CaDHN3, a Pepper (Capsicum annuum L.) Dehydrin Gene Enhances the Tolerance against Salt and Drought Stresses by Reducing ROS Accumulation
Abstract
1. Introduction
2. Results
2.1. CaDHN3 Sub-Cellular Localization
2.2. Silencing of CaDHN3 Reduces Salt Stress Tolerance in Pepper
2.3. Knockdown of CaDHN3 Reduced Drought-Stress Tolerance in Pepper
2.4. Overexpression of CaDHN3 in Arabidopsis Increased Salt Stress Tolerance
2.5. Overexpression of CaDHN3 in Arabidopsis Increased Drought Tolerance
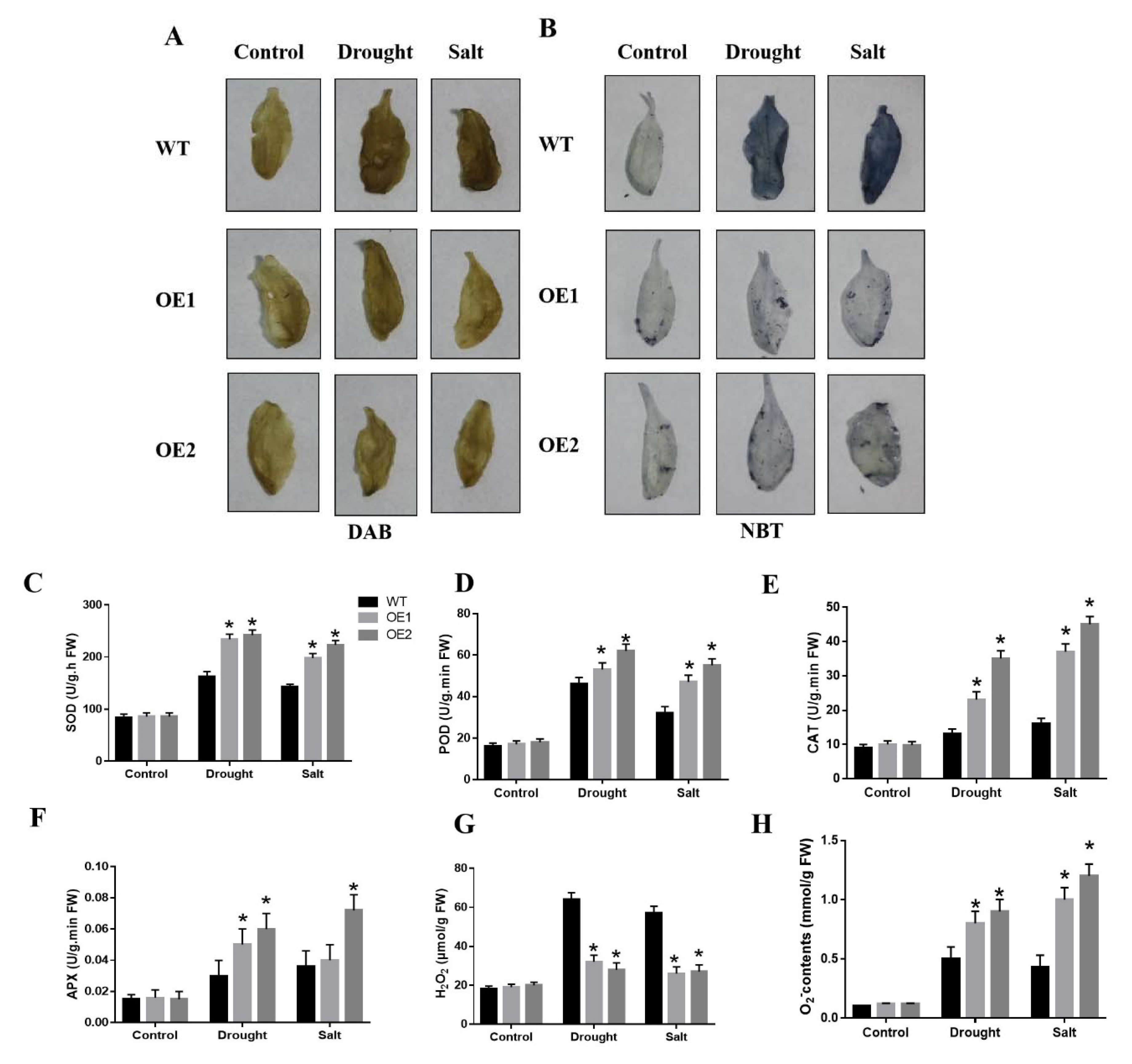
2.6. CaDHN3 Overexpression Relieves ROS Accumulation
2.7. CaDHN3 Overexpression Activated the Expression of Relative Stress-Responsive Genes
2.8. CaDHN3 Interact with CaHIRD11 Protein
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction and QPCR Analysis
4.3. VIGS of Pepper CaDHN3
4.4. Transient Expression in Arabidopsis Plants
4.5. Salt and Drought Assays
4.6. Measurement REL, Chlorophyll Content and MDA Content
4.7. Measurement of H2O2 and O2•− Contents, Histochemical Detection of ROS and Antioxidant Enzymes Activities
4.8. NBT and DAB Staining
4.9. Measurement of Stomatal Aperture
4.10. Sub-Cellular Localization, Yeast Two-Hybrid (Y2H) and Bimolecular Fluorescence Complementation (BiFC) Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.I. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum spp. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Lin, H.H. Role of plant dehydrins in antioxidation mechanisms. Biologia 2010, 65, 755–759. [Google Scholar] [CrossRef]
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 576. [Google Scholar] [CrossRef] [PubMed]
- Enebe, M.C.; Babalola, O.O. The influence of plant growth-promoting rhizobacteria in plant tolerance to abiotic stress: A survival strategy. Appl. Microbiol. Biotechnol. 2018, 102, 7821–7835. [Google Scholar] [CrossRef] [PubMed]
- Allagulova, C.R.; Gimalov, F.R.; Shakirova, F.M.; Vakhitov, V.A. The Plant Dehydrins: Structure and Putative Functions. Biochemistry 2003, 68, 945–951. [Google Scholar] [CrossRef]
- Koag, M.C.; Wilkens, S.; Fenton, R.D.; Resnik, J.; Vo, E.; Close, T.J. The K-Segment of Maize DHN1 Mediates Binding to Anionic Phospholipid Vesicles and Concomitant Structural Changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef]
- Hughes, S.L.; Schart, V.; Malcolmson, J.; Hogarth, K.A.; Martynowicz, D.M.; Tralman-Baker, E. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013, 163, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Close, T.J. Dehydrins: Emergence of a biochemical role of a family of plant dehydration proteins. Plant Physiol. 1996, 97, 795–803. [Google Scholar] [CrossRef]
- Close, T.J. Dehydrins: A commonality in the response of plants to dehydration and low temperature. Plant Physiol. 1997, 100, 291–296. [Google Scholar] [CrossRef]
- Nguyen, P.N.; Tossounian, M.-A.; Kovacs, D.S.; Thu, T.T.; Stijlemans, B.; Vertommen, D.; Pauwels, J.; Gevaert, K.; Angenon, G.; Messens, J.; et al. Dehydrin ERD14 activates glutathione transferase Phi9 in Arabidopsis thaliana under osmotic stress. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129506. [Google Scholar] [CrossRef]
