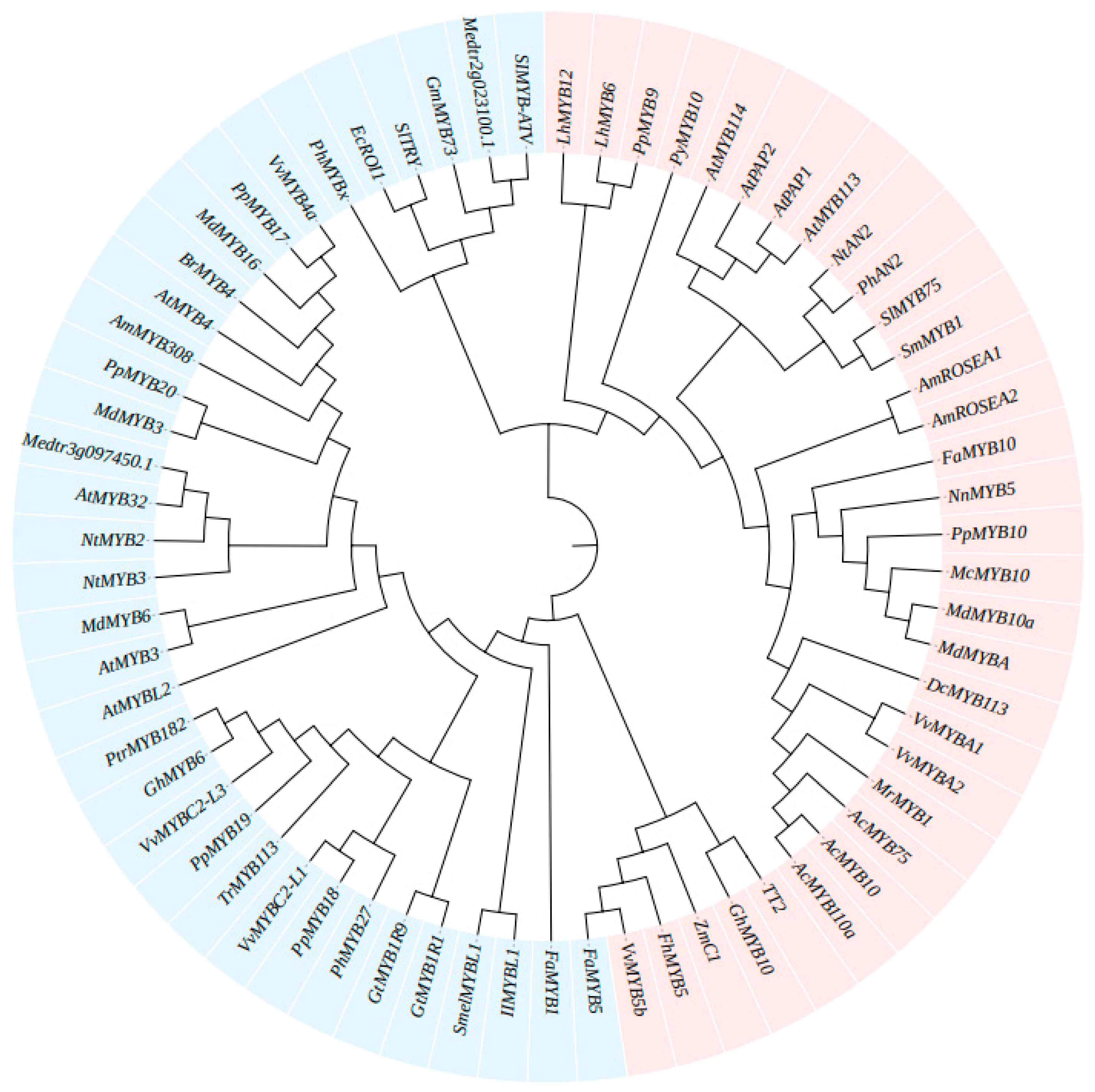
R2R3-MYBs and R3-MYBs play crucial roles as transcriptional regulators in the production of anthocyanins. The recent view of regulation of anthocyanin synthesis includes control by single TFs, as well as structured regulation by an anthocyanin biosynthesis network [20,72,73] in which the ternary MBW complex of transcription factors, involving a variety of interactions and mechanisms, plays a key role in anthocyanin and proanthocyanin accumulation and inhibition. This network promotes the synthesis of anthocyanin through positive regulation and inhibits or reduces anthocyanin accumulation by negative regulation [74,75,76]. Gene homologs related to anthocyanin biosynthesis regulated by MYB-TFs were found by sequence searches and a proposed evolutionary tree was constructed (Table 1 and Figure 3).

Table 1.
MYB-TFs in Regulation of Anthocyanins Biosynthesis in Plants.

Figure 3.
Phylogenetic tree of the regulation of anthocyanin biosynthesis by MYB genes. The tree was constructed based on the entire protein sequences using MEGA 6 software. Blue represents the antagonistic effects of MYB factors in the anthocyanin biosynthetic pathway, and red represents the MYBs as activators to promote the biosynthesis of anthocyanin.
4.1. Mechanisms of Positive Regulation
Most of the genes with positive regulatory effects on anthocyanin biosynthesis encoded R2R3-MYB factor. Sequence motifs encoding three potential alpha helixes are found in each incomplete repeated (R1, R2, R3), of which the second and third helixes could form a helix-turn-helix (HTH) structure [110]. Structural studies of the MYB DNA-binding domain interacting with a double DNA strand reveal that the third helices of both the R2 and R3 repeats are considered recognition helices, which bind directly to specific cis-acting regulatory DNA elements [110]. The third helix of R2 and R3 repeating sequences is the key to modulating the binding affinity of DNA, and the variation of their repeating sequences will affect the specific binding of R2R3-MYB to DNA [111,112]. The Leu→Glu substitution in the third helix of the R2 repeat of MYB protein P1 in maize causes a loss of DNA binding [113]. The DNA binding domain is conserved within the separate classes of TF and contain trans-acting motifs that bind to the cis-acting sequences of target gene promoters. TFs are identified by their distinct DNA binding domains [114]. Therefore, the MYB-TF is able to bind directly to the promoter of the gene key enzyme gene for anthocyanin biosynthesis, affecting the final enrichment of anthocyanin. A database of nucleotide sequence motif of cis-acting elements in plants was discovered using PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/) (accessed on 1 February 2021). Flavonoid-related MYB binding elements were identified, including MYB26PS [115], MYB core [116], MYBPLANT [43], and MYBPZM [117]. In tomato (S. lycopersicum), SlMYB75 is able to directly bind to the MYBPLANT and MYBPZM -regulatory elements and to activate the promoters of the structure genes, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits [118]. The MYB-TFs bind indirectly to promoter-specific cis-acting elements by interacting with other DNA-binding proteins [119]. The MYB10 can regulate skin color of apples varieties, which has been shown to auto-activate its own expression by binding a cis-element in its own promoter, then activating the expression of anthocyanin structural genes [114]. Synthetic promoter engineering may find targeted enhancers more precisely through the presence of cis-regulatory elements.
4.1.1. Expression Patterns of MYB Activators
Expression patterns of MYB activators vary in vegetative and reproductive plant tissues. MdMYB110a controls ‘red meat’ coloring in red heart apples, and this pigment is only expressed in the fruit cortex late in fruit development [12], which indicates that MYB-TFs have different targeting effects on anthocyanin pigment deposition in different tissues. PhAN2 is responsible for flower pigmentation in Petunia hybrida. The apparent orthologs in (Lilium davidii), LdMYB12 and LdMYB6 showed high amino acid sequence similarity with PhAN2. LdMYB12 is involved in anthocyanin accumulation in sepals and petals (perianth) sheets, filaments and stigma, while LdMYB6 is mainly responsible for perianth sheet spots and dominant expression in flowers and bulbs [120]. Therefore, anthocyanin deposition in different parts of the plant is influenced by different gene activators. In kyoho grape (Vitis labruscana), MybA gene expression increases strongly with the commencement of coloring and berry softening and is detected only in berry skin and flesh. When MYBA was introduced into somatic embryo of grape, led to reddish-purple spots in non-colored embryos [70]. In nectarine (P. persica), PpMYB10.1, PpMYB10.4, and PpMYB9/PpMYB10 were positive regulators of anthocyanin accumulation in fruits, leaves and flowers, respectively [25]. The expression level brought about by activators also depends on the predetermined level of anthocyanin in different genotypes [109,121]. In white clover (Trifolium repens), the pigmentation of red stripes on the leaves is regulated by the R2R3-MYB family and anthocyanins are found at the R and V pigmentation sites [107]. Moreover, the atv (atroviolacium), Aft (Aubergine fruit) and Abg (Aubergine) loci of tomato can also promote the pigmentation resulting from anthocyanins in fruits. After binding with Aft or Abg loci, atv loci can significantly increase the anthocyanin content in cultivated tomato fruit [104,122]. But in Arabidopsis, activation tagging induced to identification of a bright-purple mutant (pap1-D) in which overexpression of a MYB factor led to massive accumulation of anthocyanins in the entire plant [123,124]. By linking with enhancer sequences, indicates that activation tagging can be used to overcome the stringent genetic controls regulating the developmental accumulation of specific natural products [123]. According to this situation, anthocyanin color vision may be used to isolate candidate regulatory genes and express easily screened marker genes under the control of promoters from genes encode enzymes involved in the biosynthesis of natural products of interest [123].
