The Landscape of the Genomic Distribution and the Expression of the F-Box Genes Unveil Genome Plasticity in Hexaploid Wheat during Grain Development and in Response to Heat and Drought Stress
Abstract
:1. Introduction
2. Results
2.1. Structure, Organization and Genomic Distribution of the TaFBX Family in Bread Wheat
2.1.1. Identification of Wheat TaFBX Gene Family
2.1.2. Genomic Distribution of TaFBX among Subgenomes, Chromosomes and Chromosomal Regions
2.1.3. Structuration of the TaFBX Genes into Subfamilies
2.1.4. Lineage-Specific Expansion of Wheat TaFBX Genes
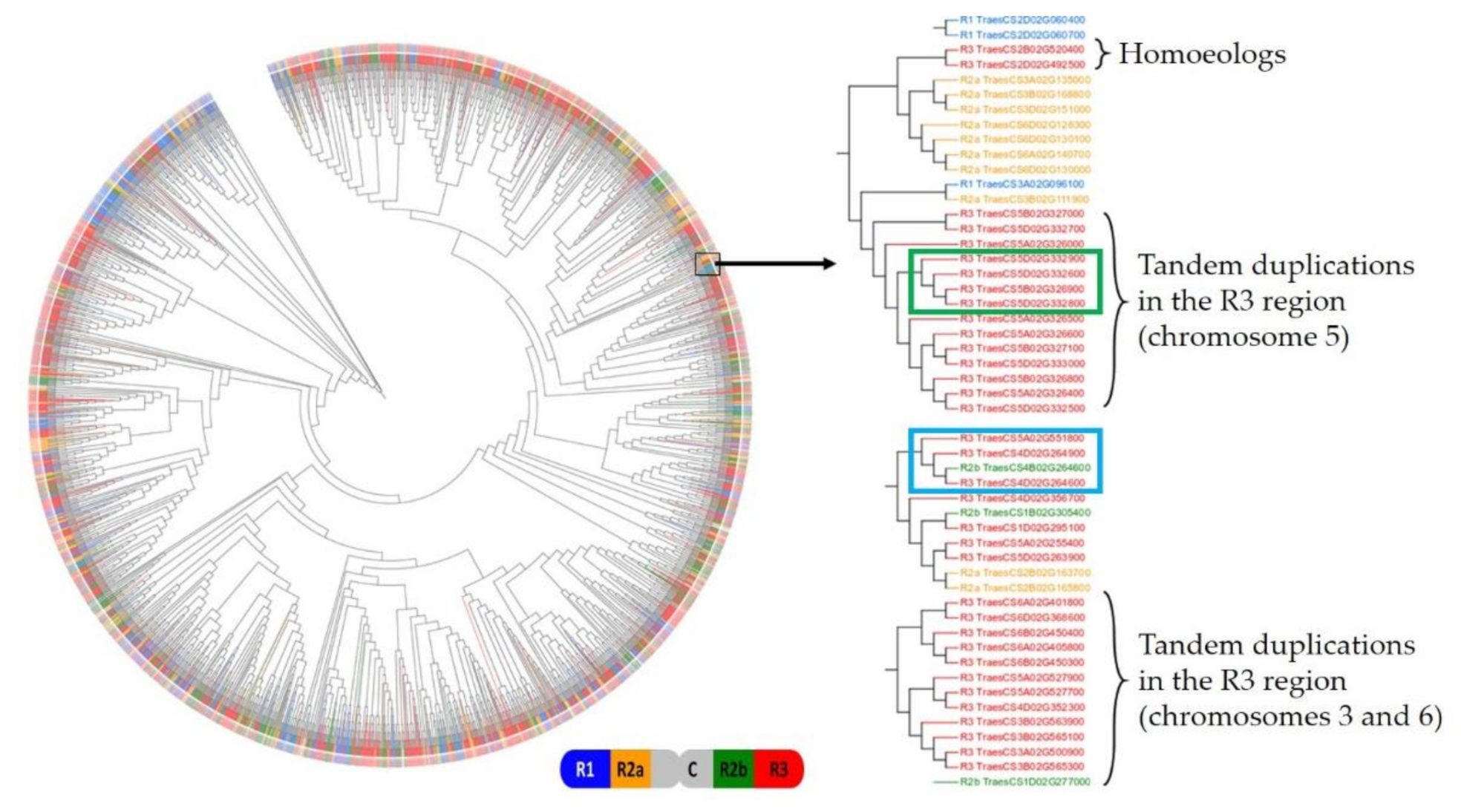
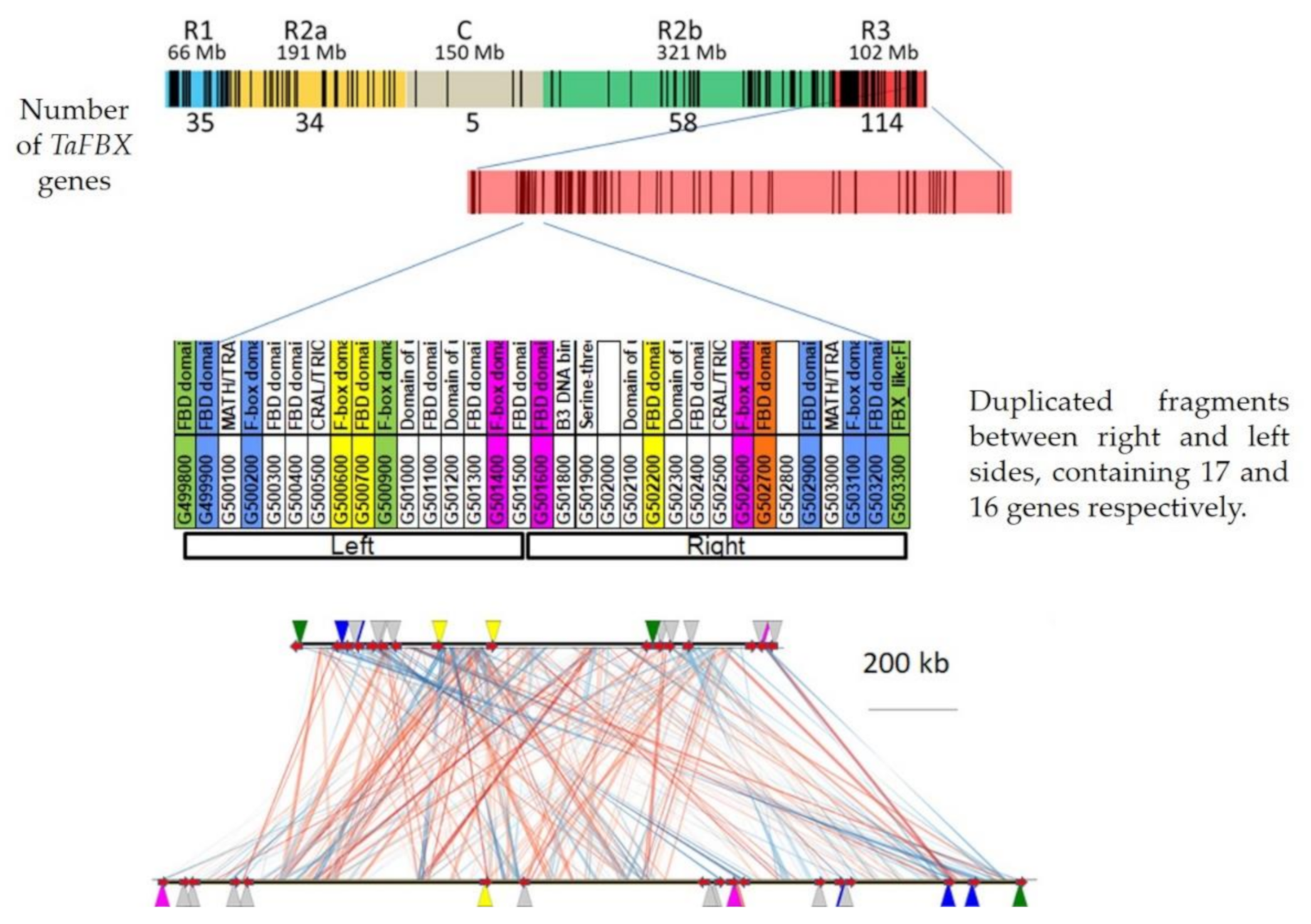
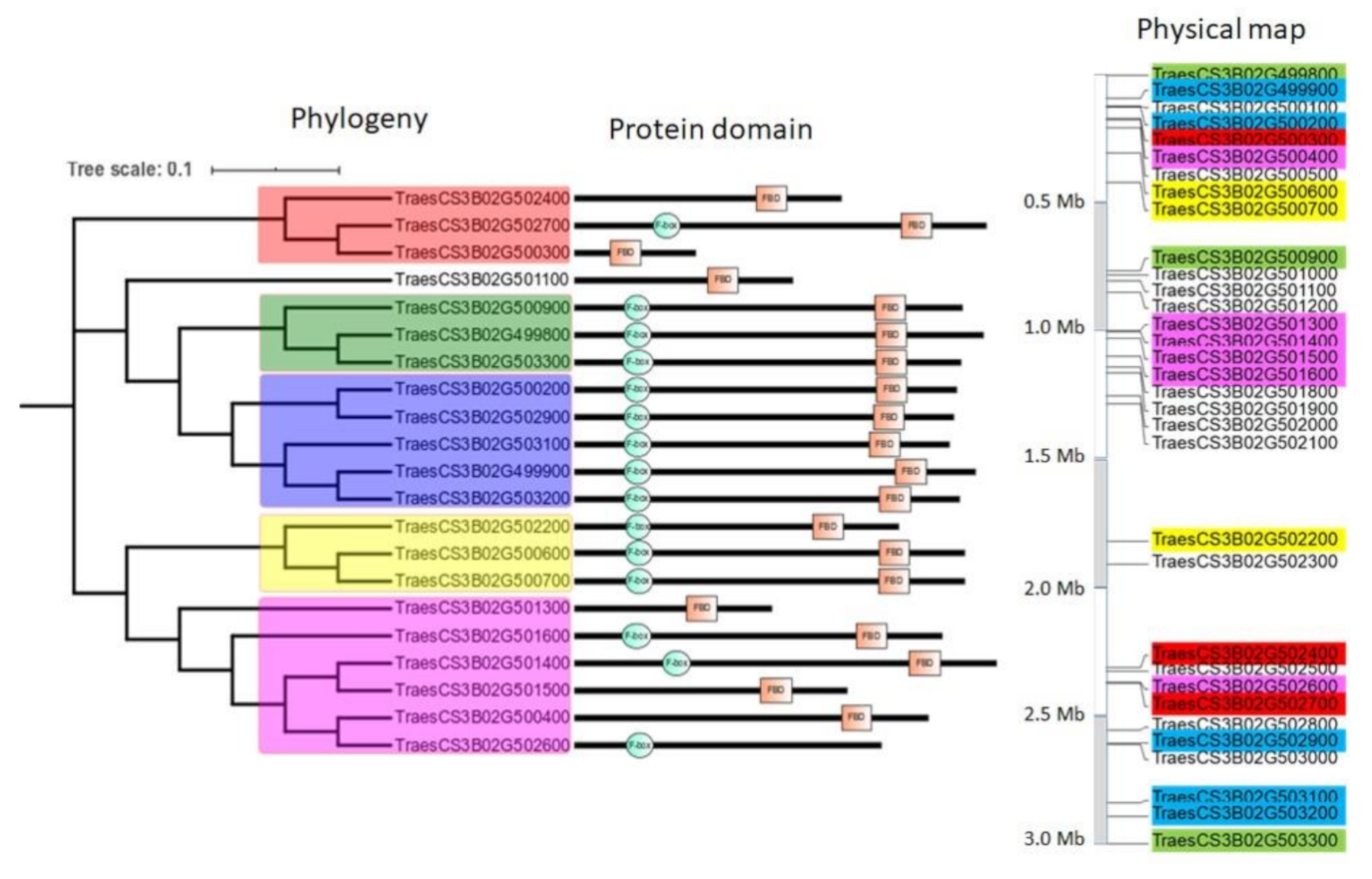
2.1.5. Genomic Structuration and Duplication Fates of TaFBX Genes
2.2. TaFBX Family Expression Pattern
2.2.1. TaFBX Expression during Wheat Embryogenesis and Grain Development
2.2.2. TaFBX Gene Expression in Response to Heat and Drought Stress
3. Discussion
3.1. FBX Family Size Is Larger in Wheat Than in Other Known Plant Species
3.2. FBX Gene Family Has Experienced Local Expansion through Retrotransposition, Tandem and Interchromosomal Duplications
3.3. The TaFBX Family Structured into Subfamilies Is Unequally Expanded in Wheat
3.4. The FBX Gene Distribution Is Not Homogeneous in the Wheat Genome
3.5. FBX Gene Distribution Is Not Homogeneous among the Five Chromosomal Regions of the Bread Wheat Genome
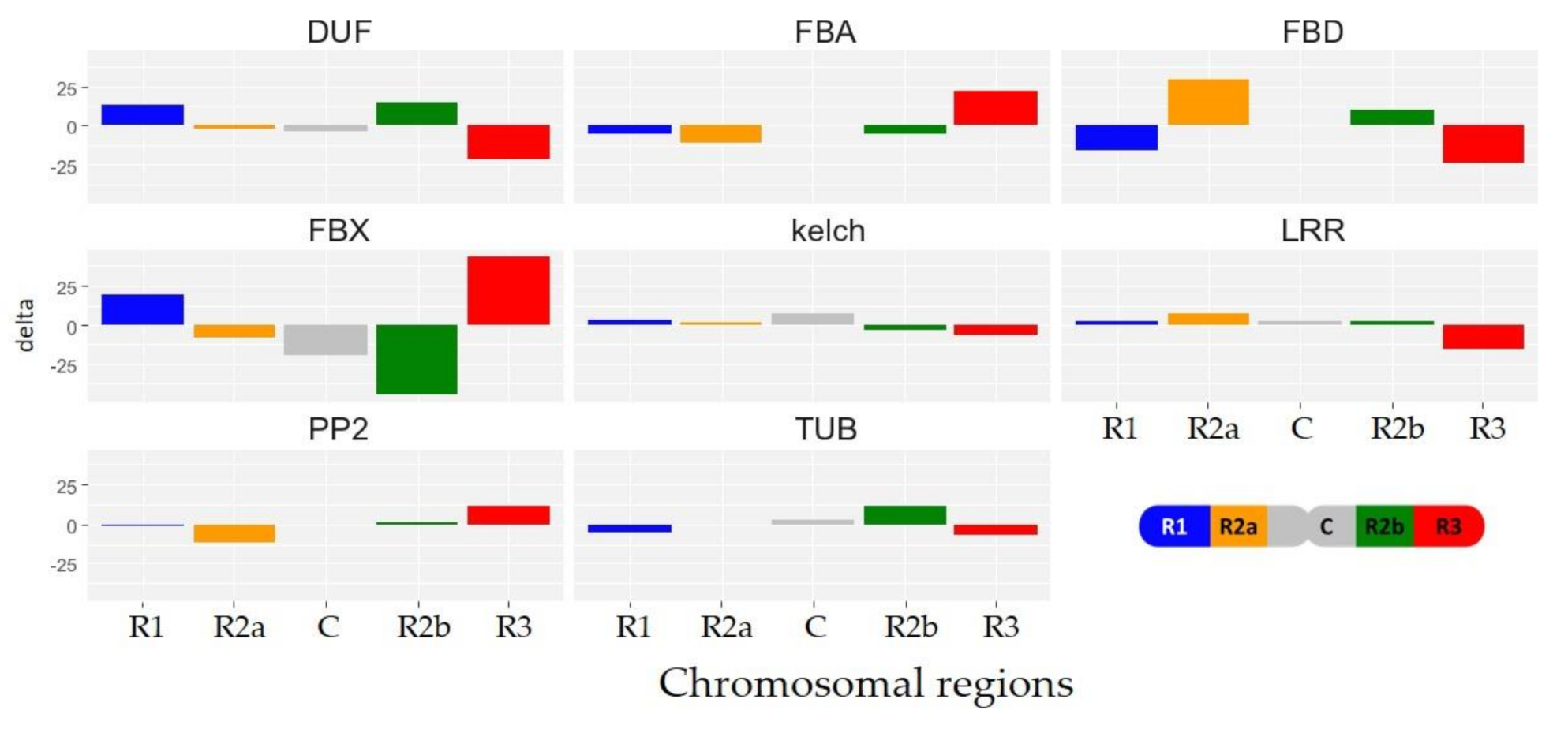
3.6. Differentially Expressed TaFBX Are Concentrated in the R2b Region
4. Materials and Methods
4.1. Survey of TaFBX in the Wheat Sequence
4.2. Phylogenetic Analysis of the FBX Family
4.3. Chromosomal Distribution of TaFBX Coding Genes
4.4. Evolutionary Study of the R3 Region of the Chromosome 3B
4.5. Expression of TaFBX Genes
4.5.1. TaFBX Gene Expression during Embryogenesis and Grain Development
4.5.2. TaFBX Gene Expression in Response to Heat and Drought Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEG | Differentially Expressed Gene |
| FBX | F-box proteins |
| Ta | Triticum aestivum, bread wheat |
| HC | High Confidence |
References
- Parzych, K.R.; Klionsky, D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [Green Version]
- Cundiff, M.D.; Hurley, C.M.; Wong, J.D.; Boscia, J.A.; Bashyal, A.; Rosenberg, J.; Reichard, E.L.; Nassif, N.D.; Brodbelt, J.S.; Kraut, D.A. Ubiquitin Receptors Are Required for Substrate-Mediated Activation of the Proteasome’s Unfolding Ability. Sci. Rep. 2019, 9, 14506. [Google Scholar] [CrossRef]
- Groll, M.; Huber, R. Substrate Access and Processing by the 20S Proteasome Core Particle. Int. J. Biochem. Cell Biol. 2003, 35, 606–616. [Google Scholar] [CrossRef]