- Lv, A.; Fan, N.; Xie, J.; Yuan, S.; An, Y.; Zhou, P. Expression of CdDHN4, a Novel YSK2-Type Dehydrin Gene from Bermudagrass, Responses to Drought Stress through the ABA-Dependent Signal Pathway. Front. Plant Sci. 2017, 8, 748. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Wang, X.; Zhang, M.; Xi, Y.; Wang, A.; Zhu, J. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef] [PubMed]
- Aduse Poku, S.; Nkachukwu Chukwurah, P.; Aung, H.H.; Nakamura, I. Over-Expression of a Melon Y3SK2-Type LEA Gene Confers Drought and Salt Tolerance in Transgenic Tobacco Plants. Plants 2020, 9, 1749. [Google Scholar] [CrossRef]
- Kumar, M.; Lee, S.C.; Kim, J.Y.; Kim, S.J.; Aye, S.S.; Kim, S.R. Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 383–393. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2019, 16, 862–877. [Google Scholar] [CrossRef]
- Cao, Y.; Xiang, X.; Geng, M.; You, Q.; Huang, X. Effect of HbDHN1 and HbDHN2 Genes on Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2017, 8, 470. [Google Scholar] [CrossRef]
- Halder, T.; Upadhyaya, G.; Ray, S. YSK2 Type Dehydrin (SbDhn1) from Sorghum bicolor Showed Improved Protection under High Temperature and Osmotic Stress Condition. Front. Plant Sci. 2017, 8, 918. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Li, C.; Ma, F.; Ma, J.H.; Abid, K.; Wang, X.; Zhao, L.Y.; Gong, Z.H.; Chen, R.G. Genome-Wide Identification, Expression Diversification of Dehydrin Gene Family and Characterization of CaDHN3 in Pepper (Capsicum annuum L.). PLoS ONE 2016, 8, e0161073. [Google Scholar]
- Alexandersson, E.; Saalbach, G.; Larsson, C.; Kjellbom, P. Arabidopsis Plasma Membrane Proteomics Identifies Components of Transport, Signal Transduction and Membrane Trafficking. Plant Cell Physiol. 2004, 45, 1543–1556. [Google Scholar] [CrossRef] [PubMed]
- Borovskii, G.B.; Stupnikova, I.V.; Antipina, A.I.; Vladimirova, S.V.; Voinikov, V.K. Accumulation of dehydrin-like proteins in the mitochondria of cereals in response to cold, freezing, drought and ABA treatment. BMC Plant Biol. 2002, 2, 5. [Google Scholar] [CrossRef]
- Salleh, F.M.; Evans, K.; Goodall, B.; Machin, H.; Mowla, S.B.; Mur, L.A.J.; Runions, J.; Theodoulou, F.L.; Foyer, C.H.; Rogers, H.J. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 2012, 35, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Mueller, J.K.; Heckathorn, S.A.; Fernando, D. Identification of a Chloroplast Dehydrin in Leaves of Mature Plants. Int. J. Plant Sci. 2003, 164, 535–542. [Google Scholar] [CrossRef]
- Xing, X.; Liu, Y.; Kong, X.; Liu, Y.; Li, D. Overexpression of a maize dehydrin gene, ZmDHN2b, in tobacco enhances tolerance to low temperature. Plant Growth Regul. 2011, 65, 109–118. [Google Scholar] [CrossRef]
- Lee, S.C.; Kim, S.H.; Kim, S.R. Drought inducible OsDhn1 promoter is activated by OsDREB1A and OsDREB1D. J. Plant Biol. 2013, 56, 115–121. [Google Scholar] [CrossRef]