4.1.2. Regulation of the Structural Gene Network
MYB-TFs regulate anthocyanin structural genes that then activate anthocyanin biosynthesis. In the R2R3-MYB-TFs, the highly conserved N-terminus binds to the bHLH TF and WD40 repeat (WDR) to form the MYB-bHLH-WD40 (MBW) complex. The accepted view is that the MYB protein determines the specific activation of target genes [30]. In painter’s pallet (Anthurium andraeanum), AaMYB2 is considered to be the R locus ortholog that controls corolla color inheritance. As a potential target of coding TFs, the locus is involved in the joint expression of AaCHS, AaF3H and AaANS. AaMYB2 is highly expressed in the spathes (sheathing bract) of red, pink and purple cultivars but hardly detected in the spathes of white and green cultivars [125]. As a late synthesis gene of anthocyanin biosynthesis, anthocyanidin synthase (ANS) is an important target gene. In a study of binding specificity of the kiwifruit (Actinidia chinensis) AcMYB75 protein, the promoter of ANS was identified in a yeast single hybrid experiment [78]. When the MYB-TF bound to the promoters of structural genes in anthocyanin biosynthesis pathway, the expression of structural genes was increased so that the entire downstream network of anthocyanin biosynthesis was activated. This illustrates that the expression of structural genes in the anthocyanin synthesis pathway was positively correlated with the accumulation of anthocyanin [126]. Activators can combine with other TFs to form complexes that participate in the expression of anthocyanin structural genes and stimulate the anthocyanin synthesis system. In addition to direct action on target gene sites, expression of anthocyanin MYB-TFs may be controlled by binding with other TFs. The R3 domain of positive regulatory factor shows a conserved bHLH binding motif, [D/E]Lx2[R/K]x3Lx6Lx3R (Figure S1). R2R3-MYBs usually interact with bHLH1 and WD40 to create an MBW activation complex, thereby augmenting the expression of bHLH2 and the structural genes to promote the accumulation of anthocyanin [44,106,112]. In tomato (S. lycopersicum)), the Aft (Anthocyanin fruit: MYB) protein interacts with SlJAF13 (bHLH) and SlAN11 (WDR) to form an MBW-activated complex and activate the expression of SlAN1 (bHLH). With the formation of a core MBW-activated complex of SlAN1, Aft and SlAN11, the expression of the SlAN1 gene and most of the anthocyanin structural genes were activated, and anthocyanin pigments were enhanced in fruits [127]. MrMYB1 from Chinese bayberry (Myricarubra Chinese) when genetically transformed into N. tabacum, the activation of the NtDFR promoter by MrMYB1 depends on the expression of bHLH TFs. When MrMYB1 is co-expressed with bHLH, the formation of anthocyanins is promoted [96]. Similarly, the heterologous expression of MYBA from tobacco can independently regulate AtDFR in grape hyacinth (Muscari armeniacum), evidenced by strong purple anthocyanin pigment accumulated in leaves, petals, anthers and calyx, while MaAN2 relies on MabHLH1 to control the expression of AtDFR to promote the synthesis of anthocyanin [128]. In orchids (Phalaenopsis equestris), PeMYB2, PeMYB11, and PeMYB12 can independently activate PeDFR expression, when the presence of PeBHLH1 the red pigmentation of butterfly orchids can be increased on this basis [129]. Similar results were found in peach (P. persica) [130], which indicates that the bHLH TF does not act as a restrictive regulator of anthocyanin accumulation in some plants but acts as a cofactor that promotes the biosynthesis of anthocyanin. In tree peony (Paeonia × suffruticosa), PsMYB12 interacts with bHLH and WD40 in the protein complex to directly activate the expression of PsCHS, showing pigmentation in the form of a blotch, while the loss of CHS activity leads to albino flowers [75,131,132]. The activity of the MdDFR promoter was enhanced significantly when MdMYB10 was co-transformed with apple bHLHs [133]. Meanwhile, Pr-D (Pr: Purple, encoded a R2R3-MYB-TF) overexpressed in wild cabbage (Brassica oleracea) it activates bHLH TFs acting on BoF3H, BoDFR, and BoANS [36,134]. Therefore, many lines of evidence support the conclusion that MYB activators control the anthocyanin synthesis pathway by regulating the expression of anthocyanin structural and the control of anthocyanin accumulation.
4.2. Negative Regulatory Mechanism
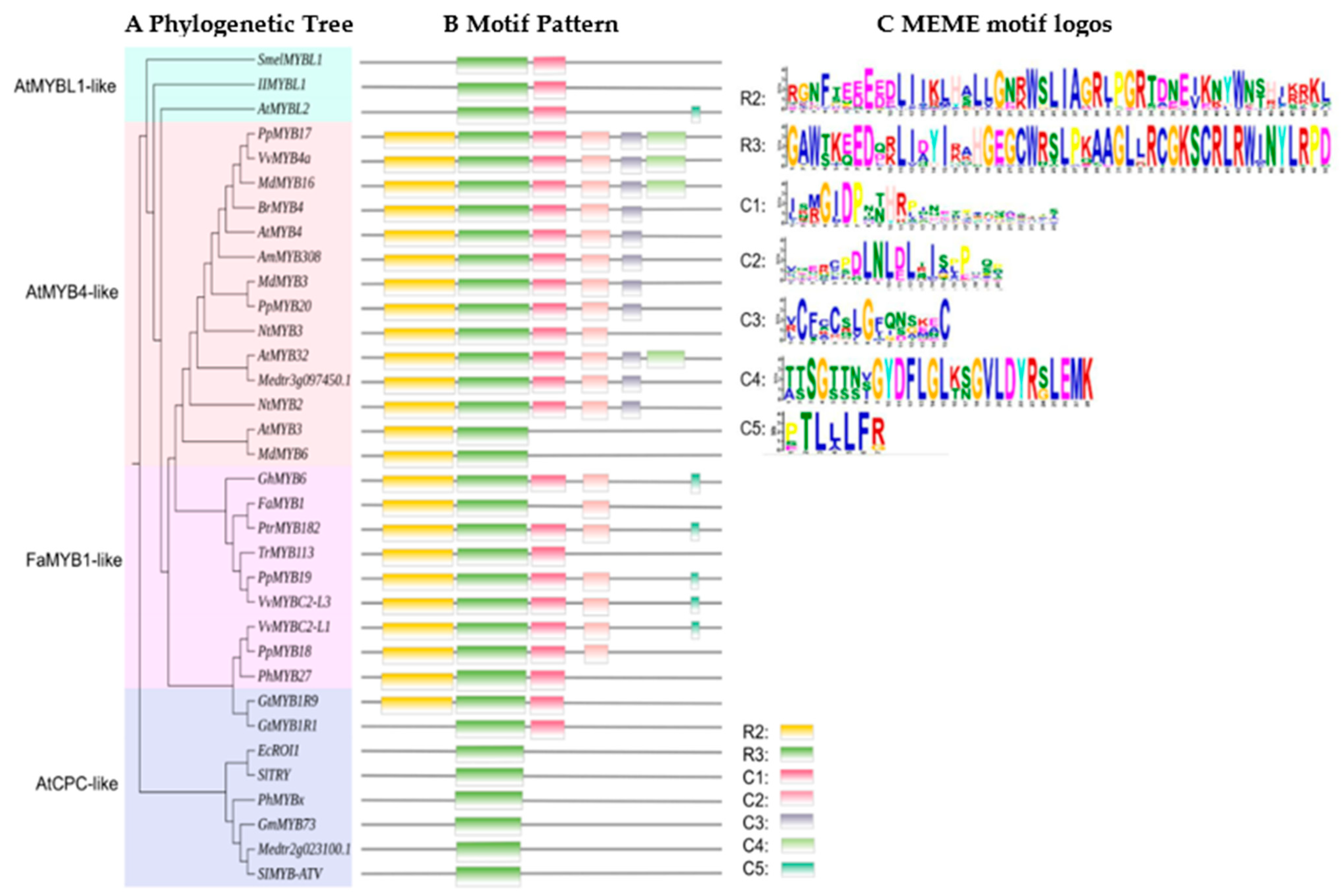
MYB-TFs can also repress anthocyanin biosynthesis. In the transcriptional structure of MYB factors, there are not only transcriptional activation regions but also transcriptional inhibition regions. The first negative regulatory MYB gene (AmMYB308) was identified in snapdragon (Antirrhinum majus) [43]. Overexpression of AmMYB308 in older leaves of tobacco resulted in the virtual absence of the soluble phenylpropanoid derivatives including two flavonoids. Subsequently, MYB-TFs that reduced the accumulation of anthocyanins in various plant tissues or organs were found in species, such as peach, PpMYB19 (P. persica), poplar, PtrMYB182 (P. tremula × Populus), daffodil, NtMYB2 (Narcissus tazetta), apple, MdMYB6 (M. domestica), grape, VVMYBC2-L1 and VVMYBC2-L3 (Vitis vinifera), and strawberry, FaMYB1 (Fragaria × ananassa) [11,44,47,98,101,110]. MYB-TFs are known to have one or more domains near in the C-terminus, which can induce transcription through specific binding to target sites [84,135]. Based on the clustering of the conserved motif (a fragment of a protein that is unable to interact) (in Figure 4), it was confirmed that the conserved motif was found in most plants. The C1 motif may have an assumed activation function but the role of C1 motif has not been characterized yet [11,135]. The C2 motif is also known as the EAR (for ethylene-responsive factor [ERF]-associated amphiphilic repression) motif, and it is the most common C-terminal restraint motif. The C2 region with the core EAR motif is considered to be the predominant transcriptional repression motif in plants [136,137,138]. C3 and C4 motifs exist in a small segment of the R2R3-MYB structural domain, and the motif inhibits benzene propane biosynthesis [84,139,140]. The C5 (TLLLFR) motif in the AtMYB-like of R3-MYB, participates in the formation of the MBW complex in bHLHs. The fourth subgroup of R2R3-MYB is defined by the existence of C1, C2, and C3 motifs [98,110]. In the regulation of flavonoid biosynthesis, MYB repressors mainly exist in the DNA binding domain with two repeats of R2R3-MYB and a single repeat of R3-MYB.

Figure 4.
Phylogenetic relationships of conserved protein motifs in MYB transcription factor repressors related to anthocyanin biosynthesis [74]. (A) The phylogenetic tree was constructed based on the entire protein sequences using MEGA 6 software. Accession numbers are listed in Table 1. (B) The motif composition on every branch of the phylogenetic tree is represented by colored boxes. (C) The sequence for motifs R1-2, C1-5 identified by MEME.
4.2.1. R2R3-MYB Repressors
Whereas R2R3-MYBs can act as activators or repressors of anthocyanin biosynthesis, with other TF binding transcription repressors act through a variety of mechanisms [11,141]. In a R2R3-MYB of Brassica rapa, BrMYB4 can inhibit the expression of BrC4H directly to the promoter, thus inhibiting anthocyanin accumulation [85]. The poplar MYB, PtrMYB57 (P. tremula × Populus) interacts with bHLH131 (bHLH) and PtrTTG1 (WDR) to form an MBW complex, which competes with the activation complex to bind the promoter and inhibits the of activate the anthocyanin regulatory network [142]. AtMYB7 (A. thaliana) and FaMYB1 (Fragaria × ananassa) were identified and found to downregulate anthocyanin biosynthesis [37,143]. The proposed phylogeny (Figure 4), for R2R3-MYB-TF, suggests two ways to regulate flavonoid metabolism: AtMYB4-like (direct binding) [74,92,99] and FaMYB1-like (combined with other proteins) [44,74,104].
