1. Introduction
Breast cancer (BC) is the most common gynaecological cancer, and it was identified in 2018 as the main cause of female cancer death in 11 of the 20 major global regions [
1]. There was recorded annual increase to 3.1% between 1980 and 2015, and the incidence continues to rise [
2]. While this incidence is 92 in 100,000 in North America [
3] and 144 in 100,000 in European Union countries [
4], BC is diagnosed in Asian countries approximately 20 years earlier, at an average 40–50 years compared to 50–70 in European countries. Africa and Central Asia, however, record the lowest BC incidence, but the incidence there is most likely influenced by insufficient screening and diagnostic programmes [
5,
6]. In addition, BC is diagnosed in the earlier stages in the more developed countries and although it is more common there, the overall mortality is lower. Finally, the highest BC mortality is recorded in sub-Saharan Africa [
7], where women of African origin are more prone to rapid metastasising BC with lower survival rate [
8].
BC comprises variable tumours with different histological, clinical and molecular-biological manifestations [
9]. The basic histological division of BC tumours is into invasive and pre-invasive types. The latter has two distinct entities, lobular carcinoma in-situ (LCIS) and ductal carcinoma in-situ (DCIS). The risk of LCIS progression is very low, and these lesions are considered more a risk factor than an invasive lesion precursor. This type can be bilateral, but it does not usually distort lobular architecture. In contrast, DCIS are unilateral and can spread through ducts, thus disturbing ductal architecture. This type comprises more than 80% of all pre-invasive lesions and it can progress to invasive forms [
7,
9,
10].
The invasive BC subtypes spread into surrounding breast tissue, and the most frequent of these are invasive carcinoma of no special type (NST; formerly known as invasive ductal carcinoma) and invasive lobular carcinoma. NST accounts for 70–75% of all BCs, followed by lobular carcinomas with 10–15% and the remaining BC tumours form 17 rare histological subtypes [
9,
11]. Finally, only up to 10% of all diagnosed BC are non-invasive, and 20–53% of women with DCIS develop invasive carcinoma within 10 years [
12].
Initial BC molecular classification was introduced by Perou et Sørlie [
13] at the beginning of the millennium, and this enabled BC tumour classification into Luminal, HER2 positive, basal and ‘normal-like’ subtypes through differences in gene expression pattern. Perou and Sørlie’s pilot study inspired further research in molecular BC classification and several variations of molecular classification based on gene expression or mutations in particular genes have been created. However, molecular classification is still developing and cannot be considered established [
14,
15,
16,
17,
18,
19]. Current molecular classification is typically based on PAM50, which is a 50-gene expression signature that classifies BC in five molecular intrinsic subtypes: Luminal A, Luminal B, HER2-enriched, Basal-like, and Normal-like. Moreover, additional subtypes have been identified, including claudin-low, molecular apocrine and interferon-rich subtypes [
14,
15,
16,
17,
18,
19]. However, there is still no consensus whether to consider these “additional” subtypes analogous to the intrinsic subtypes, or if tumours carry these phenotypes only in addition to their intrinsic subtype [
10,
16].
In summary, although molecular classifications provide important characteristics of particular tumour types and can be beneficial in predicting tumour progression, molecular classifications are often too methodologically and financially demanding for daily clinical use [
14,
15,
16,
17,
18,
19]. These problems mean that current clinical practice typically uses a surrogate classification of five subtypes based on histology and immunohistochemistry This particularly involves assessing the expression of key proteins: oestrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and the proliferation marker Ki67. These subtypes are: Luminal A-like, Luminal B-like HER2-, Luminal B-like Her2+, HER2-enriched, and Triple negative (TNBC) [
9,
10,
20].
(1). Luminal A-like is the most common subtype, accounting for 60–70% of all BC tumours. These tumours have the best prognosis and overall survival rates. They are low grade tumours, ER and PR positive and HER2 and Ki67 negative (ER+; PR+; HER2−; Ki67− (<14%)).
(2). Luminal B-like HER2- is the second most common subtype, comprising almost 10–20% of BC tumours. These have intermediate prognosis, and although their ER and PR positivity is lower than Luminal A, they are still classified as ER+; PR+; HER2- and Ki67+ (>14%).
(3). Luminal B-like HER2+ have similar characteristics to the luminal-B like subtype, but are also positive for HER2. Their prognosis is worse than luminal B-like and often have higher grade.
(4). HER2-enriched are typically (ER−; PR−; HER2+, Ki67+). Although these tumours are considered aggressive, they respond reasonably well to therapy and their prognosis therefore remains intermediate. Together with the Luminal B -like HER2+ subtype they cover 15–20% of all BC tumours.