- Vierstra, R.D. The Ubiquitin–26S Proteasome System at the Nexus of Plant Biology. Nat. Rev. Mol. Cell. Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Li, L.; Su, Z. PlantsUPS: A Database of Plants’ Ubiquitin Proteasome System. BMC Genom. 2009, 10, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Wang, C.; Higgins, J.D.; Yu, J.; Zong, J.; Lu, P.; Zhang, D.; Liang, W. MEIOTIC F-BOX Is Essential for Male Meiotic DNA Double-Strand Break Repair in Rice[OPEN]. Plant Cell 2016, 28, 1879–1893. [Google Scholar] [CrossRef] [Green Version]
- Woo, H.R.; Chung, K.M.; Park, J.-H.; Oh, S.A.; Ahn, T.; Hong, S.H.; Jang, S.K.; Nam, H.G. ORE9, an F-Box Protein That Regulates Leaf Senescence in Arabidopsis. Plant Cell 2001, 13, 1779–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Luong, P.; Huq, E. The F-Box Protein MAX2 Functions as a Positive Regulator of Photomorphogenesis in Arabidopsis. Plant Physiol. 2007, 145, 1471–1483. [Google Scholar] [CrossRef] [Green Version]
- Stirnberg, P.; van de Sande, K.; Leyser, H.M.O. MAX1 and MAX2 Control Shoot Lateral Branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar]
- Dieterle, M.; Zhou, Y.-C.; Schäfer, E.; Funk, M.; Kretsch, T. EID1, an F-Box Protein Involved in Phytochrome A-Specific Light Signaling. Genes Dev. 2001, 15, 939–944. [Google Scholar] [CrossRef] [Green Version]
- Somers, D.E.; Schultz, T.F.; Milnamow, M.; Kay, S.A. ZEITLUPE Encodes a Novel Clock-Associated PAS Protein from Arabidopsis. Cell 2000, 101, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Samach, A.; Klenz, J.E.; Kohalmi, S.E.; Risseeuw, E.; Haughn, G.W.; Crosby, W.L. The UNUSUAL FLORAL ORGANS Gene of Arabidopsis Thaliana Is an F-Box Protein Required for Normal Patterning and Growth in the Floral Meristem. Plant J. 1999, 20, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Chae, E.; Tan, Q.K.-G.; Hill, T.A.; Irish, V.F. An Arabidopsis F-Box Protein Acts as a Transcriptional Co-Factor to Regulate Floral Development. Development 2008, 135, 1235–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, B.M.; Walker, J.M.; Gagne, J.M.; Emborg, T.J.; Hemmann, G.; Bleecker, A.B.; Vierstra, R.D. The Arabidopsis EIN3 Binding F-Box Proteins EBF1 and EBF2 Have Distinct but Overlapping Roles in Ethylene Signaling. Plant Cell 2007, 19, 509–523. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, L.E.; Keller, K.; Chan, K.X.; Gessel, M.M.; Thines, B.C. Transcriptome Analysis Uncovers Arabidopsis F-BOX STRESS INDUCED 1 as a Regulator of Jasmonic Acid and Abscisic Acid Stress Gene Expression. BMC Genom. 2017, 18, 533. [Google Scholar] [CrossRef]
- Xu, L.; Liu, F.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.; Xie, D. The SCFCOI1 Ubiquitin-Ligase Complexes Are Required for Jasmonate Response in Arabidopsis. Plant Cell 2002, 14, 1919–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate Perception by Inositol-Phosphate-Potentiated COI1–JAZ Co-Receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Xie, D.-X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis Gene Required for Jasmonate-Regulated Defense and Fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of Auxin Perception by the TIR1 Ubiquitin Ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. The Arabidopsis Kelch Repeat F-Box E3 Ligase ARKP1 Plays a Positive Role for the Regulation of Abscisic Acid Signaling. Plant Mol. Biol. Rep. 2016, 34, 582–591. [Google Scholar] [CrossRef]