AtMYB4-Like Regulation
AtMYB4-like regulation acts directly on the promoters of anthocyanin structural genes to inhibit their expression [74,92,99]. In the negative regulation of anthocyanins, each repressor usually contains a small repression domain. Each type of repression motif is conserved and contacts specific promoter targets resulting in inhibition of gene expression [144]. The C1 motif and EAR(C2) repressor motifs are contained in the C-terminus of the AtMYB4-like regulators. A small number of C3 and C4 motifs are also present [52,145,146]. For example, NtMYB2 (daffodil, N. tazetta) contains three typical conserved motifs, C1, C2, and C3, at the C-terminus, which participate in the inhibition of anthocyanin deposition [99]. In apple, MdMYB16 directly inhibits the expression of MdUFGT and MdANS via a C-terminal EAR motif [46], which inhibits the biosynthesis of anthocyanins. If the EAR motif at the MdMYB16 C-terminus is removed, anthocyanins increase [99]. In the AtMYB4-like evolutionary branch, VvMYB4a reduces anthocyanin by inhibiting the expression of the DFR, ANS and UFGT genes [99]. AtMYB7 of Arabidopsis also uses DFR and UFGT as early targets to inhibit flavonoid pathways [147]. Overexpression of the apple homolog MdMYB6 in calli of apple red fruit also reduces the anthocyanin content. MdMYB6 binds directly to the promoters of MdANS and MdGSTF12, thus inhibiting the synthesis of anthocyanin [86]. Inhibition of ANS, DFR, and UFGT is a common feature of all transgenic tobacco plants and other plants transformed with AtMYB4-like type repressors [46,74]. The AtMYB4-like repressor inhibits anthocyanin accumulation by binding to the promoter of the terminal structural gene of the anthocyanin pathway (in Figure 5) [11,74]. In Arabidopsis, this mechanism of action extended our understanding of the AtMYB4-like repressor explains at least in part the complexity of the negative regulation.

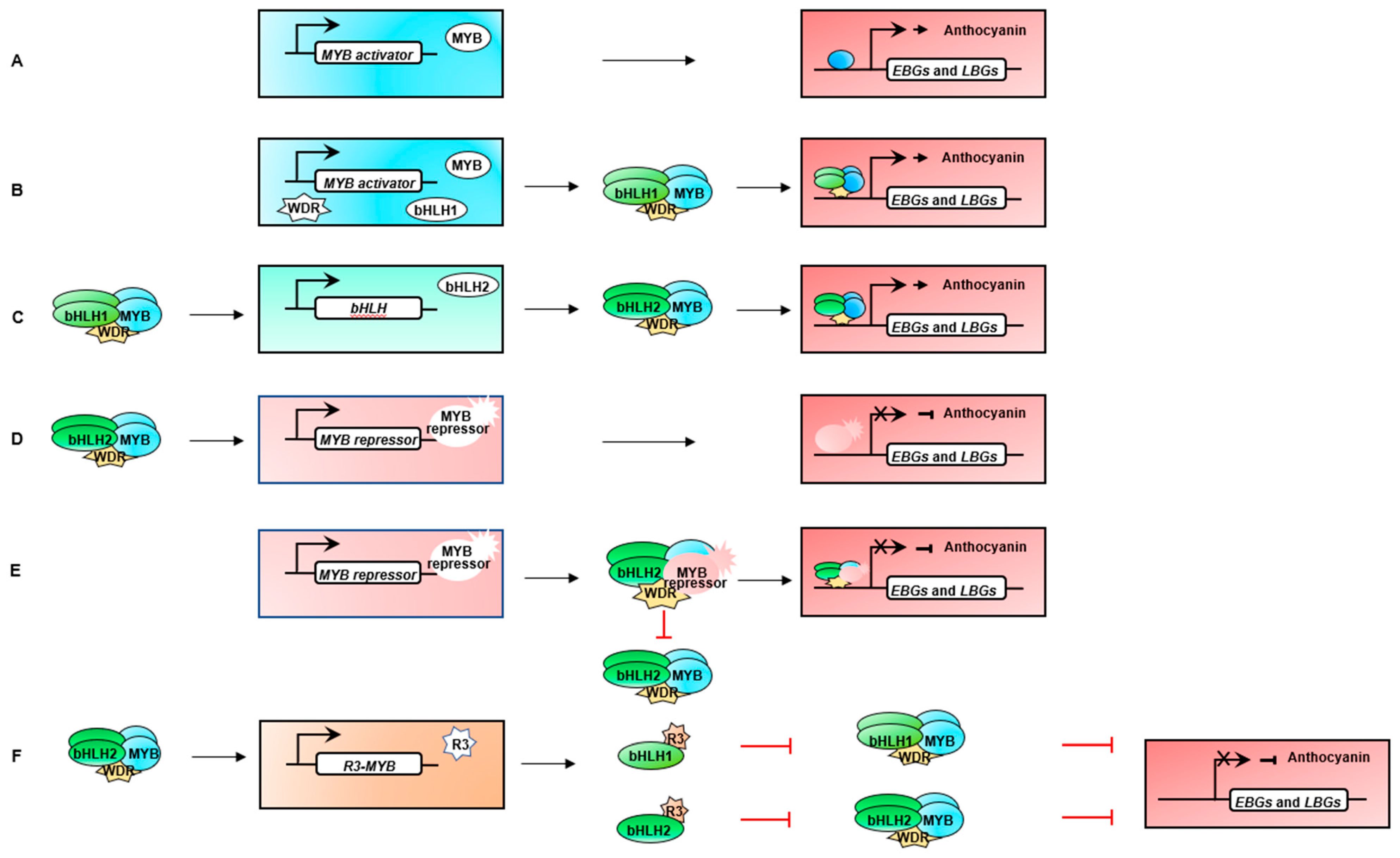
Figure 5.
Proposed molecular mechanisms of MYB transcription factor in the regulation of expression of target genes [148]. (A) Anthocyanin synthesis is initiated in binding directly to target gene by the MYB activator. (B) The MYB activator, WD40 repeat (WDR), and bHLH1 proteins form an MBW activation complex that activated structural genes and promoted the accumulation of anthocyanin. (C) The MYB activator, WDR, and bHLH1 proteins form an MBW activation complex that activated the expression of bHLH2 and structural genes and promote the accumulation of anthocyanin. (D) MYB repressors are activated by the MBW complex, that binding to the promoter of the terminal structural gene in the anthocyanin biosynthesis pathway. (E) MYB repressors could inhibit anthocyanin biosynthesis by incorporating into the MBW activation complex as a corepressor or by binding to the promoter of target genes directly. (F) Feedback inhibition is provided by R3-MYB repressors; these are activated by the MBW complex and inhibit the formation of new MBW complexes by titrating bHLH factors.
FaMYB1-Like Regulation
FaMYB1-like transcription factors bind to other proteins to repress the expression of core activating complexes or to compete with activator complexes to suppress the expression of structural genes by promoters [44,74,104]. Overexpression of FaMYB1 in tobacco inhibited anthocyanin synthesis [11]. FaMYB1 does not contain a functional activation domain and the expression of anthocyanin structural genes was not affected by the overexpression of FaMYB1. Therefore, FaMYB1 inhibits anthocyanin synthesis by a different mechanism than the AtMYB4-like class. Yeast two-hybrid binding indicated that FaMYB1 interacts with two bHLH proteins (JAF13 and AN1), which can act as repressors of anthocyanin biosynthesis when interacting with the core MBW activated complex [44,74,104]. Similar effects were confirmed observed in petunia PhMYB27 (P. hybrida), grape VvMYBC2 (V. vinifera) and VvMYBC2-L3, apple MdMYB15L (M. domestica), and poplar PtrMYB182 (P. tremula) [42,46,47]. This regulatory mechanism is related to the action of the conserved C-terminal motif of R2R3-MYB. In the conserved bHLH region in the R3 domain of PtrMYB182, this MYB evolutionary branch has neither a zinc finger structure nor a C4 motif, but it does contain conserved C1 and C2 motifs and bHLH binding regions [47]. The absence of the bHLH binding site apparently eliminates the repressor activity of PtrMYB182, indicating that its activity depends on the interaction with the bHLH domain. The same result was obtained with PhMYB27 and VVMYB4-like regulators [47,146]. The C2 motif is associated with the anthocyanin repressor PhMYB27, but the addition of C2 motif mutations in PtrMYB182 does not reduce its repressor activity, indicating that the activity of such repressors is independent of the C2 motif [144,149] and suggesting that other yet unknown regulatory mechanisms are important. The characteristic action of FaMYB1-like factors is that they cannot directly bind to the promoter of the target gene but that they are integrated into the MBW complex interfere with the correct assembly (in Figure 5) [47]. According to this proposed regulatory mechanism, the genes targeted by FaMYB1-like repressors are the same as those targeted by the anthocyanin MBW activation complex, but that the role of the complex is changed change the complex activity and transform from activation to inhibition [47,74].
4.2.2. R3-MYB Repressors
The MYB family also includes R3-MYB repressors containing a single MYB R3 repeat. In the R3-MYB-TFs, there are two domain types involved in anthocyanin inhibition. In one type, there is only one R3 domain, which does not directly contact DNA and has a conserved motif, [D/E]Lx2[R/K]x3Lx6Lx3R, that participates in interactions with BHLH-TFs, namely, AtCPC-like (cpc: caprice) activity [83,92,111]. The other domain contains a part of R2 and a complete R3 domain, which also interacts with the bHLH TF to inhibit anthocyanin biosynthesis, specifically AtMYBL1-like activity [106].
The AtCPC-Like Regulation
The AtCPC-like regulator is a single MYB-type protein that acts as a repressor in anthocyanin biosynthesis [150]. Six gene models of the R3-MYB-TFs were found in the Arabidopsis genome: CPC, TRY, TCL, ETC1, ETC2 and ETC3. These genes play an important role in regulating root hair differentiation and trichome germination [151]. CPC and TRY (a negative regulator of trichome initiation in Arabidopsis) both inhibit the synthesis of anthocyanin. CPC is not conserved motif that can act as a direct activator of transcription [151], but CPC retains the binding motif of bHLH and exerts passive inhibition by competing with the R2R3-MYB activator for bHLH [74]. The homologs PhMYBx of petunia and SlMYBATV of tomato also use this regulatory mechanism to regulate anthocyanins. In petunia, PhMYBx is able to bind the bHLH component from the MBW complex, thereby inhibiting anthocyanin biosynthesis [44,152]. Yeast two-hybrid and transient expression in tobacco both showed that the GtMYB1R1 ((from Japanese gentian; Gentiana triflora) and GtMYB1R9 proteins interact with GtbHLH1 proteins and control anthocyanin accumulation by binding to GtDFR promoters to inhibit their expression. Overexpression of GtMYB1R1 and GtMYB1R9 significantly reduces the anthocyanin pigmentation in tobacco petals, resulting in a white-flower phenotype [90]. Similarly, in poplar mutants, R3-MYB repressors block anthocyanin accumulation together with bHLH binding to exert passive inhibition or binding to the MBW-activated complex to trigger active inhibition (Figure 5). This competitive repressor eventually depletes the required TF and is known as a “squeeze” [44,153]. Many mechanisms have been proposed for the action of this repressor, in which the activator activates the repressor, the repressor suppresses the activator, and the activator self-activates [44,154]. This model is consistent with the anthocyanin-related MBW regulatory network of petunia [148]: PhAN1 (ANTHOCYANIN1) can activate the regulatory mechanism of PhMYBx (R3-repressor); PhAN1 activates its own expression; and PhMYBx, in turn, inhibits PhAN1 activity. The competitiveness of AtCPC-like repressors is affected by binding protein affinity, suggesting that regulation of binding proteins might alter anthocyanin inhibition. The outcome of such a mechanism can be homeostasis.
