Shake It Off: The Elimination of Erroneous Kinetochore-Microtubule Attachments and Chromosome Oscillation
Abstract
1. Introduction
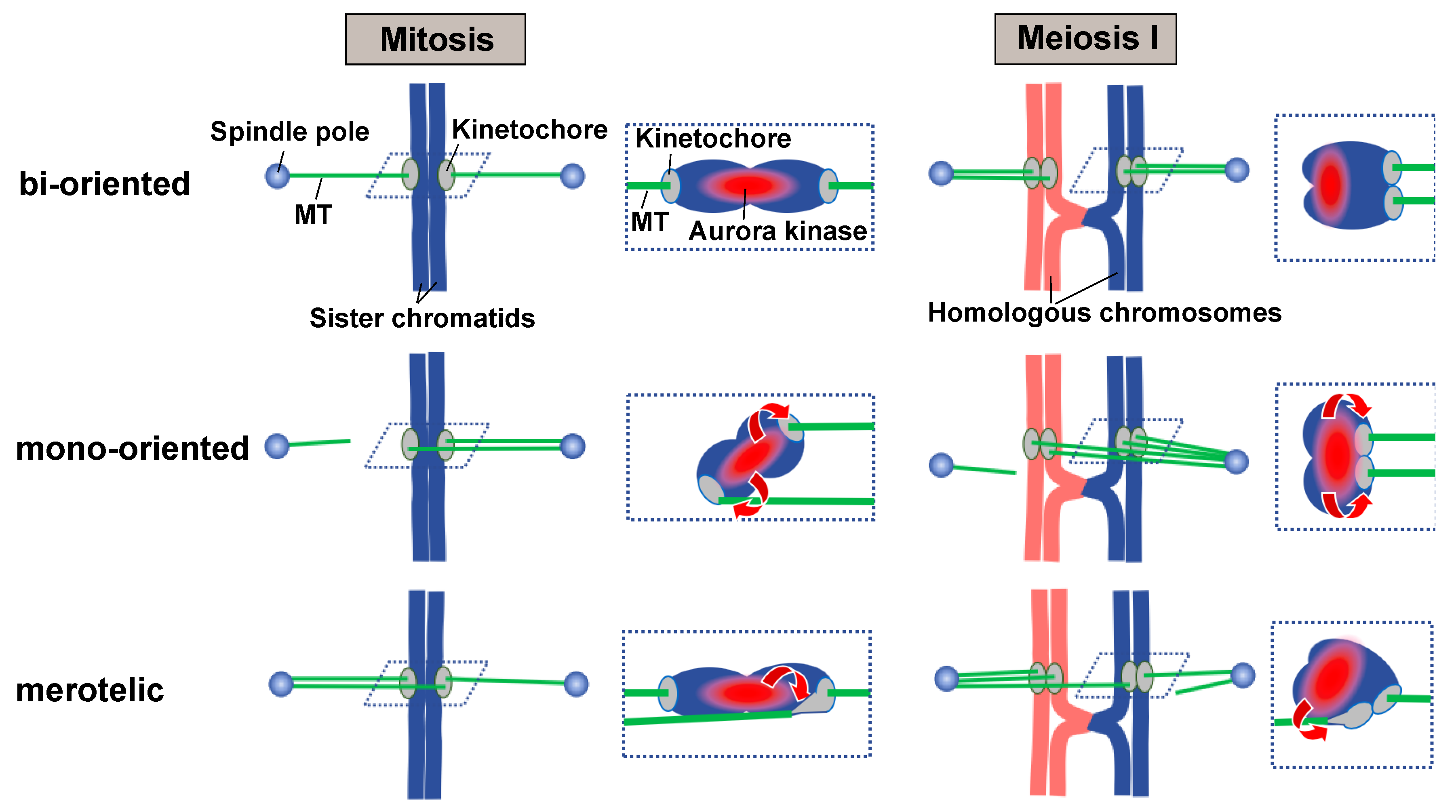
2. Tension-Dependent Attachment Establishment and Chromosome Dynamics during the Establishment
2.1. The Current Model for the Tension-Dependent Attachment Establishment
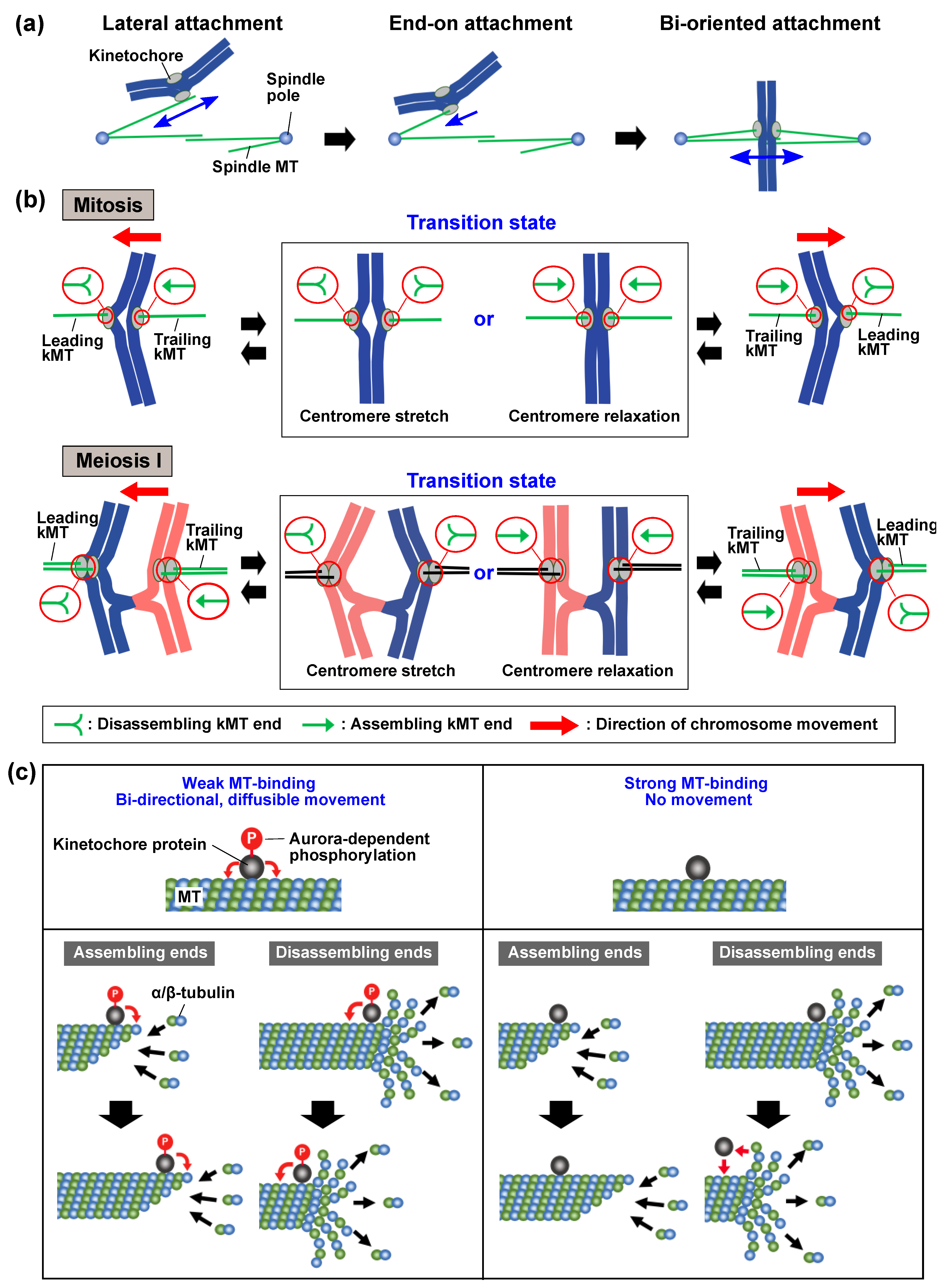
2.2. Chromosome Oscillation during Attachment Establishment
3. Problems with the Tension-Dependent Spatial Separation Model during Meiosis
4. The Intimate Relationship between Attachment Elimination and Centromere Oscillation
5. The Possible Contribution of Centromere Oscillation to Attachment Elimination
6. Future Direction
Funding
Data Availability Statement
Conflicts of Interest
References
- McIntosh, J.; Hays, T. A Brief History of Research on Mitotic Mechanisms. Biology 2016, 5, 55. [Google Scholar] [CrossRef]
- Nicklas, R.B. How Cells Get the Right Chromosomes. Science 1997, 275, 632–637. [Google Scholar] [CrossRef]
- Lampson, M.; Grishchuk, E. Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B.; Koch, C.A. Chromosome Micromanipulation. 3. Spindle Fiber Tension and the Reorientation of Mal-Oriented Chromosomes. J. Cell Biol. 1969, 43, 40–50. [Google Scholar] [CrossRef]
- Nezi, L.; Musacchio, A. Sister Chromatid Tension and the Spindle Assembly Checkpoint. Curr. Opin. Cell Biol. 2009, 21, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Lampson, M.A.; Cheeseman, I.M. Sensing Centromere Tension: Aurora B and the Regulation of Kinetochore Function. Trends Cell Biol. 2011, 21, 133–140. [Google Scholar] [CrossRef]
- Onn, I.; Heidinger-Pauli, J.M.; Guacci, V.; Ünal, E.; Koshland, D.E. Sister Chromatid Cohesion: A Simple Concept with a Complex Reality. Annu. Rev. Cell Dev. Biol. 2008, 24, 105–129. [Google Scholar] [CrossRef]
- Skibbens, R.V.; Skeen, V.P.; Salmon, E.D. Directional Instability of Kinetochore Motility during Chromosome Congression and Segregation in Mitotic Newt Lung Cells: A Push-Pull Mechanism. J. Cell Biol. 1993, 122, 859–875. [Google Scholar] [CrossRef] [PubMed]
- Inoué, S.; Salmon, E.D. Force Generation by Microtubule Assembly/Disassembly in Mitosis and Related Movements. Mol. Biol. Cell 1995, 6, 1619–1640. [Google Scholar] [CrossRef]
- Maiato, H.; Gomes, A.; Sousa, F.; Barisic, M.; Maiato, H.; Gomes, A.M.; Sousa, F.; Barisic, M. Mechanisms of Chromosome Congression during Mitosis. Biology 2017, 6, 13. [Google Scholar] [CrossRef]
- Auckland, P.; McAinsh, A.D. Building an Integrated Model of Chromosome Congression. J. Cell Sci. 2015, 128, 3363–3374. [Google Scholar] [CrossRef] [PubMed]
- Wakiya, M.; Nishi, E.; Kawai, S.; Yamada, K.; Katsumata, K.; Hirayasu, A.; Itabashi, Y.; Yamamoto, A. Chiasmata and the Kinetochore Component Dam1 Are Crucial for Elimination of Erroneous Chromosome Attachments and Centromere Oscillation at Meiosis I. Open Biol. 2021, 11, 200308. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The Chromosomal Passenger Complex (CPC): From Easy Rider to the Godfather of Mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef]
- Cheerambathur, D.K.; Desai, A. Linked in: Formation and Regulation of Microtubule Attachments during Chromosome Segregation. Curr. Opin. Cell Biol. 2014, 26, 113–122. [Google Scholar] [CrossRef]
- Krenn, V.; Musacchio, A. The Aurora B Kinase in Chromosome Bi-Orientation and Spindle Checkpoint Signaling. Front. Oncol. 2015, 5, 225. [Google Scholar] [CrossRef]
- Tanaka, K. Regulatory Mechanisms of Kinetochore–Microtubule Interaction in Mitosis. Cell. Mol. Life Sci. 2013, 70, 559–579. [Google Scholar] [CrossRef]
- Tanaka, T.U. Kinetochore-Microtubule Interactions: Steps towards Bi-Orientation. EMBO J. 2010, 29, 4070–4082. [Google Scholar] [CrossRef]