- Xu, G.; Cui, Y.; Wang, M.; Li, M.; Yin, X.; Xia, X. OsMsr9, a Novel Putative Rice F-Box Containing Protein, Confers Enhanced Salt Tolerance in Transgenic Rice and Arabidopsis. Mol. Breed. 2014, 34, 1055–1064. [Google Scholar] [CrossRef]
- Yan, Y.-S.; Chen, X.-Y.; Yang, K.; Sun, Z.-X.; Fu, Y.-P.; Zhang, Y.-M.; Fang, R.-X. Overexpression of an F-Box Protein Gene Reduces Abiotic Stress Tolerance and Promotes Root Growth in Rice. Mol. Plant 2011, 4, 190–197. [Google Scholar] [CrossRef] [Green Version]
- Iantcheva, A.; Boycheva, I.; Vassileva, V.; Revalska, M.; Zechirov, G. Cyclin-like F-Box Protein Plays a Role in Growth and of the Three Model Species Medicago Truncatula, Lotus Japonicus, and Arabidopsis Thaliana. Res. Rep. Biol. 2015, 6, 117–130. [Google Scholar] [CrossRef] [Green Version]
- Kipreos, E.T.; Pagano, M. The F-Box Protein Family. Genome Biol. 2000, 1, reviews3002.1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elzanati, O.; Roche, J.; Boulaflous-Stevens, A.; Mouzeyar, S.; Bouzidi, M.F. Genome-Wide Analysis, Classification, Expression and Interaction of Physcomitrella Patens SKP1-like (PpSKP) and F-Box (FBX) Genes. Plant Gene 2017, 12, 13–22. [Google Scholar] [CrossRef]
- HajSalah El Beji, I.; Mouzeyar, S.; Bouzidi, M.-F.; Roche, J. Expansion and Functional Diversification of SKP1-Like Genes in Wheat (Triticum Aestivum L.). Int. J. Mol. Sci. 2019, 20, 3295. [Google Scholar] [CrossRef] [Green Version]
- Skowyra, D.; Craig, K.L.; Tyers, M.; Elledge, S.J.; Harper, J.W. F-Box Proteins Are Receptors That Recruit Phosphorylated Substrates to the SCF Ubiquitin-Ligase Complex. Cell 1997, 91, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Cardozo, T.; Lovering, R.C.; Elledge, S.J.; Pagano, M.; Harper, J.W. Systematic Analysis and Nomenclature of Mammalian F-Box Proteins. Genes Dev. 2004, 18, 2573–2580. [Google Scholar] [CrossRef] [Green Version]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. SnapShot: F Box Proteins I. Cell 2009, 137, 1160–1160.e1. [Google Scholar] [CrossRef] [Green Version]
- Hua, Z.; Zou, C.; Shiu, S.-H.; Vierstra, R.D. Phylogenetic Comparison of F-Box (FBX) Gene Superfamily within the Plant Kingdom Reveals Divergent Evolutionary Histories Indicative of Genomic Drift. PLoS ONE 2011, 6, e16219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Garg, V.; Kant, C.; Bhatia, S. Genome-Wide Survey and Expression Analysis of F-Box Genes in Chickpea. BMC Genom. 2015, 16, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Tian, Z.; Li, H.; Guo, Y.; Zhang, Y.; Roberts, J.A.; Zhang, X.; Miao, Y. Genome-Wide Analysis and Characterization of F-Box Gene Family in Gossypium Hirsutum L. BMC Genom. 2019, 20, 993. [Google Scholar] [CrossRef] [PubMed]