- Zhu, W.N.; Zhang, D.P.; Lu, X.X.; Zhang, L.S.; Yu, Z.Y.; Lv, H. Characterization of an SKn-type dehydrin promoter from wheat and its responsiveness to various abiotic and biotic stresses. Plant Mol. Biol. Rep. 2014, 32, 664–678. [Google Scholar] [CrossRef]
- Qin, Y.X.; Qin, F. Dehydrins from wheat x Thinopyrum ponticum amphiploid increase salinity and drought tolerance under their own inducible promoters without growth retardation. Plant Physiol. Biochem. 2016, 99, 142–149. [Google Scholar] [CrossRef]
- Wisniewski, M.; Webb, R.; Balsamo, R.; Close, T.J.; Yu, X.M.; Griffith, M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: A dehydrin from peach (Prunus persica). Physiol. Plant. 1999, 105, 600–608. [Google Scholar] [CrossRef]
- Lin, C.H.; Peng, P.H.; Ko, C.Y.; Markhart, A.H.; Lin, T.Y. Characterization of a Novel Y2K-type Dehydrin VrDhn1 from Vigna radiata. Plant Cell Physiol. 2012, 53, 930–942. [Google Scholar] [CrossRef]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Mafakheri, A.; Siosemardeh, A.; Bahramnejad, B.; Struik, P.C.; Sohrabi, Y. Effect of drought stress on yield, proline and chlorophyll contents in three chick pea cultivars. Aust. J. Crop Sci. 2010, 4, 580–585. [Google Scholar]
- Moore, K.; Roberts, L.J. Measurement of Lipid Peroxidation. Free Radic. Res. 1998, 28, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Zhang, R.; Qu, Y.; Miao, Z.; Zhang, Y.; Shen, X.; Wang, T.; Dong, J. Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 2012, 195, 124–135. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Zhang, T.; Yang, X.; Li, D. Functional characterization of KS-type dehydrin ZmDHN13 and its related conserved domains under oxidative stress. Sci. Rep. 2017, 7, 7361. [Google Scholar] [CrossRef]
- Yokoyama, T.; Ohkubo, T.; Kamiya, K.; Hara, M. Cryoprotective activity of Arabidopsis KS-type dehydrin depends on the hydrophobic amino acids of two active segments. Arch. Biochem. Biophys. 2020, 691, 108510. [Google Scholar] [CrossRef]
- Guo, W.L.; Chen, R.G.; Gong, Z.H.; Yin, Y.X.; Li, D.W. Suppression Subtractive Hybridization Analysis of Genes Regulated by Application of Exogenous Abscisic Acid in Pepper Plant (Capsicum annuum L.) Leaves under Chilling Stress. PLoS ONE 2013, 8, e66667. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Khan, A.; Li, R.J.; Sun, J.T.; Ma, F.; Zhang, H.X.; Jin, J.H.; Ali, M.; Haq, S.U.; Wang, J.E.; Gong, Z.-H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef] [PubMed]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Campos, P.S.; Quartin, V.N.; Ramalho, J.C.; Nunes, M.A. Electrolyte leakage and lipid degradation account for cold sensitivity in leaves ofCoffea sp. plants. J. Plant Physiol. 2003, 160, 283–292. [Google Scholar] [CrossRef]
- Chakrabarty, D.; Datta, S.K. Micro propagation of gerbera: Lipid peroxidation and antioxidant enzyme activities during acclimatization process. Plant Physiol. 2008, 30, 325–331. [Google Scholar]