- Funabiki, H. Correcting Aberrant Kinetochore Microtubule Attachments: A Hidden Regulation of Aurora B on Microtubules. Curr. Opin. Cell Biol. 2019, 58, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.E.; Funabiki, H. Correcting Aberrant Kinetochore Microtubule Attachments: An Aurora B-Centric View. Curr. Opin. Cell Biol. 2009, 21, 51–58. [Google Scholar] [CrossRef]
- Gorbsky, G.J. Mitosis: MCAK under the Aura of Aurora B. Curr. Biol. 2004, 14, R346–R348. [Google Scholar] [CrossRef]
- Lan, W.; Zhang, X.; Kline-Smith, S.L.; Rosasco, S.E.; Barrett-Wilt, G.A.; Shabanowitz, J.; Hunt, D.F.; Walczak, C.E.; Stukenberg, P.T. Aurora B Phosphorylates Centromeric MCAK and Regulates Its Localization and Microtubule Depolymerization Activity. Curr. Biol. 2004, 14, 273–286. [Google Scholar] [CrossRef]
- Andrews, P.D.; Ovechkina, Y.; Morrice, N.; Wagenbach, M.; Duncan, K.; Wordeman, L.; Swedlow, J.R. Aurora B Regulates MCAK at the Mitotic Centromere. Dev. Cell 2004, 6, 253–268. [Google Scholar] [CrossRef]
- Watanabe, Y. Geometry and Force behind Kinetochore Orientation: Lessons from Meiosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.U.; Rachidi, N.; Janke, C.; Pereira, G.; Galova, M.; Schiebel, E.; Stark, M.J.R.; Nasmyth, K. Evidence That the Ipl1-Sli15 (Aurora Kinase-INCENP) Complex Promotes Chromosome Bi-Orientation by Altering Kinetochore-Spindle Pole Connections. Cell 2002, 108, 317–329. [Google Scholar] [CrossRef]
- Liu, D.; Vader, G.; Vromans, M.J.M.; Lampson, M.A.; Lens, S.M.A. Sensing Chromosome Bi-Orientation by Spatial Separation of Aurora B Kinase from Kinetochore Substrates. Science 2009, 323, 1350–1353. [Google Scholar] [CrossRef]
- Sassoon, I.; Severin, F.F.; Andrews, P.D.; Taba, M.R.; Kaplan, K.B.; Ashford, A.J.; Stark, M.J.; Sorger, P.K.; Hyman, A.A. Regulation of Saccharomyces Cerevisiae Kinetochores by the Type 1 Phosphatase Glc7p. Genes Dev. 1999, 13, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Vanoosthuyse, V.; Hardwick, K.G. A Novel Protein Phosphatase 1-Dependent Spindle Checkpoint Silencing Mechanism. Curr. Biol. 2009, 19, 1176–1181. [Google Scholar] [CrossRef] [PubMed]
- Posch, M.; Khoudoli, G.A.; Swift, S.; King, E.M.; DeLuca, J.G.; Swedlow, J.R. Sds22 Regulates Aurora B Activity and Microtubule–Kinetochore Interactions at Mitosis. J. Cell Biol. 2010, 191, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, B.A.; Kotwaliwale, C.V.; Tatsutani, S.Y.; Breed, C.A.; Biggins, S. Glc7/Protein Phosphatase 1 Regulatory Subunits Can Oppose the Ipl1/Aurora Protein Kinase by Redistributing Glc7. Mol. Cell. Biol. 2006, 26, 2648–2660. [Google Scholar] [CrossRef]
- Ye, A.A.; Deretic, J.; Hoel, C.M.; Hinman, A.W.; Cimini, D.; Welburn, J.P.; Maresca, T.J. Aurora A Kinase Contributes to a Pole-Based Error Correction Pathway. Curr. Biol. 2015, 25, 1842–1851. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, K.F.; Meppelink, A.; Broad, A.J.; Mick, J.E.; Peersen, O.B.; Pektas, S.; Lens, S.M.A.; DeLuca, J.G. Aurora A Kinase Phosphorylates Hec1 to Regulate Metaphase Kinetochore–Microtubule Dynamics. J. Cell Biol. 2018, 217, 163–177. [Google Scholar] [CrossRef]
- Cimini, D.; Howell, B.; Maddox, P.; Khodjakov, A.; Degrassi, F.; Salmon, E.D. Merotelic Kinetochore Orientation Is a Major Mechanism of Aneuploidy in Mitotic Mammalian Tissue Cells. J. Cell Biol. 2001, 153, 517–528. [Google Scholar] [CrossRef]
- Kitajima, T.S.; Ohsugi, M.; Ellenberg, J. Complete Kinetochore Tracking Reveals Error-Prone Homologous Chromosome Biorientation in Mammalian Oocytes. Cell 2011, 146, 568–581. [Google Scholar] [CrossRef]