The AtMYBL1-Like Regulation
In contrast to the AtCPC-like, the AtMYBL1-like factor has a conserved EAR or “TLLLFR” motif of anthocyanin synthesis [150]. The R2R3-MYB repressor contains the structural domains at its C-terminus required for repressor activity [43,44]. In contrast, PhMYB27, as a negative regulator of R2R3-MYB, has a dual locking mechanism, which achieves repression by preventing the formation of the MBW complex or by competing with the activator of MBW to bind to the MBW-activated complex and convert it into an repressory complex. This repression requires a conserved EAR motif to achieve repression [44]. To date, researchers have found that in the R3 -MYB-TFs of an ortholog (Lilium hybrid), there is a C2 motif downstream of the MYB repeats of two genes, LhR3MYB1 and LhR3MYB2, and the C2 motif in R2R3-MYBs can directly inhibit the expression of their target genes [92,104,114]. Transient expression of the two genes, LhR3MYB1 and LhR3MYB2, showed that LhR3MYB2 inhibits the expression of the GUS gene driven by the DFR promoter in L. hybrid, while LhR3MYB1 does not. Whether the two can interact with the bHLH protein is not clear. However, AtMYBL2 is also a R3-MYB-TF and does not have a C2 suppression motif but does have a conserved TLLLFR motif, which is functionally similar to the factor of the subtribe R2R3-MYB [44,155]. The repressor functions of AtMYBL2 can be demonstrated by competing with the R2R3-MYB-TF, which activates anthocyanin synthesis with bHLH [84,150]. Unlike AtCPC-like regulation, AtMYBL2 has a conserved repression motif. Similar results were observed in IlMYBL1 (from the purple tube flower; Iochroma loxense), AtMYBL2, and SmelMYBL1 (eggplant: S. melongena L.)) belonging to the same evolutionary branch, showing high similarity to the anthocyanin inhibition mechanism [90,92,106]. The homolog of IlMYBL1 [92] in tomatoes strongly inhibits the accumulation of anthocyanin, leading to a deficiency of anthocyanin in fruits and flowers [155]. Overexpression of SmelANT1 and SmelAN2 results in a red phenotype in poplar leaves, while their co-expression with SmelMYBL1 prevents anthocyanin accumulation, showing that SmelMYBL1 inhibits the MBW complex by competing with the MYB activator for bHLH binding sites [106]. However, the inhibition of R3-MYB is rare, and the role of the conserved repression motif in this process deserves verification.
5. Environment Affects MYB Gene Regulation of Anthocyanin Biosynthesis
Environmental conditions affect plant growth and development. Plants respond to abiotic stress by production of protective metabolites. The regulation of anthocyanin biosynthesis by MYB factors is known to be affected by environmental stress or other external signals [103,156]. Genes associated with light stress or other abiotic stress responses (such as Ultraviolet-B light (UV-B) radiation, low temperature or drought) may exert strong effects. Under blue and far-red light, AtMYB75 expression increased at low temperature while drought enhanced AtMYB114 expression [37]. Environmentally induced mediation of MYB-TFs in anthocyanin biosynthesis may reveal fundamental features of the hierarchical genetic regulatory networks active during responses to environmental and developmental signals.
5.1. Light
Plant maturation requires appropriate light stimulation. UV-B is one environmental signal perceived by plants that affects the flavonoid pathway and influences the level of anthocyanins [157,158]. Apple trees were grown under spectral filters that altered transmission of solar UV light, ripening was delayed, fruit size decreased, and anthocyanin and flavonols were reduced [158]. Thotoreceptor scryptochrome 1 (CRY1) and phytochrome (PHY) can also induce the accumulation of anthocyanins [24]. Anthocyanin accumulates when the intensity of light is too high because anthocyanin protects photosynthetic organs [51,159]. Leucine Zip TF elongated hypocotyl 5 (HY5) is part of the light signaling pathway downstream of PHY, CRY and UV-B photoreceptors, which regulating the regulatory network of PAP1 activates anthocyanin biosynthesis [24]. In Arabidopsis, hy5 regulated pap1 expression by directly combining G-box and ACE elements, while pap1 acted on the promoters of EBGs and LBGs to induce the downstream network activation of anthocyanins [24,160]. PAP1 expression was highly dependent on the presence of the HY5 TF, and this optical signaling pathway was affected by photosynthesis [24]. In the Asian pear (Pyrus pyrifolia), PyHY5 was also determined to directly recognize and combine with PyMYB10 and PyWD40 to activate anthocyanin biosynthesis [103]. Photosensitivity plays an important role in the regulation of anthocyanins. In Arabidopsis, the blue light photoreceptor CRY1, red light photoreceptor phyB and far-red light photoreceptor phyA can promote anthocyanin accumulation under blue, red and far-red light, respectively [82]. When Arabidopsis was exposed to far-red light, phyA could promote anthocyanin accumulation by enhancing AtMYB75 abundance and AtDFR, AtLDOX and AtUFGT expression [82]. To maintain the balance of anthocyanins, light signaling pathways downstream of constitutively photomorphogenic 1 (COP1: ubiquitin E3 ligase morphogenesis protein) TF suppress the accumulation of anthocyanins [161]. COP1 is found in the nucleus under dark conditions, where it targets photomorphogenetic activating TFs and mediates their degradation [162]. Apple MdMYB1 is ubiquinated and degraded by MdCOP1 dependent activity and fruit coloration in the dark is inhibited [162]. COP1 can act as a central switch of optical signal transduction, directly interacting with the photoreceptor and downstream target proteins [163,164]. CRY1-COP1-MYB75 and phyB-COP1-MYB75 signal transduction modules inhibit the accumulation of anthocyanins under blue and red light [82]. COP1 and HY5 anthocyanins are regulatory factors under the control of light. The HY5 TF is negatively regulated by COP1 [165], because COP1 can target the degradation of many TFs [164]. The genomic expression of anthocyanins may be regulated by the control of COP1 activity [164] to maintain an appropriate balance [161].
5.2. Temperature
Changes in temperature affect the regulation of anthocyanin biosynthesis by MYB-TFs. Strong light affects the accumulation of reactive oxygen species (ROS), a major component of oxidative stress. To ameliorate this stress, plants produce antioxidants such as ascorbic acid and a diverse group of polyphenols [28,166]. The protective effect of ascorbic acid in plants is inhibited at low temperatures, leading to the synthesis of additional antioxidants and the activation of anthocyanin biosynthesis [28,167]. Low temperature increases the accumulation of anthocyanin, which has been confirmed in apple (M. domestica) [168], rapeseed (Brassica rapa) [169] and eggplant (Solanum melongena) [39]. The expression of CHS1 and MYB1 at 6 °C was upregulated in gerbera daisy (Gerbera jamesonii), and the anthocyanin content was increased [143]. Similarly, in apples, the combination of MdbHLH3 and MdMYB1 increased the expression of MdDFR and MdUFGT, and low temperature improved the binding ability between bHLH and MYB and thereby promoting the biosynthesis of anthocyanin [39,168]. For anthocyanin activation by cold stress, C repeated binding factor (CBF) was found to play a central regulatory role in eggplant under cold stress [39]. A yeast two-hybrid experiment found that SmCBF2 and SmCBF3 upregulated the expression of SmCHS and SmDFR through the physical action of SmMYB113 [168]. When SmTT8 (a bHLH-TF partner of SmMYB113) were expressed together with SmCBFs and SmMYB113, the anthocyanin content was significantly higher than when SmMYB113 was expressed alone [39]. Therefore, SmMYB113 is the bridge for the mutual interaction between SmCBFs and SmTT8 [39]. Two bZIP-TFs, HY5 and a HY5 homolog HYH, induced anthocyanin accumulation at low temperature in Arabidopsis [170]. These two factors require light to upregulate DFR at low temperature. They can also activate the downstream network of anthocyanins.
5.3. Other Factors Regulation Anthocyanin
Other environmental factors also affect the regulation of anthocyanin biosynthesis by MYB-TFs. Not only do light factors and low temperature regulate anthocyanin accumulation, but phytohormones, drought and high sucrose stress also induce MYBs that activate the regulatory network of anthocyanin biosynthesis. Drought stress that induces excessive ROS, can damage the cellular structure and metabolism of plants. To alleviate the damage caused by ROS stress, the antioxidant capacity is improved by activating the regulatory network and biosynthesis of anthocyanins [138,171]. Under drought stress, HD-ZIP II TF HAT1 negatively regulates (inhibits) anthocyanin accumulation by binding to the MBW protein complex [138]. In addition, the phytohormone abscisic acid (ABA) activates MYB75, a possible anthocyanin accumulation-induced TF, by inhibiting the physical interaction between HAT1 and MYB75 [138]. Similarly, IbMYB1 from sweet potato (Ipomoea batatas) can activate the accumulation of anthocyanin, improve the antioxidant capacity and enhance resistance to drought stress. IbMYB1 overexpressed in Arabidopsis can also increase the tolerance to osmotic stress [172]. While the external environment may regulate the activation of anthocyanins by MYB- TFs, internal environmental factors (conditions within the plant) such as phytohormones and pH may affect expression of the regulatory network of MYB-TFs. In the MBW complex, JAZ proteins (repressors of the jasmonate signaling cascade) can interact with bHLH and MYB members to inhibit MBW formation and consequently inhibit anthocyanin biosynthesis. To activate the anthocyanin synthesis pathway, the degradation of JAZ proteins through the synthesis of jasmonic acid (JA) reduces its inhibition of MBW [82]. Moreover, JA relies on the photoreceptor encoded by phyA to induce MYB75 expression and to consequently activate DFR, UFGT and LDOX and to regulate anthocyanin enrichment when Arabidopsis is exposed to far-red light [82]. The color of anthocyanins is pH dependent and therefore the color observed in plant tissues is affected by the pH in the vacuole [173]. As described in the Introduction of this review, the major anthocyanins cyanidin and peonidin are red at low pH and blue at high pH. Cyanidin, peonidin and delphinidin are all blue at high pH [32]. Genes controlling blue color expression may reduce internal vacuolar acidity by transporting sodium ions outside the vacuoles, intensifying the color of the blue pigments [174]. In petunia (Petunia × hybrida), PhPH4 is a member of the R2R3MYB family, encoding a MYB domain protein expressed in the red epidermis of petals. Mutation of PhPH4 causes the color to turn blue, presumably due to an increase in the internal pH, as reflected by an increase in the pH of petal extracts [173]. Anthocyanin accumulates under acidic pH conditions. In Malus Crabapple, low pH promoted the expression of the McANS and McUFGT genes, which increased anthocyanins and the red color of the leaves. The color changed from red to green with increasing pH [85]. The mechanisms of anthocyanin regulation in the internal and external environment are complex and multifactorial.
6. Epigenetic Effects of MYB-TFs on Anthocyanin Synthesis
R2R3-MYB-TFs change the conformation and stability of proteins through posttranslational modifications such as methylation, phosphorylation and ubiquitination [128,175].