- Semple, C.A.M. The Comparative Proteomics of Ubiquitination in Mouse. Genome Res. 2003, 13, 1389–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional Analysis of the RING-Type Ubiquitin Ligase Family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.H. Evolution of Duplicate Genes and Pseudogenes. In Evolution of Genes and Proteins; Sinauer Associates: Sunderland, MA, USA, 1983; pp. 14–37. [Google Scholar]
- Thomas, J.H. Adaptive Evolution in Two Large Families of Ubiquitin-Ligase Adapters in Nematodes and Plants. Genome Res 2006, 16, 1017–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide Analysis of MIKC-type MADS -box Genes in Wheat: Pervasive Duplications, Functional Conservation and Putative Neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef] [Green Version]
- Guérin, C.; Roche, J.; Allard, V.; Ravel, C.; Mouzeyar, S.; Bouzidi, M.F. Genome-Wide Analysis, Expansion and Expression of the NAC Family under Drought and Heat Stresses in Bread Wheat (T. Aestivum L.). PLoS ONE 2019, 14, e0213390. [Google Scholar] [CrossRef] [Green Version]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling Angiosperm Genome Evolution by Phylogenetic Analysis of Chromosomal Duplication Events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Simillion, C.; Vandepoele, K.; Montagu, M.C.E.V.; Zabeau, M.; de Peer, Y.V. The Hidden Duplication Past of Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2002, 99, 13627–13632. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-Box Proteins in Rice. Genome-Wide Analysis, Classification, Temporal and Spatial Gene Expression during Panicle and Seed Development, and Regulation by Light and Abiotic Stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.-H. Importance of Lineage-Specific Expansion of Plant Tandem Duplicates in the Adaptive Response to Environmental Stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Hong, M.J.; Kim, J.-B.; Seo, Y.W.; Kim, D.Y. F-Box Genes in the Wheat Genome and Expression Profiling in Wheat at Different Developmental Stages. Genes 2020, 11, 1154. [Google Scholar] [CrossRef]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. ExpVIP: A Customizable RNA-Seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Wheat Genome Sequencing Consortium(IWGSC); Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; et al. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, eaar7191. [Google Scholar]