- Sakuno, T.; Tanaka, K.; Hauf, S.; Watanabe, Y. Repositioning of Aurora B Promoted by Chiasmata Ensures Sister Chromatid Mono-Orientation in Meiosis I. Dev. Cell 2011, 21, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Alexander, S.P. Kinetochores are Transported Poleward along a Single Astral Microtubule during Chromosome Attachment to the Spindle in Newt Lung Cells. J. Cell Biol. 1990, 110, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Mukae, N.; Dewar, H.; Van Breugel, M.; James, E.K.; Prescott, A.R.; Antony, C.; Tanaka, T.U. Molecular Mechanisms of Kinetochore Capture by Spindle Microtubules. Nature 2005, 434, 987–994. [Google Scholar] [CrossRef]
- Tanaka, K.; Kitamura, E.; Kitamura, Y.; Tanaka, T.U. Molecular Mechanisms of Microtubule-Dependent Kinetochore Transport toward Spindle Poles. J. Cell Biol. 2007, 178, 269–281. [Google Scholar] [CrossRef]
- Akera, T.; Goto, Y.; Sato, M.; Yamamoto, M.; Watanabe, Y. Mad1 Promotes Chromosome Congression by Anchoring a Kinesin Motor to the Kinetochore. Nat. Cell Biol. 2015, 17, 1124–1133. [Google Scholar] [CrossRef]
- Yang, Z.; Tulu, U.S.; Wadsworth, P.; Rieder, C.L. Kinetochore Dynein Is Required for Chromosome Motion and Congression Independent of the Spindle Checkpoint. Curr. Biol. 2007, 17, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.J.; Rogers, G.C.; Scholey, J.M. Cytoplasmic Dynein Is Required for Poleward Chromosome Movement during Mitosis in Drosophila Embryos. Nat. Cell Biol. 2000, 2, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Varma, D.; Monzo, P.; Stehman, S.A.; Vallee, R.B. Direct Role of Dynein Motor in Stable Kinetochore-Microtubule Attachment, Orientation, and Alignment. J. Cell Biol. 2008, 182, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Yen, T.J.; Li, G.; Schaar, B.T.; Szilak, I.; Cleveland, D.W. CENP-E Is a Putative Kinetochore Motor That Accumulates Just before Mitosis. Nature 1992, 359, 536–539. [Google Scholar] [CrossRef]
- Wood, K.W.; Sakowicz, R.; Goldstein, L.S.; Cleveland, D.W. CENP-E Is a Plus End–Directed Kinetochore Motor Required for Metaphase Chromosome Alignment. Cell 1997, 91, 357–366. [Google Scholar] [CrossRef]
- Kapoor, T.M.; Lampson, M.A.; Hergert, P.; Cameron, L.; Cimini, D.; Salmon, E.D.; McEwen, B.F.; Khodjakov, A. Chromosomes Can Congress to the Metaphase Plate before Biorientation. Science 2006, 311, 388–391. [Google Scholar] [CrossRef]
- Asbury, C.L.; Gestaut, D.R.; Powers, A.F.; Franck, A.D.; Davis, T.N. The Dam1 Kinetochore Complex Harnesses Microtubule Dynamics to Produce Force and Movement. Proc. Natl. Acad. Sci. USA 2006, 103, 9873–9878. [Google Scholar] [CrossRef]
- Grishchuk, E.L.; Efremov, A.K.; Volkov, V.A.; Spiridonov, I.S.; Gudimchuk, N.; Westermann, S.; Drubin, D.; Barnes, G.; McIntosh, J.R.; Ataullakhanov, F.I. The Dam1 Ring Binds Microtubules Strongly Enough to Be a Processive as Well as Energy-Efficient Coupler for Chromosome Motion. Proc. Natl. Acad. Sci. USA 2008, 105, 15423–15428. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, G.J.; Dong, Y.; McEwen, B.F.; DeLuca, J.G. Kinetochore-Microtubule Attachment Relies on the Disordered N-Terminal Tail Domain of Hec1. Curr. Biol. 2008, 18, 1778–1784. [Google Scholar] [CrossRef]
- DeLuca, J.G.; Gall, W.E.; Ciferri, C.; Cimini, D.; Musacchio, A.; Salmon, E.D. Kinetochore Microtubule Dynamics and Attachment Stability Are Regulated by Hec1. Cell 2006, 127, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Powers, A.F.; Franck, A.D.; Gestaut, D.R.; Cooper, J.; Gracyzk, B.; Wei, R.R.; Wordeman, L.; Davis, T.N.; Asbury, C.L. The Ndc80 Kinetochore Complex Forms Load-Bearing Attachments to Dynamic Microtubule Tips via Biased Diffusion. Cell 2009, 136, 865–875. [Google Scholar] [CrossRef]