6.1. Effect of MYB-TF Methylation on Anthocyanins
DNA methylation has played an important role in genetic regulatory mechanisms such as transposon silencing and gene expression [176]. DNA methylation and demethylation are essential for normal eukaryotic development. DNA methylation has been found at redundant cytosine residues (CG, CHH and CHG), where H equals “not G” [177]. In the WEREWOLF (WER) (AtMYB66) structural model of Arabidopsis MYB-TFs [178], R2 in the R2R3-MYB protein was associated with redundant GC elements and R3 specifically recognized AAC elements [178,179]. The methylation of the DNA at the 5th position of cytosine (5 mC) and the 6th position of adenine (6 mA) in the AAC element blocked the interaction between WER and DNA so that the accumulation of anthocyanin in apple peel is inhibited. The methylation level in the promoter region of MYB-TFs is related to anthocyanin accumulation [180]. The ‘Royal Gala’ apple has red stripes on the peel, whereas ‘Honeycrisp’ has green stripes. The green stripes are positively correlated with increased anthocyanin content, which is correlated with increased major anthocyanin biosynthetic gene transcript levels including MdMYB10. The MdMYB10 promoter is highly methylated and methylation enrichment is observed in the green stripes [180]. In pears, methylation of three cytosines in PcMYB10, inhibits the accumulation of anthocyanin [179]. The relationship of DNA methylation and TF binding remains complex and is not sufficient to explain the contradictory correlations. Further exploration is needed.
6.2. Phosphorylation and Ubiquitination
Phosphorylation and ubiquitination also affect the regulation of anthocyanin biosynthesis by MYB-TFs. In Arabidopsis the light-induced accumulation of anthocyanins is dependent on MYB75, which is functionally dependent on phosphorylation by MPK4, a multifunctional MAP kinase [51,181]. MPK4 is a mitogen-activated protein (MAP) and its interaction with MYB75 and phosphorylation, is required for MYB75 regulation. This interaction signifies an important role for a MAPK pathway in light signal transduction [51]. When MPK4 phosphorylates MYB75, it improves the stability of MYB75 but does not affect the binding of MYB75 to the anthocyanin target promoter [51]. Both R2R3MYBs and MPK4 can function as positive regulators of anthocyanin biosynthesis [181].
When plants are harmed by cold stress or osmotic pressure, they activate MPK4 kinase, suggesting that MPK4 plays an important role in the interaction of the different stress response pathways induced by these signals leading to the accumulation of anthocyanins [148,182]. COP1 is known to be a molecular switch that regulates plant development and inhibits anthocyanin accumulation downstream of the light photoreceptor. When plants are exposed to light, light inactivates COP1 [82]. COP1 contains an N-terminal WD40 domain and has a conserved ring finger domain in the ubiquitin protein ligase subclass [51]. Under dark conditions, the COP1/SPAE3 (Suppressor of phyA E3 ubiquitin ligase) ubiquitin ligase degradation, targets gene MYB75 and inhibits anthocyanin synthesis [138]. This is contrary to the effect of MPK4 on the stability of MYB75, indicating that MPK4 and COP1 jointly regulate the accumulation of photoinduced anthocyanins. The mechanisms of phosphorylation and ubiquitination in anthocyanins are complex and merit further investigation.
7. Integrated Control Network
The role of MYB-TFs in the regulation of anthocyanin biosynthesis depends on multiple regulatory factors that operate in a hierarchical manner [44]. The expression level of activator PhAN1 (from Petunia) is low and that of MYB repressor PhMYBx is high under normal growth (noninductive) conditions [74]. MYBx repressors inhibit the formation of the MBW complex by binding to bHLH proteins, preventing the activation of the target gene DFR [44]. When the mature flowers and fruits of Petunia are photoinduced, the light signaling pathway activates R2R3-MYBs and MBW (including bHLH1) to form an activation complex, inducing the expression of bHLH [103,106,160]. Consequent bHLH2 forms an MBW complex with the activator and WDR protein and acts on the promoter of the late acting anthocyanin structural gene DFR to regulate the downstream network of anthocyanin biosynthesis [160]. PhMYBx repressors are also activated by MBW activating complexes, competing with activators for target gene sites for feedback inhibition [44]. To promote the biosynthesis of anthocyanins, plants prevent the activation of repressors by activating complexes through the mechanisms induced by environmental conditions. bHLH2 enhances inhibition [44]. Studies investigating the control of Arabidopsis hairy cells also confirmed this regulatory mechanism. Several environmental factors play an important role in the regulation of anthocyanins by MYB-TFs (Figure 6). The regulation of anthocyanin species by various MYB-TFs, appear to demonstrate this environmental tendency because the bHLH factor can bridge interactive regulation between MYB activators and MYB repressors. The overexpression of MdMYB16 in apple calli inhibits the synthesis of anthocyanin, and the overexpression of MdbHLH33 in the same callus shows that the repression effect of MdMYB16 on anthocyanin biosynthesis is weakened [46,74]. Different plants exhibit different functional zoning of bHLH factors and play different roles in modulating regulation. In summary, the biosynthetic pathway of anthocyanins is regulated by the environment, repressors and activators, and the interaction of the MBW complex and various regulatory factors to maintain the balance of anthocyanins in plants and ensure the stability of plant pigment production.

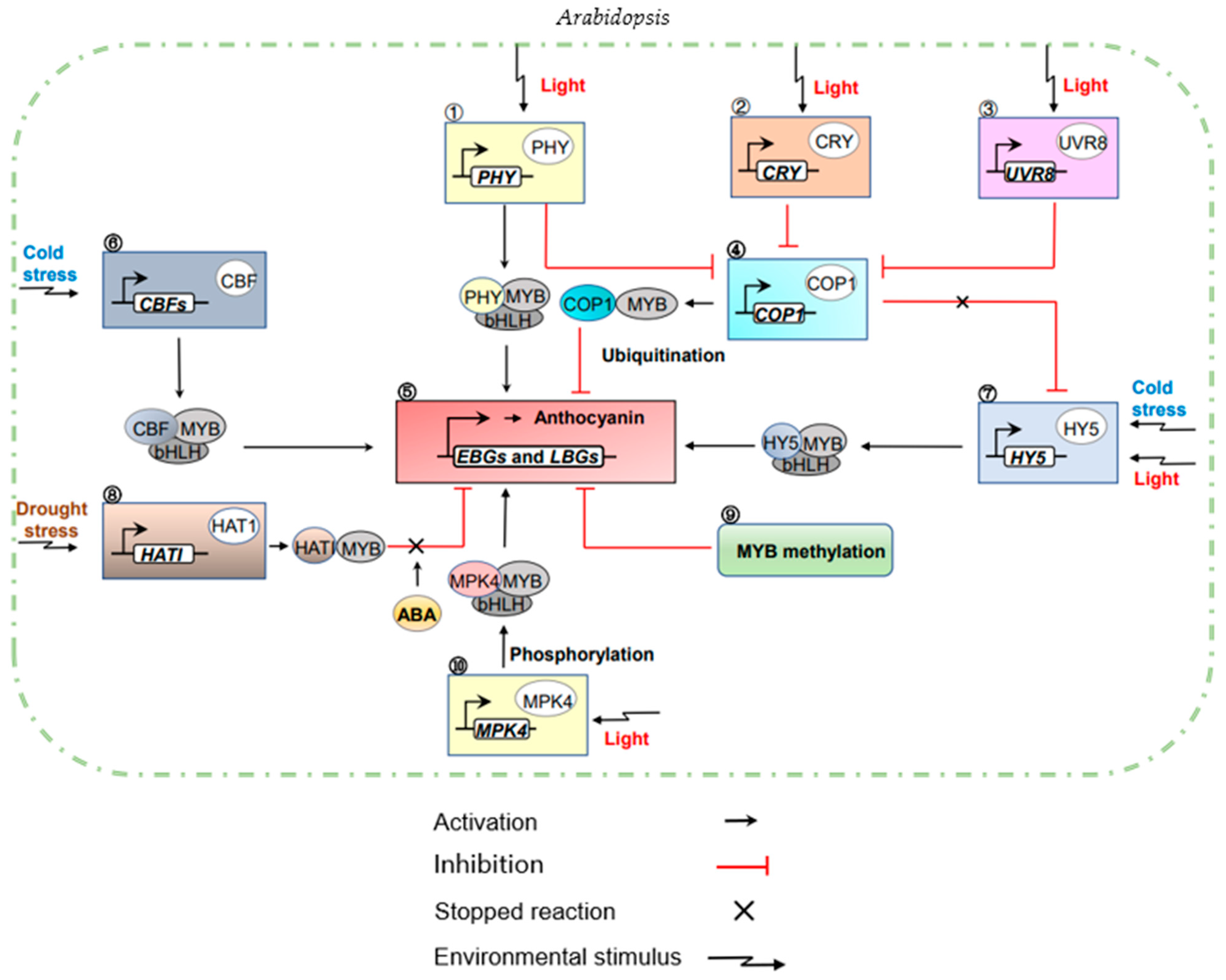
Figure 6.
MYB transcription factors regulates the action network of anthocyanin in plants’ internal and external environment. Under light conditions, anthocyanin biosynthesis is initiated in developing leaves or developing flowers/fruits by PHY photoreceptors (1) binding directly to MYB and bHLH. At the same time, CRY (2) and UVR8 (3) photoreceptors could inhibit (4) COP1 factor. In the dark, COP1 degraded MYB transcription factors that reduce the expression of genes activating anthocyanin biosynthesis (5), ultimately inhibiting anthocyanin accumulation. Under cold stress, C repeated binding factor (CBF factor) (6) activated anthocyanin biosynthesis through physical interaction with MYB and bHLH. At the same time, HY5 factor (7) also induces anthocyanin accumulation so that the anthocyanin structural gene could be up-regulated under low temperature under light conditions to activate the downstream network of anthocyanins. Under drought conditions, HAT1 factor (8) negatively regulates anthocyanin accumulation by binding to MBW protein complexes, while in the presence of the plant hormone ABA, ABA could inhibit the physical interaction between HAT1 and MYB75 and activate MYB75 to induce anthocyanin accumulation. When the MYB target sequence was methylated (9), anthocyanin accumulation was also inhibited. Under light conditions, MPK4 (10) interacts with MYB and bHLH and phosphorylates MYB, then participating in the accumulation of light-induced anthocyanins.
8. Future Prospects
MYB-TFs play an important role in the regulation of anthocyanin biosynthesis and accumulation. Environmental factors fine-tune the control of anthocyanin biosynthesis to maintain the normal plant growth and development. Elucidating the regulatory network of MYB-TFs and its effects on anthocyanins will help us to better understand the activity of MYBs and their interactions. The expression patterns of the presumed functional MYB-TF orthologs in different plants are not identical and this variation enriches the network of plant phenotypes and environmental responses. There are still questions about the activity and evolution of this extensive regulatory network, which deserve further in-depth description and analysis. Many features of these cis-acting elements on their target gene promoters and their role in regulation of the anthocyanin biosynthetic pathway remain unknown. Similarly, the mechanisms by which environmental factors affect the regulation of activators and repressors in the anthocyanin regulatory hierarchy are also not established. The mechanisms by which specific TFs such as R3-MYB-TFs affect anthocyanins are poorly understood. What is the strength of MYB activator that is needed to activate the repressor? How is the strength of the activators and repressors determined? Does the MBW protein complex regulating anthocyanins, bind to other TFs and MYB-TFs in the same way? Exploring the regulatory mechanisms of MYB-TFs could help us to more fully understand the mechanisms of anthocyanin biosynthesis and the specific features of the major interactions of hierarchical genetic regulatory networks in the growth, development and adaptation of higher plants.