- Ke, D.; Sun, G.; Wang, Z. Effects of superoxide radicals on ACC synthase activity in chilling-stressed etiolated mungbean seedlings. Plant Growth Regul. 2007, 51, 83–91. [Google Scholar] [CrossRef]
- Stengel, F.; Baldwin, A.J.; Painter, A.J.; Jaya, N.; Basha, E.; Kay, L.E.; Vierling, E.; Robinson, C.V.; Benesch, J.L.P. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc. Natl. Acad. Sci. USA 2010, 107, 2007–2012. [Google Scholar] [CrossRef]
- Jariteh, M.; Ebrahimzadeh, H.; Niknam, V. Developmental changes of protein, proline and some antioxidant enzymes activities in somatic and zygotic embryosof Persian walnut (Juglans regia L.). Plant Cell Tissue Organ Cult. 2015, 122, 101–115. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. Meth. Enzymol. 1984, 105, 121–126. [Google Scholar]
- Mittova, V.; Volokita, M.; Guy, M.; Tal, M. Activities of SOD and the ascorbate-glutathione cycle enzymes in subcellular compartments in leaves and roots of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii. Physiol. Plant. 2000, 110, 42–51. [Google Scholar] [CrossRef]
- Able, A.J. Role of reactive oxygen species in the response of barley to necrotrophic pathogens. Protoplasma 2003, 221, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Sekulska-Nalewajko, J.; Gocławski, J.; Chojak-Kozniewska, J. Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 2016, 109, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, F.; Wang, X.; Liu, S.; Saeed, U.H.; Hou, X.; Zhang, Y.; Luo, D.; Meng, Y.; Zhang, W.; et al. Molecular and Functional Characterization of CaNAC035, an NAC Transcription Factor From Pepper (Capsicum annuum L.). Front. Plant Sci. 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, Y.-C.; Zhang, H.-F.; Pan, X.-X.; Chen, N.; Hu, H.-F.; Haq, S.u.; Khan, A.; Chen, R.-G. CaDHN3, a Pepper (Capsicum annuum L.) Dehydrin Gene Enhances the Tolerance against Salt and Drought Stresses by Reducing ROS Accumulation. Int. J. Mol. Sci. 2021, 22, 3205. https://doi.org/10.3390/ijms22063205
Meng Y-C, Zhang H-F, Pan X-X, Chen N, Hu H-F, Haq Su, Khan A, Chen R-G. CaDHN3, a Pepper (Capsicum annuum L.) Dehydrin Gene Enhances the Tolerance against Salt and Drought Stresses by Reducing ROS Accumulation. International Journal of Molecular Sciences. 2021; 22(6):3205. https://doi.org/10.3390/ijms22063205
Chicago/Turabian StyleMeng, Yuan-Cheng, Hua-Feng Zhang, Xiao-Xiao Pan, Nan Chen, Hui-Fang Hu, Saeed ul Haq, Abid Khan, and Ru-Gang Chen. 2021. "CaDHN3, a Pepper (Capsicum annuum L.) Dehydrin Gene Enhances the Tolerance against Salt and Drought Stresses by Reducing ROS Accumulation" International Journal of Molecular Sciences 22, no. 6: 3205. https://doi.org/10.3390/ijms22063205
APA StyleMeng, Y.-C., Zhang, H.-F., Pan, X.-X., Chen, N., Hu, H.-F., Haq, S. u., Khan, A., & Chen, R.-G. (2021). CaDHN3, a Pepper (Capsicum annuum L.) Dehydrin Gene Enhances the Tolerance against Salt and Drought Stresses by Reducing ROS Accumulation. International Journal of Molecular Sciences, 22(6), 3205. https://doi.org/10.3390/ijms22063205