- McIntosh, J.R.; Grishchuk, E.L.; Morphew, M.K.; Efremov, A.K.; Zhudenkov, K.; Volkov, V.A.; Cheeseman, I.M.; Desai, A.; Mastronarde, D.N.; Ataullakhanov, F.I. Fibrils Fonnect Microtubule Tips with Kinetochores: A Mechanism to Couple Tubulin Dynamics to Chromosome Motion. Cell 2008, 135, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.-S.; Toda, T. Ndc80 Internal Loop Interacts with Dis1/TOG to Ensure Proper Kinetochore-Spindle Attachment in Fission Yeast. Curr. Biol. 2011, 21, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.H.; Takada, H.; Hsu, K.S.; Toda, T. The Internal Loop of Fission Yeast Ndc80 Binds Alp7/TACC-Alp14/TOG and Ensures Proper Chromosome Attachment. Mol. Biol. Cell 2013, 24, 1095–1251. [Google Scholar] [CrossRef]
- Huisin’t, V.P.J.; Volkov, V.A.; Stender, I.D.; Musacchio, A.; Dogterom, M. Molecular Determinants of the Ska-Ndc80 Interaction and Their Influence on Microtubule Tracking and Force-Coupling. Elife 2019, 8, 8. [Google Scholar] [CrossRef]
- Auckland, P.; Clarke, N.I.; Royle, S.J.; McAinsh, A.D. Congressing Kinetochores Progressively Load Ska Complexes to Prevent Force-Dependent Detachment. J. Cell Biol. 2017, 216, 1623–1639. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, A.; Silljé, H.H.W.; Nigg, E.A. Timely Anaphase Onset Requires a Novel Spindle and Kinetochore Complex Comprising Ska1 and Ska2. EMBO J. 2006, 25, 5504–5515. [Google Scholar] [CrossRef]
- Helgeson, L.A.; Zelter, A.; Riffle, M.; MacCoss, M.J.; Asbury, C.L.; Davis, T.N. Human Ska Complex and Ndc80 Complex Interact to Form a Load-Bearing Assembly That Strengthens Kinetochore-Microtubule Attachments. Proc. Natl. Acad. Sci. USA 2018, 115, 2740–2745. [Google Scholar] [CrossRef]
- Gachet, Y.; Reyes, C.; Courthéoux, T.; Goldstone, S.; Gay, G.; Serrurier, C.; Tournier, S. Sister Kinetochore Recapture in Fission Yeast Occurs by Two Distinct Mechanisms, Both Requiring Dam1 and Klp2. Mol. Biol. Cell 2008, 19, 1646–1662. [Google Scholar] [CrossRef] [PubMed]
- Jeyaprakash, A.A.; Santamaria, A.; Jayachandran, U.; Chan, Y.W.; Benda, C.; Nigg, E.A.; Conti, E. Structural and Functional Organization of the Ska Complex, a Key Component of the Kinetochore-Microtubule Interface. Mol. Cell 2012, 46, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Welburn, J.P.I.; Grishchuk, E.L.; Backer, C.B.; Wilson-Kubalek, E.M.; Yates, J.R.; Cheeseman, I.M. The Human Kinetochore Ska1 Complex Facilitates Microtubule Depolymerization-Coupled Motility. Dev. Cell 2009, 16, 374–385. [Google Scholar] [CrossRef]
- Franco, A.; Meadows, J.C.; Millar, J.B.A. The Dam1/DASH Complex Is Required for the Retrieval of Unclustered Kinetochores in Fission Yeast. J. Cell Sci. 2007, 120, 3345–3351. [Google Scholar] [CrossRef]
- Lampert, F.; Hornung, P.; Westermann, S. The Dam1 Complex Confers Microtubule plus End–Tracking Activity to the Ndc80 Kinetochore Complex. J. Cell Biol. 2010, 189, 641–649. [Google Scholar] [CrossRef]
- Westermann, S.; Wang, H.-W.; Avila-Sakar, A.; Drubin, D.G.; Nogales, E.; Barnes, G. The Dam1 Kinetochore Ring Complex Moves Processively on Depolymerizing Microtubule Ends. Nature 2006, 440, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V.A.; Zaytsev, A.V.; Gudimchuk, N.; Grissom, P.M.; Gintsburg, A.L.; Ataullakhanov, F.I.; McIntosh, J.R.; Grishchuk, E.L. Long Tethers Provide High-Force Coupling of the Dam1 Ring to Shortening Microtubules. Proc. Natl. Acad. Sci. USA 2013, 110, 7708–7713. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Perez, I.; Renwick, S.J.; Crawley, K.; Karig, I.; Buck, V.; Meadows, J.C.; Franco-Sanchez, A.; Fleig, U.; Toda, T.; Millar, J.B.A. The DASH Complex and Klp5/Klp6 Kinesin Coordinate Bipolar Chromosome Attachment in Fission Yeast. EMBO J. 2005, 24, 2931–2943. [Google Scholar] [CrossRef] [PubMed]