As more is known about the mechanisms of regulation and targeting of MYB-TFs, the elements and domains of regulation increase in value as components in the genetic engineers toolbox for the construction of novel regulatory sequences in synthetic biology. History of molecular biology shows that the best understood molecules of present studies become the reagents and building blocks of the future biochemical technology. Anthocyanins have great value in agriculture, nutriceuticals, medicine and basic plant sciences. Therefore, the design of pigments in advanced breeding technology is likely to make use of the intricate mechanisms of MYB-TFs and their roles in the hierarchical genetic regulatory networks of transcriptional control.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/6/3103/s1.
Author Contributions
Conceptualization, V.L.C., and X.Z. (Xiyang Zhao); writing—original draft preparation, H.Y., X.P. and H.Z.; writing—review and editing, X.L., X.Z. (Xinxin Zhang); and M.Z.; supervision, R.R.S. and X.Z. (Xiyang Zhao).; project administration, X.Z. (Xiyang Zhao).; funding acquisition, X.Z. (Xiyang Zhao). All authors have read and agreed to the published version of the manuscript.
Funding
We are grateful to the National Natural Science Foundation of China (31770712), Fundamental Research Funds for the Central Universities (2572020DR01) and Heilongjiang Touyan Innovation team program (Tree Genetics and Breeding Innovation Team) for supporting this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Campanella, J.J.; Smalley, J.V.; Dempsey, M.E. A phylogenetic examination of the primary anthocyanin production pathway of the Plantae. Bot. Stud. 2014, 55, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cui, B.; Tan, Z.; Song, B.; Cao, H.; Zong, C. RNA sequencing and anthocyanin synthesis-related genes expression analyses in white-fruited Vaccinium uliginosum. BMC Genom. 2018, 19, 930. [Google Scholar] [CrossRef]
- Bendz, G.; Mårtensson, O.; Sillén, L.G. Moss Anthocyanins. Acta Chem. Scand. 1961, 15, 1185. [Google Scholar] [CrossRef]
- Harborne, J.B. Anthocyanins in ferns. Nature 1965, 207, 984. [Google Scholar] [CrossRef]
- Bu, C.; Zhang, Q.; Zeng, J.; Cao, X.; Hao, Z.; Qiao, D.; Cao, Y.; Xu, H. Identification of a novel anthocyanin synthesis pathway in the fungus Aspergillus sydowii H-1. BMC Genom. 2020, 21, 29. [Google Scholar] [CrossRef]
- Zuo, R.; Xu, M.L. Function and regulation mechanism of plant MYB transcription factors. Life Sci. 2012, 24, 1133–1140. [Google Scholar] [CrossRef]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Lev-Yadun, S.; Gould, K.S. Role of Anthocyanins in plant defence. In Anthocyanins; Winefield, C., Davies, K., Eds.; Springer: New York, NY, USA, 2008; pp. 22–28. [Google Scholar]
- Feild, T.S.; Lee, D.W. Why leaves turn red in autumn. The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol. 2002, 128, 783. [Google Scholar] [CrossRef]
- Gai, Z.; Wang, Y.; Ding, Y.; Qian, W.; Qiu, C.; Xie, H.; Sun, L.; Jiang, Z.; Ma, Q.; Wang, L.; et al. Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Sci. Rep. 2020, 10, 12275. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; De Vos, C.H.R.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.M.; O’Connell, A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N.; et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Ren, L.; Lian, H.; Liu, Y.; Chen, H. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 2016, 249, 46–58. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Z.; Zeng, Q.; Wang, S.; Luo, Y.; Huang, Y.; Xin, Y.; He, N. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 2020, 7, 1–19. [Google Scholar] [CrossRef]
- Tominaga-Wada, R.; Nukumizu, Y.; Wada, T. Tomato (Solanum lycopersicum) Homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal. Behav. 2013, 8, e24575. [Google Scholar] [CrossRef]
- Wang, H.; Guan, S.; Zhu, Z.; Wang, Y.; Lu, Y. A valid strategy for precise identifications of transcription factor binding sites in combinatorial regulation using bioinformatic and experimental approaches. Plant Methods 2013, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, J.; Guan, M.; Zhao, C.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef]
- Wu, W.C. Research progress of plant anthocyanin. Mod. Chem. Res. 2018, 9, 183–185. [Google Scholar]
- Zhou, M.; Sun, Z.; Wang, C.; Zhang, X.; Tang, Y.; Zhu, X.; Shao, J.; Wu, Y. Changing a conserved amino acid in R2R3-MYB transcription repressors results in cytoplasmic accumulation and abolishes their repressive activity in Arabidopsis. Plant J. 2015, 84, 395–403. [Google Scholar] [CrossRef]
- Pelletier, M.K.; Burbulis, I.E.; Winkel-Shirley, B. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol. Biol. 1999, 40, 45–54. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.-B.; Choi, G.; Park, Y.-I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin-Wang, K.; Wang, F.; Espley, R.V.; Ren, F.; Zhao, J.; Ogutu, C.; He, H.; Jiang, Q.; Allan, A.C.; et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019, 221, 1919–1934. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Cominelli, E.; Gusmaroli, G.; Allegra, D.; Galbiati, M.; Wade, H.K.; Jenkins, G.I.; Tonelli, C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008, 165, 886–894. [Google Scholar] [CrossRef]
- Ilk, N.; Ding, J.; Ihnatowicz, A.; Koornneef, M.; Reymond, M. Natural variation for anthocyanin accumulation under high-light and low-temperature stress is attributable to the ENHANCER OF AG -4 2 (HUA 2) locus in combination with PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP 1) and PAP2. New Phytol. 2015, 206, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shan, X.; Zhou, L.; Gao, R.; Yang, S.; Wang, S.; Wang, L.; Gao, X. The R2R3-MYB factor FhMYB5 from Freesia hybrida contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis. Front. Plant Sci. 2019, 9, 1935. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef]
- Romero, I.; Fuertes, A.; Benito, M.J.; Malpica, J.M.; Leyva, A.; Paz-Ares, J. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998, 14, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Lila, M.A. Anthocyanins and human health: An in vitro investigative approach. J. Biomed. Biotechnol. 2004, 2004, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB transcription factors as regulators of secondary metabolism in plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Mondal, S.K.; Roy, S. Genome-wide sequential, evolutionary, organizational and expression analyses of phenylpropanoid biosynthesis associated MYB domain transcription factors in Arabidopsis. J. Biomol. Struct. Dyn. 2018, 36, 1577–1601. [Google Scholar] [CrossRef]
- Schwinn, K.; Venail, J.; Shang, Y.; Mackay, S.; Alm, V.; Butelli, E.; Oyama, R.; Bailey, P.; Davies, K.; Martin, C. A Small Family of MYB-Regulatory Genes Controls Floral Pigmentation Intensity and Patterning in the Genus Antirrhinum. Plant Cell 2006, 18, 831–851. [Google Scholar] [CrossRef]
- Zhou, L.; He, Y.; Li, J.; Liu, Y.; Chen, H. CBFs Function in Anthocyanin Biosynthesis by Interacting with MYB113 in Eggplant (Solanum melongena L.). Plant Cell Physiol. 2020, 61, 416–426. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Lin-Wang, K.; McGhie, T.K.; Wang, M.Y.; Liu, Y.; Warren, B.; Storey, R.; Espley, R.V.; Allan, A.C. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 2014, 5, 651. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.H.; Liu, C.; Xiao, X.; Dixon, R.A. The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell 2015, 27, 2860–2879. [Google Scholar] [CrossRef]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef]
- Yoshida, K.; Ma, D.; Constabel, C.P. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015, 167, 693–710. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Broeckling, B.E.; Watson, R.A.; Steinwand, B.; Bush, D.R. Intronic sequence regulates sugar-dependent expression of Arabidopsis thaliana production of anthocyanin pigment-1/MYB75. PLoS ONE 2016, 11, e0156673. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, W.; Gao, J.; Yin, K.; Wang, R.; Wang, C.; Petersen, M.; Mundy, J.; Qiu, J.-L. MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in arabidopsis. Plant Cell 2016, 28, 2866–2883. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef]
- Kortstee, A.J.; Khan, S.A.; Helderman, C.; Trindade, L.M.; Wu, Y.; Visser, R.G.F.; Brendolise, C.; Allan, A.; Schouten, H.J.; Jacobsen, E. Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res. 2011, 20, 1253–1264. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3- MYB transcription factor MYB 6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Klempnauer, K.-H.; Gonda, T.J.; Bishop, J.M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef]
- Wang, X.Q.; Chen, B.J. The olant MYB transcription factors. Biotechnol. Bull. 2003, 22–25. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb protooncogene products and with structural similarities to transcriptional activators. EMBO J. 1988, 6, 3553–3558. [Google Scholar] [CrossRef]
- Rosinski, J.A.; Atchley, W.R. Molecular Evolution of the myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Feng, G.; Burleigh, J.G.; Braun, E.L.; Mei, W.; Barbazuk, W.B. Evolution of the 3R-MYB gene family in plants. Genome Biol. Evol. 2017, 9, 1013–1029. [Google Scholar] [CrossRef]
- Jiang, C.; Gu, J.; Chopra, S.; Gu, X.; Peterson, T. Ordered origin of the typical two- and three-repeat myb genes. Gene 2004, 326, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Tombuloglu, H. Genome-wide identification and expression analysis of R2R3, 3R- and 4R-MYB transcription factors during lignin biosynthesis in flax (Linum usitatissimum). Genomics 2020, 112, 782–795. [Google Scholar] [CrossRef]
- Pelt, J.L.; Downes, W.A.; Schoborg, R.V.; McIntosh, C.A. Flavanone 3-hydroxylase expression in Citrus paradisi and Petunia hybrida seedlings. Phytochemistry 2003, 64, 435–444. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2002, 55, 481–504. [Google Scholar] [CrossRef]
- Zhao, M.-H.; Li, X.; Zhang, X.-X.; Zhang, H.; Zhao, X.-Y. Mutation mechanism of leaf color in plants: A review. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Reif, H.J.; Niesbach, U.; Deumling, B.; Saedler, H. Cloning and analysis of two genes for chalcone synthase from Petunia hybrida. Mol. Gen. Genet. 1985, 199, 208–215. [Google Scholar] [CrossRef]