- Xu, G.; Ma, H.; Nei, M.; Kong, H. Evolution of F-Box Genes in Plants: Different Modes of Sequence Divergence and Their Relationships with Functional Diversification. Proc. Natl. Acad. Sci. USA 2009, 106, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, D.; Quilichini, T.D.; Liu, Z.; Gao, P.; Pan, Y.; Li, Q.; Nilsen, K.T.; Venglat, P.; Esteban, E.; Pasha, A.; et al. The Transcriptional Landscape of Polyploid Wheats and Their Diploid Ancestors during Embryogenesis and Grain Development. Plant Cell 2019, 31, 2888–2911. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal Transcriptome Profiling Reveals Expression Partitioning of Homeologous Genes Contributing to Heat and Drought Acclimation in Wheat (Triticum Aestivum L.). BMC Plant Biol. 2015, 15, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Majee, M.; Kumar, S.; Kathare, P.K.; Wu, S.; Gingerich, D.; Nayak, N.R.; Salaita, L.; Dinkins, R.; Martin, K.; Goodin, M.; et al. KELCH F-BOX Protein Positively Influences Arabidopsis Seed Germination by Targeting PHYTOCHROME-INTERACTING FACTOR1. Proc. Natl. Acad. Sci. USA 2018, 115, E4120–E4129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jayaweera, D.; Peters, J.L.; Szecsi, J.; Bendahmane, M.; Roberts, J.A.; González-Carranza, Z.H. The Arabidopsis Thaliana F-Box Gene HAWAIIAN SKIRT Is a New Player in the MicroRNA Pathway. PLoS ONE 2017, 12, e0189788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McSteen, P.; Zhao, Y. Plant Hormones and Signaling: Common Themes and New Developments. Dev. Cell 2008, 14, 467–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kepinski, S.; Leyser, O. The Arabidopsis F-Box Protein TIR1 Is an Auxin Receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ Repressor Proteins Are Targets of the SCF(COI1) Complex during Jasmonate Signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Scaffidi, A.; Dun, E.A.; Waters, M.T.; Flematti, G.R.; Dixon, K.W.; Beveridge, C.A.; Ghisalberti, E.L.; Smith, S.M. F-Box Protein MAX2 Has Dual Roles in Karrikin and Strigolactone Signaling in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 8897–8902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gusti, A.; Baumberger, N.; Nowack, M.; Pusch, S.; Eisler, H.; Potuschak, T.; De Veylder, L.; Schnittger, A.; Genschik, P. The Arabidopsis Thaliana F-Box Protein FBL17 Is Essential for Progression through the Second Mitosis during Pollen Development. PLoS ONE 2009, 4, e4780. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 Conveys Crucial Timing Information for CONSTANS Stabilization in the Photoperiodic Flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Gou, M.; Guo, C.; Yang, H.; Liu, C.-J. Down-Regulation of Kelch Domain-Containing F-Box Protein in Arabidopsis Enhances the Production of (Poly)Phenols and Tolerance to Ultraviolet Radiation. Plant Physiol. 2015, 167, 337–350. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Abrahan, C.; Colquhoun, T.A.; Liu, C.-J. A Proteolytic Regulator Controlling Chalcone Synthase Stability and Flavonoid Biosynthesis in Arabidopsis. Plant Cell 2017, 29, 1157–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.Y.; Jung, K.W.; Jeung, J.U.; Shin, J.S. A Novel F-Box Protein Represses Endothecial Secondary Wall Thickening for Anther Dehiscence in Arabidopsis Thaliana. J. Plant Physiol. 2012, 169, 212–216. [Google Scholar] [CrossRef]