- Kalantzaki, M.; Kitamura, E.; Zhang, T.; Mino, A.; Novák, B.; Tanaka, T.U. Kinetochore-Microtubule Error Correction Is Driven by Differentially Regulated Interaction Modes. Nat. Cell Biol. 2015, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Cheerambathur, D.K.; Prevo, B.; Hattersley, N.; Lewellyn, L.; Corbett, K.D.; Oegema, K.; Desai, A. Dephosphorylation of the Ndc80 Tail Stabilizes Kinetochore-Microtubule Attachments via the Ska Complex. Dev. Cell 2017, 41, 424–437.e4. [Google Scholar] [CrossRef]
- Grissom, P.M.; Fiedler, T.; Grishchuk, E.L.; Nicastro, D.; West, R.R.; McIntosh, J.R. Kinesin-8 from Fission Yeast: A Heterodimeric, plus-End–Directed Motor That Can Couple Microtubule Depolymerization to Cargo Movement. Mol. Biol. Cell 2009, 20, 963–972. [Google Scholar] [CrossRef]
- Garcia, M.A.; Koonrugsa, N.; Toda, T. Two Kinesin-like Kin I Family Proteins in Fission Yeast Regulate the Establishment of Metaphase and the Onset of Anaphase A. Curr. Biol. 2002, 12, S0960–S9822. [Google Scholar] [CrossRef]
- Garcia, M.A.; Koonrugsa, N.; Toda, T. Spindle-Kinetochore Attachment Requires the Combined Action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in Fission Yeast. EMBO J. 2002, 21, 6015–6024. [Google Scholar] [CrossRef] [PubMed]
- Klemm, A.H.; Bosilj, A.; Gluncˇic´, M.; Pavin, N.; Tolic, I.M. Metaphase Kinetochore Movements Are Regulated by Kinesin-8 Motors and Microtubule Dynamic Instability. Mol. Biol. Cell 2018, 29, 1332–1345. [Google Scholar] [CrossRef]
- Mary, H.; Fouchard, J.; Gay, G.; Reyes, C.; Gauthier, T.; Gruget, C.; Pécréaux, J.; Tournier, S.; Gachet, Y. Fission Yeast Kinesin-8 Controls Chromosome Congression Independently of Oscillations. J. Cell Sci. 2015, 128, 3720–3730. [Google Scholar] [CrossRef]
- West, R.R.; Malmstrom, T.; McIntosh, J.R. Kinesins Klp5+ and Klp6+ Are Required for Normal Chromosome Movement in Mitosis. J. Cell Sci. 2002, 115, 931–940. [Google Scholar]
- Rieder, C.L.; Davison, E.A.; Jensen, L.C.; Cassimeris, L.; Salmon, E.D. Oscillatory Movements of Monooriented Chromosomes and Their Position Relative to the Spindle Pole Result from the Ejection Properties of the Aster and Half-Spindle. J. Cell Biol. 1986, 103, 581–591. [Google Scholar] [CrossRef]
- Ke, K.; Cheng, J.; Hunt, A.J. The Distribution of Polar Ejection Forces Determines the Amplitude of Chromosome Directional Instability. Curr. Biol. 2009, 19, 807–815. [Google Scholar] [CrossRef]
- Stumpff, J.; Wagenbach, M.; Franck, A.; Asbury, C.L.; Wordeman, L. Kif18A and Chromokinesins Confine Centromere Movements via Microtubule Growth Suppression and Spatial Control of Kinetochore Tension. Dev. Cell 2012, 22, 1017–1029. [Google Scholar] [CrossRef]
- Cane, S.; Ye, A.A.; Luks-Morgan, S.J.; Maresca, T.J. Elevated Polar Ejection Forces Stabilize Kinetochore–Microtubule Attachments. J. Cell Biol. 2013, 200, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Takagi, J.; Itabashi, T.; Suzuki, K.; Ishiwata, S. Chromosome Position at the Spindle Equator Is Regulated by Chromokinesin and a Bipolar Microtubule Array. Sci. Rep. 2013, 3, 2808. [Google Scholar] [CrossRef] [PubMed]
- Wandke, C.; Barisic, M.; Sigl, R.; Rauch, V.; Wolf, F.; Amaro, A.C.; Tan, C.H.; Pereira, A.J.; Kutay, U.; Maiato, H.; et al. Human Chromokinesins Promote Chromosome Congression and Spindle Microtubule Dynamics during Mitosis. J. Cell Biol. 2012, 198, 847–863. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, N.J.; Harry, E.F.; McAinsh, A.D. Super-Resolution Kinetochore Tracking Reveals the Mechanisms of Human Sister Kinetochore Directional Switching. Elife 2015, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Desai, A.; Onuchic, J.N.; Hwa, T. An Integrated Mechanobiochemical Feedback Mechanism Describes Chromosome Motility from Prometaphase to Anaphase in Mitosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13752–13757. [Google Scholar] [CrossRef] [PubMed]