- Van der Krol, A.R.; Mur, L.A.; de Lange, P.; Mol, J.N.M.; Stuitje, A.R. Inhibition of flower pigmentation by antisense CHS genes: Promoter and minimal sequence requirements for the antisense effect. Plant Mol. Biol. 1990, 14, 457–466. [Google Scholar] [CrossRef]
- Froemel, S.; De Vlaming, P.; Stotz, G.; Wiering, H.; Forkmann, G.; Schram, A.W. Genetic and biochemical studies on the conversion of flavanones to dihydroflavonols in flowers of Petunia hybrida. Theor. Appl. Genet. 1985, 70, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Beld, M.; Martin, C.; Huits, H.; Stuitje, A.R.; Gerats, A.G.M. Flavonoid synthesis in Petunia hybrida: Partial characterization of dihydroflavonol-4-reductase genes. Plant Mol. Biol. 1989, 13, 491–502. [Google Scholar] [CrossRef]
- Kobayashi, S.; Ishimaru, M. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 2002, 215, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Broun, P. Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005, 8, 272–279. [Google Scholar] [CrossRef]
- Dong, N.; Lin, H. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.; et al. Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef]
- Durbin, M.L.; Learn, G.H.; Huttley, G.A.; Clegg, M.T. Evolution of the chalcone synthase gene family in the genus Ipomoea. Proc. Natl. Acad. Sci. USA 1995, 92, 3338–3342. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Peng, Y.; Lin-Wang, K.; Cooney, J.M.; Wang, T.; Espley, R.V.; Allan, A.C. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic. Res. 2019, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xiaoling, Y.; Ding, Z.; Ruan, M.; Yu, X.; Peng, M.; Liu, Y. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Dou, M.; Fan, S.; Yang, S.; Huang, R.; Yu, H.; Feng, X. Overexpression of AmRosea1 gene confers drought and salt tolerance in rice. Int. J. Mol. Sci. 2016, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.; Wheeler, J.; Heazlewood, J.; Li, S.F.; Parish, R.W. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004, 40, 979–995. [Google Scholar] [CrossRef]
- Shin, D.H.; Cho, M.; Choi, M.G.; Das, P.K.; Lee, S.-K.; Choi, S.-B.; Park, Y.-I. Identification of genes that may regulate the expression of the transcription factor production of anthocyanin pigment 1 (PAP1)/MYB75 involved in Arabidopsis anthocyanin biosynthesis. Plant Cell Rep. 2015, 34, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jia, K.-P.; Lian, H.-L.; Yang, X.; Li, L.; Yang, H.-Q. Jasmonic acid enhancement of anthocyanin accumulation is dependent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 454, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.-W.; Sagawa, J.M.; Young, R.C.; Christensen, B.J.; Bradshaw, H.D. Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of mimulus. Genetics 2013, 194, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Sun, M.; Wang, J.; Kawabata, S.; Li, Y. BrMYB4, a suppressor of genes for phenylpropanoid and anthocyanin biosynthesis, is down-regulated by UV-B but not by pigment-inducing sunlight in Turnip cv. Tsuda. Plant Cell Physiol. 2014, 55, 2092–2101. [Google Scholar] [CrossRef]
- Xu, H.; Zou, Q.; Yang, G.; Jiang, S.; Fang, H.; Wang, Y.; Zhang, J.; Zhang, Z.; Wang, N.; Chen, X. MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 2020, 7, 1–17. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; De La Fuente, I.R.; Van Houwelingen, A.M.M.L.; De Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.-M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2012, 197, 454–467. [Google Scholar] [CrossRef]
- Loguercio, L.L.; Zhang, J.-Q.; Wilkins, T.A. Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.). Mol. Gen. Genet. 1999, 261, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 1–13. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Yamada, E.; Saito, M.; Fujita, K.; Nishihara, M. Heterologous expression of gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers. Plant Cell Rep. 2013, 32, 1925–1937. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Li, Q.-T.; Lu, X.; Song, Q.-X.; Lam, S.-M.; Zhang, W.-K.; Ma, B.; Lin, Q.; Man, W.-Q.; Du, W.-G.; et al. Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol. 2014, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Gates, D.J.; Olson, B.J.S.C.; Clemente, T.E.; Smith, S.D. A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma. New Phytol. 2018, 217, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.Y.; Ahn, Y.O.; Kim, S.H.; Kim, Y.-H.; Lee, H.-S.; Catanach, A.S.; Jacobs, J.M.E.; Conner, A.J.; Kwak, S.-S. The sweet potato IbMYB1 gene as a potential visible marker for sweet potato intragenic vector system. Physiol. Plant 2020, 139, 229–240. [Google Scholar] [CrossRef]
- Pang, H.; Tian, J. The study on promoter structure of MYB10 in different crabapple culativars. J. Beijing Univ. Agric. 2016, 31, 76–80. [Google Scholar] [CrossRef]
- Chiu, L.-W.; Li, L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 2012, 236, 1153–1164. [Google Scholar] [CrossRef]
- Niu, S.-S.; Xu, C.-J.; Zhang, W.-S.; Zhang, B.; Li, X.; Lin-Wang, K.; Ferguson, I.B.; Allan, A.C.; Chen, K.-S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 2010, 231, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-S.; Gugger, P.F.; Wang, Q.-F.; Chen, J.-M. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus (Nelumbo Adans.). PeerJ 2016, 4, e2369. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.; Wang, G.; Wu, J.; Waheed, S.; Allan, A.C.; Zeng, L. Ectopic overexpression of a novel R2R3-MYB, NtMYB2 from Chinese Narcissus represses anthocyanin biosynthesis in tobacco. Molecules 2018, 23, 781. [Google Scholar] [CrossRef]
- Anwar, M.; Yu, W.; Yao, H.; Zhou, P.; Allan, A.C.; Zeng, L. NtMYB3, an R2R3-MYB from Narcissus, regulates flavonoid biosynthesis. Int. J. Mol. Sci. 2019, 20, 5456. [Google Scholar] [CrossRef]
- Huang, Z.-A.; Zhao, T.; Fan, H.-J.; Wang, N.; Zheng, S.-S.; Ling, H.-Q. The upregulation of NtAN2 expression at low temperature is required for anthocyanin accumulation in juvenile leaves of Lc-transgenic tobacco (Nicotiana tabacum L.). J. Genet. Genom. 2012, 39, 149–156. [Google Scholar] [CrossRef]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef] [PubMed]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Zhao, Y.; Yang, J.; He, Y.; Li, G.; Ma, W.; Huang, X.; Su, J. Transcription factor PyHY5 binds to the promoters of PyWD40 and PyMYB10 and regulates its expression in red pear ‘Yunhongli No. 1’. Plant Physiol. Biochem. 2020, 154, 665–674. [Google Scholar] [CrossRef]
- Cao, X.; Qiu, Z.; Wang, X.; Van Giang, T.; Liu, X.; Wang, J.; Wang, X.; Gao, J.; Guo, Y.; Du, Y.; et al. A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J. Exp. Bot. 2017, 68, 5745–5758. [Google Scholar] [CrossRef]
- Zuluaga, D.L.; Gonzali, S.; Loreti, E.; Pucciariello, C.; Degl’Innocenti, E.; Guidi, L.; Alpi, A.; Perata, P. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct. Plant Biol. 2008, 35, 606–618. [Google Scholar] [CrossRef]
- Andrea, M.; Francesco, E.F.; Sergio, I.; Alessandra, G.; Maria, A.M.; Cinzia, C.; Lorenzo, B.; Arianna, M.; Cecilia, C.; Patrizia, R.; et al. Identification of a N. R3 MYB type repressor and functional characterization of the members of the MBW transcriptional complex involved in anthocyanin biosynthesis in eggplant (S. melongena L.). PLoS ONE 2020, 15, e0232986. [Google Scholar] [CrossRef]
- Albert, N.W.; Griffiths, A.G.; Cousins, G.R.; Verry, I.M.; Williams, W.M. Anthocyanin leaf markings are regulated by a family of R2R3-MYBgenes in the genus Trifolium. New Phytol. 2014, 205, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Mathews, H.; Clendennen, S.K.; Caldwell, C.G.; Liu, X.L.; Connors, K.; Matheis, N.; Schuster, D.K.; Menasco, D.J.; Wagoner, W.; Lightner, J.; et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 2003, 15, 1689–1703. [Google Scholar] [CrossRef]
- Deluc, L.; Barrieu, F.; Marchive, C.; Lauvergeat, V.; Decendit, A.; Richard, T.; Carde, J.-P.; Mérillon, J.-M.; Hamdi, S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006, 140, 499–511. [Google Scholar] [CrossRef]
- Cavallini, E.; Matus, J.T.; Finezzo, L.; Zenoni, S.; Loyola, R.; Guzzo, F.; Schlechter, R.; Ageorges, A.; Arce-Johnson, P.; Tornielli, G.B. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015, 167, 1448–1470. [Google Scholar] [CrossRef]
- Tanikawa, J.; Yasukawa, T.; Enari, M.; Ogata, K.; Nishimura, Y.; Ishii, S.; Sarai, A. Recognition of specific DNA sequences by the c-myb protooncogene product: Role of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. USA 1993, 90, 9320–9324. [Google Scholar] [CrossRef]
- Zhou, H.; Liao, L.; Xu, S.; Ren, F.; Zhao, J.; Ogutu, C.; Wang, L.; Jiang, Q.; Han, Y. Two amino acid changes in the R3 repeat cause functional divergence of two clustered MYB10 genes in peach. Plant Mol. Biol. 2018, 98, 169–183. [Google Scholar] [CrossRef]
- Williams, C.E.; Grotewold, E. Differences between plant and animal myb domains are fundamental for DNA binding activity, and chimeric myb domains have novel DNA binding specificities. J. Biol. Chem. 1997, 272, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Brendolise, C.; Espley, R.V.; Lin-Wang, K.; Laing, W.; Peng, Y.; McGhie, T.; Dejnoprat, S.; Tomes, S.; Hellens, R.P.; Allan, A.C. Multiple copies of a simple MYB-binding site confers trans-regulation by specific flavonoid-related R2R3 MYBs in diverse species. Front. Plant Sci. 2017, 8, 1864. [Google Scholar] [CrossRef]
- Uimari, A.; Strommer, J. MYB26: A MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 1997, 12, 1273–1284. [Google Scholar] [CrossRef]
- Planchais, S.; Perennes, C.; Glab, N.; Mironov, V.; Inzé, D.; Bergounioux, C. Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath;CycB1;1 transcription and identification of putative regulatory proteins. Plant Mol. Biol. 2002, 50, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E.; Drummond, B.J.; Bowen, B.; Peterson, T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 1994, 76, 543–553. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 1–15. [Google Scholar] [CrossRef]
- Feldbrugge, M.; Sprenger, M.; Hahlbrock, K.; Weisshaar, B. PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J. 1997, 11, 1079–1093. [Google Scholar] [CrossRef]