- Bao, Y.; Song, W.-M.; Jin, Y.-L.; Jiang, C.-M.; Yang, Y.; Li, B.; Huang, W.-J.; Liu, H.; Zhang, H.-X. Characterization of Arabidopsis Tubby-like Proteins and Redundant Function of AtTLP3 and AtTLP9 in Plant Response to ABA and Osmotic Stress. Plant Mol. Biol. 2014, 86, 471–483. [Google Scholar] [CrossRef]
- Zhou, S.; Sun, X.; Yin, S.; Kong, X.; Zhou, S.; Xu, Y.; Luo, Y.; Wang, W. The Role of the F-Box Gene TaFBA1 from Wheat (Triticum Aestivum L.) in Drought Tolerance. Plant Physiol. Biochem. 2014, 84, 213–223. [Google Scholar] [CrossRef]
- Stefanowicz, K.; Lannoo, N.; Zhao, Y.; Eggermont, L.; Van Hove, J.; Al Atalah, B.; Van Damme, E.J.M. Glycan-Binding F-Box Protein from Arabidopsis Thaliana Protects Plants from Pseudomonas Syringae Infection. BMC Plant Biol. 2016, 16, 213. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.-M.; Kong, X.-Z.; Kang, H.-H.; Sun, X.-D.; Wang, W. The Involvement of Wheat F-Box Protein Gene TaFBA1 in the Oxidative Stress Tolerance of Plants. PLoS ONE 2015, 10, e0122117. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Zhang, G.; Zhou, S.; Ren, Y.; Wang, W. The Improvement of Salt Tolerance in Transgenic Tobacco by Overexpression of Wheat F-Box Gene TaFBA1. Plant Sci. 2017, 259, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Song, J.B.; Wang, Y.X.; Li, H.B.; Li, B.W.; Zhou, Z.S.; Gao, S.; Yang, Z.M. The F-Box Family Genes as Key Elements in Response to Salt, Heavy Mental, and Drought Stresses in Medicago Truncatula. Funct. Integr. Genom. 2015, 15, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Villalobos, L.I.A.; Nill, C.; Marrocco, K.; Kretsch, T.; Schwechheimer, C. The Evolutionarily Conserved Arabidopsis Thaliana F-Box Protein AtFBP7 Is Required for Efficient Translation during Temperature Stress. Gene 2007, 392, 106–116. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Y.; Luo, W.; Li, W.; Chen, N.; Zhang, D.; Chong, K. The F-Box Protein OsFBK12 Targets OsSAMS1 for Degradation and Affects Pleiotropic Phenotypes, Including Leaf Senescence, in Rice1[W][OPEN]. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Huo, N.; Zhang, S.; Zhu, T.; Dong, L.; Wang, Y.; Mohr, T.; Hu, T.; Liu, Z.; Dvorak, J.; Luo, M.