- Armond, J.W.; Harry, E.F.; McAinsh, A.D.; Burroughs, N.J. Inferring the Forces Controlling Metaphase Kinetochore Oscillations by Reverse Engineering System Dynamics. PLoS Comput. Biol. 2015, 11, e1004607. [Google Scholar] [CrossRef]
- Civelekoglu-Scholey, G.; He, B.; Shen, M.; Wan, X.; Roscioli, E.; Bowden, B.; Cimini, D. Dynamic Bonds and Polar Ejection Force Distribution Explain Kinetochore Oscillations in PtK1 Cells. J. Cell Biol. 2013, 201, 577–593. [Google Scholar] [CrossRef]
- Gergely, Z.R.; Crapo, A.; Hough, L.E.; McIntosh, J.R.; Betterton, M.D. Kinesin-8 Effects on Mitotic Microtubule Dynamics Contribute to Spindle Function in Fission Yeast. Mol. Biol. Cell 2016, 27, 3490–3514. [Google Scholar] [CrossRef]
- Gardner, M.K.; Bouck, D.C.; Paliulis, L.V.; Meehl, J.B.; O’Toole, E.T.; Haase, J.; Soubry, A.; Joglekar, A.P.; Winey, M.; Salmon, E.D.; et al. Chromosome Congression by Kinesin-5 Motor-Mediated Disassembly of Longer Kinetochore Microtubules. Cell 2008, 135, 894–906. [Google Scholar] [CrossRef]
- Gay, G.; Courtheoux, T.; Reyes, C.; Tournier, S.; Gachet, Y. A Stochastic Model of Kinetochore–Microtubule Attachment Accurately Describes Fission Yeast Chromosome Segregation. J. Cell Biol. 2012, 196, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Hunt, A.J. A Simple, Mechanistic Model for Directional Instability during Mitotic Chromosome Movements. Biophys. J. 2002, 83, 42–58. [Google Scholar] [CrossRef]
- Skibbens, R.V.; Rieder, C.L.; Salmon, E.D. Kinetochore Motility after Severing between Sister Centromeres Using Laser Microsurgery: Evidence That Kinetochore Directional Instability and Position Is Regulated by Tension. J. Cell Sci. 1995, 108, 2537–2548. [Google Scholar]
- Gardner, M.K.; Pearson, C.G.; Sprague, B.L.; Zarzar, T.R.; Bloom, K.; Salmon, E.D.; Odde, D.J. Tension-Dependent Regulation of Microtubule Dynamics at Kinetochores Can Explain Metaphase Congression in Yeast. Mol. Biol. Cell 2005, 16, 3764–3775. [Google Scholar] [CrossRef] [PubMed]
- Civelekoglu-Scholey, G.; Sharp, D.J.; Mogilner, A.; Scholey, J.M. Model of Chromosome Motility in Drosophila Embryos: Adaptation of a General Mechanism for Rapid Mitosis. Biophys. J. 2006, 90, 3966–3982. [Google Scholar] [CrossRef] [PubMed]
- Banigan, E.J.; Chiou, K.K.; Ballister, E.R.; Mayo, A.M.; Lampson, M.A.; Liu, A.J. Minimal Model for Collective Kinetochore–Microtubule Dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 12699–12704. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, K.F.; Lens, S.M.A.; DeLuca, J.G. Temporal Changes in Hec1 Phosphorylation Control Kinetochore-Microtubule Attachment Stability during Mitosis. J. Cell Sci. 2011, 124, 622–634. [Google Scholar] [CrossRef]
- Long, A.F.; Udy, D.B.; Dumont, S. Hec1 Tail Phosphorylation Differentially Regulates Mammalian Kinetochore Coupling to Polymerizing and Depolymerizing Microtubules. Curr. Biol. 2017, 27, 1692–1699.e3. [Google Scholar] [CrossRef] [PubMed]
- Sarangapani, K.K.; Akiyoshi, B.; Duggan, N.M.; Biggins, S.; Asbury, C.L. Phosphoregulation Promotes Release of Kinetochores from Dynamic Microtubules via Multiple Mechanisms. Proc. Natl. Acad. Sci. USA 2013, 110, 7282–7287. [Google Scholar] [CrossRef] [PubMed]
- Tien, J.F.; Umbreit, N.T.; Gestaut, D.R.; Franck, A.D.; Cooper, J.; Wordeman, L.; Gonen, T.; Asbury, C.L.; Davis, T.N. Cooperation of the Dam1 and Ndc80 Kinetochore Complexes Enhances Microtubule Coupling and Is Regulated by Aurora B. J. Cell Biol. 2010, 189, 713–723. [Google Scholar] [CrossRef]
- Akiyoshi, B.; Sarangapani, K.K.; Powers, A.F.; Nelson, C.R.; Reichow, S.L.; Arellano-Santoyo, H.; Gonen, T.; Ranish, J.A.; Asbury, C.L.; Biggins, S. Tension Directly Stabilizes Reconstituted Kinetochore-Microtubule Attachments. Nature 2010, 468, 576–579. [Google Scholar] [CrossRef]