- Xiao, W. Cloning and Preliminary Functional analysis of LdMYB6 gene from Lilium davidii var. Unicolor. Master’s Thesis, Jilin Agricultural University, Changchun, China, 2019. [Google Scholar]
- Wen, C.-H.; Chu, F.-H. A R2R3-MYB gene LfMYB113 is responsible for autumn leaf coloration in formosan sweet gum (Liquidambar formosana Hance). Plant Cell Physiol. 2017, 58, 508–521. [Google Scholar] [CrossRef]
- Mes, P.J.; Boches, P.; Myers, J.R.; Durst, R. Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hortic. Sci. 2008, 133, 262–269. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383. [Google Scholar] [CrossRef]
- Elomaa, P.; Uimari, A.; Mehto, M.; Albert, V.A.; Laitinen, R.A.; Teeri, T.H. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol. 2003, 133, 1831–1842. [Google Scholar] [CrossRef]
- Li, C.; Qiu, J.; Yang, G.; Huang, S.; Yin, J. Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum (Hort.). Plant Cell Rep. 2016, 35, 2151–2165. [Google Scholar] [CrossRef]
- Docimo, T.; Francese, G.; Ruggiero, A.; Batelli, G.; De Palma, M.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Mennella, G.; Tucci, M. Phenylpropanoids accumulation in eggplant fruit: Characterization of biosynthetic genes and regulation by a MYB transcription factor. Front. Plant Sci. 2016, 6, 1233. [Google Scholar] [CrossRef]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef]
- Chen, K.; Du, L.; Liu, H.; Liu, Y. A novel R2R3-MYB from grape hyacinth, MaMybA, which is different from MaAN2, confers intense and magenta anthocyanin pigmentation in tobacco. BMC Plant Biol. 2019, 19, 1–15. [Google Scholar] [CrossRef]
- Hsu, C.-C.; Chen, Y.-Y.; Tsai, W.-C.; Chen, W.-H.; Chen, H.-H. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis orchids. Plant Physiol. 2015, 168, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Bai, S.; Yaegaki, H.; Tamura, T.; Hihara, S.; Moriguchi, T.; Oda, K. The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol. 2015, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Durbin, M.L.; McCaig, B.; Clegg, M.T. Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol. Biol. 2000, 42, 79–92. [Google Scholar] [CrossRef]
- Gu, Z.; Zhu, J.; Hao, Q.; Yuan, Y.-W.; Duan, Y.-W.; Men, S.; Wang, Q.; Hou, Q.; Liu, Z.A.; Shu, Q.; et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 2019, 60, 599–611. [Google Scholar] [CrossRef]
- Lim, S.-H.; Song, J.-H.; Kim, D.-H.; Kim, J.K.; Lee, J.-Y.; Kim, Y.-M.; Ha, S.-H. Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep. 2016, 35, 641–653. [Google Scholar] [CrossRef]
- Chiu, L.-W.; Zhou, X.; Burke, S.; Wu, X.; Prior, R.L.; Li, L. The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 2010, 154, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Tan, W.; Yang, H.; Zhang, L.; Li, T.; Liu, B.; Zhang, D.; Lin, H. Regulation of anthocyanin accumulation via MYB75/HAT1/TPL-mediated transcriptional repression. PLoS Genet. 2019, 15, e1007993. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, Y.; Guo, L.; Gong, N.; Pang, Y.; Jiang, W.; Liu, Y.; Jiang, X.; Zhao, L.; Wang, Y.; et al. Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front. Plant Sci. 2017, 8, 943. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin-Wang, K.; Liao, L.; Gu, C.; Lu, Z.; Allan, A.C.; Han, Y. Peach MYB7 activates transcription of the proanthocyanidin pathway gene encoding leucoanthocyanidin reductase, but not anthocyanidin reductase. Front. Plant Sci. 2015, 6, 908. [Google Scholar] [CrossRef]
- Hanna-Rose, W.; Hansen, U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996, 12, 229–234. [Google Scholar] [CrossRef]
- Wan, S.; Li, C.; Ma, X.; Luo, K. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017, 36, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Park, D.Y.; Park, K.I.; Kil Kim, C. Differential expression of anthocyanin structural genes and transcription factors determines coloration patterns in gerbera flowers. 3 Biotech. 2018, 8, 393. [Google Scholar] [CrossRef]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, K.; Sun, Z.; Yan, M.; Chen, C.; Zhang, X.; Tang, Y.; Wu, Y. LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis. Plant Physiol. 2017, 174, 1348–1358. [Google Scholar] [CrossRef]
- Pérez-Díaz, J.R.; Pérez-Díaz, J.; Madrid-Espinoza, J.; González-Villanueva, E.; Moreno, Y.; Ruiz-Lara, S.N. member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 2016, 90, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Fornalé, S.; Lopez, E.; Salazar-Henao, J.E.; Fernández-Nohales, P.; Rigau, J.; Caparros-Ruiz, D. AtMYB7, a N. player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A Conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Bashandy, H.; Ainasoja, M.; Kontturi, J.; Pietiäinen, M.; Laitinen, R.A.E.; Albert, V.A.; Valkonen, J.P.T.; Elomaa, P.; Teeri, T.H. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytol. 2013, 201, 1469–1483. [Google Scholar] [CrossRef]
- Zhang, W.; Ning, G.; Lv, H.; Liao, L.; Bao, M. Single MYB-type transcription factor AtCAPRICE: A N. efficient tool to engineer the production of anthocyanin in tobacco. Biochem. Biophys. Res. Commun. 2009, 388, 742–747. [Google Scholar] [CrossRef]
- Zhu, H.-F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Colanero, S.; Perata, P.; Gonzali, S. The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 2018, 9, 830. [Google Scholar] [CrossRef]
- Hidalgo, P.; Ansari, A.Z.; Schmidt, P.; Hare, B.; Simkovich, N.; Farrell, S.; Shin, E.J.; Ptashne, M.; Wagner, G. Recruitment of the transcriptional machinery through GAL11P: Structure and interactions of the GAL4 dimerization domain. Genes Dev. 2001, 15, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, H.; Gierer, A. Applications of a theory of biological pattern formation based on lateral inhibition. J. Cell Sci. 1974, 15, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Sakai, M.; Yamagishi, M.; Matsuyama, K. Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.). Plant Cell Rep. 2019, 38, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Jeong, C.Y.; Kwon, J.; Van Kien, V.; Lee, D.; Hong, S.-W.; Lee, H. Drastic anthocyanin increase in response to PAP1 overexpression in fls1 knockout mutant confers enhanced osmotic stress tolerance in Arabidopsis thaliana. Plant Cell Rep. 2016, 35, 2369–2379. [Google Scholar] [CrossRef]
- Ban, Y.; Honda, C.; Bessho, H.; Pang, X.-M.; Moriguchi, T. Suppression subtractive hybridization identifies genes induced in response to UV-B irradiation in apple skin: Isolation of a putative UDP-glucose 4-epimerase. J. Exp. Bot. 2007, 58, 1825–1834. [Google Scholar] [CrossRef]
- Henry-Kirk, R.A.; Plunkett, B.; Hall, M.; McGhie, T.; Allan, A.C.; Wargent, J.J.; Espley, R.V. Solar UV light regulates flavonoid metabolism in apple (Malus × domestica). Plant Cell Environ. 2018, 41, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Schenke, D.; Böttcher, C.; Scheel, D. Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ. 2011, 34, 1849–1864. [Google Scholar] [CrossRef]
- Plunkett, B.J.; Henry-Kirk, R.; Friend, A.; Diack, R.; Helbig, S.; Mouhu, K.; Tomes, S.; Dare, A.P.; Espley, R.V.; Putterill, J.; et al. Apple B-box factors regulate light-responsive anthocyanin biosynthesis genes. Sci. Rep. 2019, 9, 17762. [Google Scholar] [CrossRef]
- Wu, M.; Liu, J.; Song, L.; Li, X.; Cong, L.; Yue, R.; Yang, C.; Liu, Z.; Xu, L.; Wang, Z. Differences among the anthocyanin accumulation patterns and related gene expression levels in red pears. Plants 2019, 8, 100. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Mao, K.; Zhao, C.; Zhao, X.-Y.; Zhang, H.-L.; Shu, H.-R.; Hao, Y.-J. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef] [PubMed]
- McNellis, T.W.; Torii, K.U.; Deng, X.W. Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 1996, 8, 1491–1503. [Google Scholar] [CrossRef]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Lokhande, S.D.; Ogawa, K.; Tanaka, A.; Hara, T. Effect of temperature on ascorbate peroxidase activity and flowering of Arabidopsis thaliana ecotypes under different light conditions. J. Plant Physiol. 2003, 160, 57–64. [Google Scholar] [CrossRef]
- Xie, X.-B.; Li, S.; Zhang, R.-F.; Zhao, J.; Chen, Y.-C.; Zhao, Q.; Yao, Y.-X.; You, C.-X.; Zhang, X.-S.; Hao, Y.-J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.-I.; Jung, H.-J.; Hur, Y.; Nou, I.-S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2014, 15, 383–394. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, S.; Liu, Z.; Wang, L.; Bi, Y. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 2011, 168, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Li, H.B. Overexpression of IbMYB1 gene enhanced tolerance to soil drought stress in sweet potato. Plant Physiol. J. 2015, 51, 1440–1446. [Google Scholar] [CrossRef]
- Shi, X.W. Identification and Analysis of microRNA Related to Adversity Stress and Anthocyanin Biosynthesis in Sweet Potato. Master’s Thesis, Shanxi Agricultural University, Taigu, China, 2018. [Google Scholar]
- Quattrocchio, F.; Verweij, W.; Kroon, A.; Spelt, C.; Mol, J.; Koes, R. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 2006, 18, 1274–1291. [Google Scholar] [CrossRef] [PubMed]
- Kum, Y.; Adao, K. Cause of blue petal colour. Nature 1995, 373, 291. [Google Scholar] [CrossRef]
- Zhu, T.T.; Liang, D. Progress in the study of the regulation of anthocyanin synthesis by R2R3-MYB in fruits. Genom. Appl. Biol. 2016, 35, 985–991. [Google Scholar] [CrossRef]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-F.; Ning, G.-X.; Mao, J.; Guo, Z.-G.; Zhou, Q.; Chen, B.-H. Whole-genome DNA methylation patterns and complex associations with gene expression associated with anthocyanin biosynthesis in apple fruit skin. Planta 2019, 250, 1833–1847. [Google Scholar] [CrossRef]
- Wang, B.; Luo, Q.; Li, Y.; Yin, L.; Zhou, N.; Li, X.; Gan, J.; Dong, A. Structural insights into target DNA recognition by R2R3-MYB transcription factors. Nucleic Acids Res. 2020, 48, 460–471. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef]
- Van Wersch, R.; Gao, F.; Zhang, Y. Mitogen-activated protein kinase 6 negatively regulates anthocyanin induction in Arabidopsis. Plant Signal. Behav. 2018, 213, e1526000-3. [Google Scholar] [CrossRef]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).