-C.; et al. Gene Duplication and Evolution Dynamics in the Homeologous Regions Harboring Multiple Prolamin and Resistance Gene Families in Hexaploid Wheat. Front. Plant Sci. 2018, 9, 673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, P.; Liu, C.; Kang, J.; Lv, J. Genome-Wide Analysis of WRKY Transcription Factors in Wheat (Triticum Aestivum L.) and Differential Expression under Water Deficit Condition. PeerJ 2017, 5, e3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salse, J.; Bolot, S.; Throude, M.; Jouffe, V.; Piegu, B.; Quraishi, U.M.; Calcagno, T.; Cooke, R.; Delseny, M.; Feuillet, C. Identification and Characterization of Shared Duplications between Rice and Wheat Provide New Insight into Grass Genome Evolution. Plant Cell Online 2008, 20, 11–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis Thaliana. BMC Plant Biol 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene Duplication as a Major Force in Evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J. Retroposons—Seeds of Evolution. Science 1991, 251, 753. [Google Scholar] [CrossRef]
- Glover, N.M.; Daron, J.; Pingault, L.; Vandepoele, K.; Paux, E.; Feuillet, C.; Choulet, F. Small-Scale Gene Duplications Played a Major Role in the Recent Evolution of Wheat Chromosome 3B. Genome Biol. 2015, 16, 188. [Google Scholar] [CrossRef] [Green Version]
- Wicker, T.; Buchmann, J.P.; Keller, B. Patching Gaps in Plant Genomes Results in Gene Movement and Erosion of Colinearity. Genome Res. 2010, 20, 1229–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagne, J.M.; Downes, B.P.; Shiu, S.-H.; Durski, A.M.; Vierstra, R.D. The F-Box Subunit of the SCF E3 Complex Is Encoded by a Diverse Superfamily of Genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 11519–11524. [Google Scholar] [CrossRef] [Green Version]
- Dinant, S.; Clark, A.M.; Zhu, Y.; Vilaine, F.; Palauqui, J.-C.; Kusiak, C.; Thompson, G.A. Diversity of the Superfamily of Phloem Lectins (Phloem Protein 2) in Angiosperms. Plant Physiol. 2003, 131, 114–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhunov, E.D.; Goodyear, A.W.; Geng, S.; Qi, L.-L.; Echalier, B.; Gill, B.S.; Miftahudin; Gustafson, J.P.; Lazo, G.; Chao, S.; et al. The Organization and Rate of Evolution of Wheat Genomes Are Correlated With Recombination Rates Along Chromosome Arms. Genome Res. 2003, 13, 753–763. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-González, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The Transcriptional Landscape of Polyploid Wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE team. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Gel, B.; Serra, E. KaryoploteR: An R/Bioconductor Package to Plot Customizable Genomes Displaying Arbitrary Data. Bioinformatics 2017, 33, 3088–3090. [Google Scholar] [CrossRef]
- Noé, L.; Kucherov, G. YASS: Enhancing the Sensitivity of DNA Similarity Search. Nucleic Acids Res. 2005, 33, W540–W543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]




| Subfamily (Subdomain Contained by their TaFBX Members) | Number of TaFBX Genes |
|---|---|
| ACTIN (Plant actin-related protein 8, IPR030071) | 3 |
| ARM (Armadillo, IPR000225) | 10 |
| DUF (Domain unknown function DUF295) | 234 |
| FBA (F-box associated domain, IPR007397) | 143 |
| FBD (FBD domain, IPR006566) | 359 |