- Gestaut, D.R.; Graczyk, B.; Cooper, J.; Widlund, P.O.; Zelter, A.; Wordeman, L.; Asbury, C.L.; Davis, T.N. Phosphoregulation and Depolymerization-Driven Movement of the Dam1 Complex Do Not Require Ring Formation. Nat. Cell Biol. 2008, 10, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Zaytsev, A.V.; Mick, J.E.; Maslennikov, E.; Nikashin, B.; DeLuca, J.G.; Grishchuk, E.L. Multisite Phosphorylation of the NDC80 Complex Gradually Tunes Its Microtubule-Binding Affinity. Mol. Biol. Cell 2015, 26, 1829–1844. [Google Scholar] [CrossRef]
- Ciferri, C.; Pasqualato, S.; Screpanti, E.; Varetti, G.; Santaguida, S.; Dos Reis, G.; Maiolica, A.; Polka, J.; De Luca, J.G.; De Wulf, P.; et al. Implications for Kinetochore-Microtubule Attachment from the Structure of an Engineered Ndc80 Complex. Cell 2008, 133, 427–439. [Google Scholar] [CrossRef]
- Nabeshima, K.; Nakagawa, T.; Straight, A.F.; Murray, A.; Chikashige, Y.; Yamashita, Y.M.; Hiraoka, Y.; Yanagida, M. Dynamics of Centromeres during Metaphase–Anaphase Transition in Fission Yeast: Dis1 Is Implicated in Force Balance in Metaphase Bipolar Spindle. Mol. Biol. Cell 1998, 9, 3211–3225. [Google Scholar] [CrossRef]
- Hirose, Y.; Suzuki, R.; Ohba, T.; Hinohara, Y.; Matsuhara, H.; Yoshida, M.; Itabashi, Y.; Murakami, H.; Yamamoto, A. Chiasmata Promote Monopolar Attachment of Sister Chromatids and Their Co-Segregation toward the Proper Pole during Meiosis I. PLoS Genet. 2011, 7, e1001329. [Google Scholar] [CrossRef]
- Yoshida, S.; Kaido, M.; Kitajima, T.S. Inherent Instability of Correct Kinetochore-Microtubule Attachments during Meiosis I in Oocytes. Dev. Cell 2015, 33, 589–602. [Google Scholar] [CrossRef]
- Yamamoto, A.; Hiraoka, Y. Monopolar Spindle Attachment of Sister Chromatids Is Ensured by Two Distinct Mechanisms at the First Meiotic Division in Fission Yeast. EMBO J. 2003, 22, 2284–2296. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.P. Meiotic Behavior of a Tiny Fragment Chromosome That Carries a Transposed Centromere. Genome 1987, 29, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Hunt, P.; LeMaire, R.; Embury, P.; Sheean, L.; Mroz, K. Analysis of Chromosome Behavior in Intact Mammalian Oocytes: Monitoring the Segregation of a Univalent Chromosome during Female Meiosis. Hum. Mol. Genet. 1995, 4, 2007–2012. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, E.; Arana, P. A Comparative Study of Orientation at Behavior of Univalent in Living Grasshopper Spermatocytes. Chromosoma 1995, 104, 56–67. [Google Scholar] [CrossRef]
- Miller, M.P.; Asbury, C.L.; Biggins, S. A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell 2016, 165, 1428–1439. [Google Scholar] [CrossRef]
- Trushko, A.; Schäffer, E.; Howard, J. The Growth Speed of Microtubules with XMAP215-Coated Beads Coupled to Their Ends Is Increased by Tensile Force. Proc. Natl. Acad. Sci. USA 2013, 110, 14670–14675. [Google Scholar] [CrossRef]
- Courtheoux, T.; Gay, G.; Gachet, Y.; Tournier, S. Ase1/Prc1-Dependent Spindle Elongation Corrects Merotely during Anaphase in Fission Yeast. J. Cell Biol. 2009, 187, 399–412. [Google Scholar] [CrossRef]
- Stephens, A.D.; Quammen, C.W.; Chang, B.; Haase, J.; Taylor, R.M.; Bloom, K. The Spatial Segregation of Pericentric Cohesin and Condensin in the Mitotic Spindle. Mol. Biol. Cell 2013, 24, 3909–3919. [Google Scholar] [CrossRef]
- Uchida, K.S.K.; Takagaki, K.; Kumada, K.; Hirayama, Y.; Noda, T.; Hirota, T. Kinetochore Stretching Inactivates the Spindle Assembly Checkpoint. J. Cell Biol. 2009, 184, 383–390. [Google Scholar] [CrossRef]
- Maresca, T.J.; Salmon, E.D. Intrakinetochore Stretch Is Associated with Changes in Kinetochore Phosphorylation and Spindle Assembly Checkpoint Activity. J. Cell Biol. 2009, 184, 373–381. [Google Scholar] [CrossRef]
- Cojoc, G.; Roscioli, E.; Zhang, L.; García-Ulloa, A.; Shah, J.V.; Berns, M.W.; Pavin, N.; Cimini, D.; Tolić, I.M.; Gregan, J. Laser Microsurgery Reveals Conserved Viscoelastic Behavior of the Kinetochore. J. Cell Biol. 2016, 212, 767–776. [Google Scholar] [CrossRef]
- Dumont, S.; Salmon, E.D.; Mitchison, T.J. Deformations within Moving Kinetochores Reveal Different Sites of Active and Passive Force Generation. Science 2012, 337, 355–358. [Google Scholar] [CrossRef]
- Khodjakov, A.; Cole, R.W.; McEwen, B.F.; Buttle, K.F.; Rieder, C.L. Chromosome Fragments Possessing Only One Kinetochore Can Congress to the Spindle Equator. J. Cell Biol. 1997, 136, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Hays, T.S.; Salmon, E.D. Poleward Force at the Kinetochore in Metaphase Depends on the Number of Kinetochore Microtubules. J. Cell Biol. 1990, 110, 391–404. [Google Scholar] [CrossRef]
- King, J.M.; Nicklas, R.B. Tension on Chromosomes Increases the Number of Kinetochore Microtubules but Only within Limits. J. Cell Sci. 2000, 113 Pt 21, 3815–3823. [Google Scholar]
- Fuller, B.G.; Lampson, M.A.; Foley, E.A.; Rosasco-Nitcher, S.; Le, K.V.; Tobelmann, P.; Brautigan, D.L.; Stukenberg, P.T.; Kapoor, T.M. Midzone Activation of Aurora B in Anaphase Produces an Intracellular Phosphorylation Gradient. Nature 2008, 453, 1132–1136. [Google Scholar] [CrossRef]
- Wang, E.; Ballister, E.R.; Lampson, M.A. Aurora B Dynamics at Centromeres Create a Diffusion-Based Phosphorylation Gradient. J. Cell Biol. 2011, 194, 539–549. [Google Scholar] [CrossRef]
- Saurin, A.T. Kinase and Phosphatase Cross-Talk at the Kinetochore. Front. Cell Dev. Biol. 2018, 6, 62. [Google Scholar] [CrossRef]
- Zaytsev, A.V.; Segura-Peña, D.; Godzi, M.; Calderon, A.; Ballister, E.R.; Stamatov, R.; Mayo, A.M.; Peterson, L.; Black, B.E.; Ataullakhanov, F.I.; et al. Bistability of a Coupled Aurora B Kinase-Phosphatase System in Cell Division. Elife 2016, 5, e10644. [Google Scholar] [CrossRef]
- Hart, Y.; Alon, U. The Utility of Paradoxical Components in Biological Circuits. Mol. Cell 2013, 49, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Gelens, L.; Saurin, A.T. Exploring the Function of Dynamic Phosphorylation-Dephosphorylation Cycles. Dev. Cell 2018, 44, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; McCollum, D. A Role for Metaphase Spindle Elongation Forces in Correction of Merotelic Kinetochore Attachments. Curr. Biol. 2012, 22, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Sandri, B.J.; Tank, D.; McClellan, M.; Harasymiw, L.A.; Yang, Q.; Parker, L.L.; Gardner, M.K. A Gradient in Metaphase Tension Leads to a Scaled Cellular Response in Mitosis. Dev. Cell 2019, 49, 63–76.e10. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, A. Shake It Off: The Elimination of Erroneous Kinetochore-Microtubule Attachments and Chromosome Oscillation. Int. J. Mol. Sci. 2021, 22, 3174. https://doi.org/10.3390/ijms22063174
Yamamoto A. Shake It Off: The Elimination of Erroneous Kinetochore-Microtubule Attachments and Chromosome Oscillation. International Journal of Molecular Sciences. 2021; 22(6):3174. https://doi.org/10.3390/ijms22063174
Chicago/Turabian StyleYamamoto, Ayumu. 2021. "Shake It Off: The Elimination of Erroneous Kinetochore-Microtubule Attachments and Chromosome Oscillation" International Journal of Molecular Sciences 22, no. 6: 3174. https://doi.org/10.3390/ijms22063174
APA StyleYamamoto, A. (2021). Shake It Off: The Elimination of Erroneous Kinetochore-Microtubule Attachments and Chromosome Oscillation. International Journal of Molecular Sciences, 22(6), 3174. https://doi.org/10.3390/ijms22063174