| FBX-only (F-box domain, IPR001810) | 2475 |
| Jmjc (Jmjc domain, IPR003347) | 9 |
| Kelch (Galactose oxidase/kelch, beta-propeller, IPR011043 or Kelch-type beta propeller, IPR015915) | 203 |
| LRR (Leucine-rich repeat, IPR001611) | 53 |
| LysM (LysM domain, IPR018392) | 3 |
| Other (various domains described in Table S1) | 41 |
| PP2 (Phloem protein 2-like, IPR025886) | 67 |
| QAD (Quinoprotein amine dehydrogenase, beta chain-like, IPR011044) | 17 |
| SEL1 (Sell-like repeat, IPR006597) | 3 |
| TUB (Tubby, C-terminal, IPR000007) | 31 |
| WD40 (WD40 repeat, IPR001680) | 14 |
| zf_MYND (Zinc finger, MYND-type, IPR002893) | 5 |
| Total | 3670 |
| Chromosomal Region | Number of Genes in the Whole TaFBX Family | Theroretical TaFBX DEGs | Observed TaFBX DEGs | ||
|---|---|---|---|---|---|
| Number | % | Number | % | ||
| R1 | 643 | 211.30 | 17.5 | 176 | 14.6 |
| R2a | 637 | 209.32 | 17.4 | 236 | 19.6 |
| C | 76 | 24.97 | 2.1 | 34 | 2.8 |
| R2b | 662 | 217.54 | 18 | 255 | 21.2 |
| R3 | 1518 | 498.83 | 41.4 | 486 | 40.4 |
| Unanchored | 134 | 44.03 | 3.7 | 17 | 1.4 |
| Total | 3670 | 1206 | 100% | 1206 | 100% |
| Chromosomal Region | Theroretical TaFBX DEGs | Observed TaFBX DEGs | ||
|---|---|---|---|---|
| Number | % | Number | % | |
| R1 | 92 | 17.5 | 45 | 8.6 |
| R2a | 91.1 | 17.4 | 115 | 21.9 |
| C | 10.9 | 2.1 | 22 | 4.2 |
| R2b | 94.7 | 18 | 160 | 30.5 |
| R3 | 217.2 | 41.4 | 169 | 32.2 |
| U | 19.2 | 3.7 | 14 | 2.7 |
| Total | 525 | 100% | 525 | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guérin, C.; Mouzeyar, S.; Roche, J. The Landscape of the Genomic Distribution and the Expression of the F-Box Genes Unveil Genome Plasticity in Hexaploid Wheat during Grain Development and in Response to Heat and Drought Stress. Int. J. Mol. Sci. 2021, 22, 3111. https://doi.org/10.3390/ijms22063111
Guérin C, Mouzeyar S, Roche J. The Landscape of the Genomic Distribution and the Expression of the F-Box Genes Unveil Genome Plasticity in Hexaploid Wheat during Grain Development and in Response to Heat and Drought Stress. International Journal of Molecular Sciences. 2021; 22(6):3111. https://doi.org/10.3390/ijms22063111
Chicago/Turabian StyleGuérin, Claire, Saïd Mouzeyar, and Jane Roche. 2021. "The Landscape of the Genomic Distribution and the Expression of the F-Box Genes Unveil Genome Plasticity in Hexaploid Wheat during Grain Development and in Response to Heat and Drought Stress" International Journal of Molecular Sciences 22, no. 6: 3111. https://doi.org/10.3390/ijms22063111
APA StyleGuérin, C., Mouzeyar, S., & Roche, J. (2021). The Landscape of the Genomic Distribution and the Expression of the F-Box Genes Unveil Genome Plasticity in Hexaploid Wheat during Grain Development and in Response to Heat and Drought Stress. International Journal of Molecular Sciences, 22(6), 3111. https://doi.org/10.3390/ijms22063111