(5). TNBC accounts for approximately 15% of BC tumours and they are typically (ER-; PR-; HER2-, Ki67+). These are high grade with poor prognoses and positive for basal-markers with high Ki67 proliferation index.
The method of treatment for these BC tumours is directly related to the specific subtype [
9,
21].
(1). Endocrine therapy is prescribed by default for luminal subtypes if there is minimal risk of recurrence. In greater risk, the treatment of choice is neoadjuvant or adjuvant chemotherapy with docetaxel and cyclophosphamide if the tumour burden is low (pN0-1), and anthracycline-taxane sequence is preferred in higher tumour burden (pN2-3).
(2). Adjuvant chemotherapy with paclitaxel and trastuzumab is usually applied for 1 year when HER2+ tumours are pT1/pN0. If these tumours are ≥T2, or spread to lymph nodes, chemotherapy is preferred in neoadjuvant settings with anthracycline-taxane combined with dual HER2 blockade (trastuzumab and petruzumab). Further treatment then depends on complete pathological response, and treatment continues with Anti-HER2 therapy for one year when response is positive and T-DM1 is applied when it is negative.
(3). TNBC is considered the most aggressive subtype, and initial neoadjuvant chemotherapy with anthracycline-taxane sequence is preferred (platinum agent may be added). Adjuvant chemotherapy with capecitabine is then applied in the absence of complete pathological response.
However, this is only a generalised algorithm for treatment of early BC and the preference for adjuvant or non-adjuvant therapy and the use of particular pharmaceuticals depends on the precise situation, including menopausal status, concurrent treatments and other clinical factors [
9,
21].
The human genome contains 3 × 10
9 base pairs, and the DNA in all chromosomes in the cell measures 2 metres when extended. While approximately 75% of this genetic information is further transcribed [
22,
23], only 1.5–2% of the total transcript is mRNA and this is further translated into amino acid sequence. The majority of the transcript comprises non-coding RNAs (ncRNA) which directly or indirectly regulate mRNA expression and the derived protein products. It is presumed that abnormal expression levels of the many genes involved in BC development are influenced by ncRNA activity [
24,
25]. Therefore, knowledge of particular ncRNA characteristics can help understanding complex BC cell mechanisms and improve BC subtype diagnosis and medical treatment.
The non-coding RNAs are divided into two main groups, house-keeping ncRNAs and regulatory ncRNAs. The house keeping ncRNAs are usually constitutively expressed under physiological conditions and include the following RNAs: transfer RNAs (tRNA), ribosomal RNAs (rRNA), small nuclear RNAs (snRNA), small nucleolar RNAs (snoRNA) and telomerase RNA (TERC). In contrast, the regulatory ncRNAs are expressed in a more cell-and-tissue-specific manner and often in response to environmental factors. These can regulate gene expression at the epigenetic, transcriptional, post-transcriptional, translational, and post-translational levels [
26,
27]. Regulatory non-coding RNAs are divided into short non-coding RNAs less than 200 bp and long non-coding RNAs (lncRNAs) with over 200 bp. The short ncRNAs are further sub-divided into microRNAs (miRNAs), short interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs) [
22,
23,
28,
29]. The majority of analysis and research has focused on miRNA rather than other ncRNAs, although the miRNAs constitute only a small percentage of the total transcript [
24,
25], This has inspired a summary of the effect of the more abundant, but less understood, non-coding RNAs on the development and progression of BC. This especially includes a description of the function lncRNAs, piRNAs, snRNAs and snoRNAs. Finally, the possibilities of inhibiting tumours by RNA interference (RNAi) are discussed.
2. microRNAs
Although the main goal of this article is to summarise the effects of ncRNAs other than miRNAs on BC development, miRNAs are repeatedly discussed in the following text, especially in relation to their interaction with lncRNAs. The basic characteristics and mechanisms of miRNAs are therefore included herein. miRNAs range from 21 to 25 nt in length, but are most commonly 22 nt [
30], and miRNA genes are mostly transcribed from both intragenic and intergenic regions by polymerase II. They are less often transcribed by polymerase III, and then especially from miRNA coding sequences spread among Alu repeats [
31]. In addition, miRNAs from the same family are often co-transcribed in “clusters” [
32]. miRNAs from the same family have a similar seed sequence, comprising nucleotides 2–8 from the 5′end. These sequences are essential for binding to mRNA [
33]. Approximately 1900 pre-miRNAs and over 2600 mature mRNAs have been described in the human genome [
30,
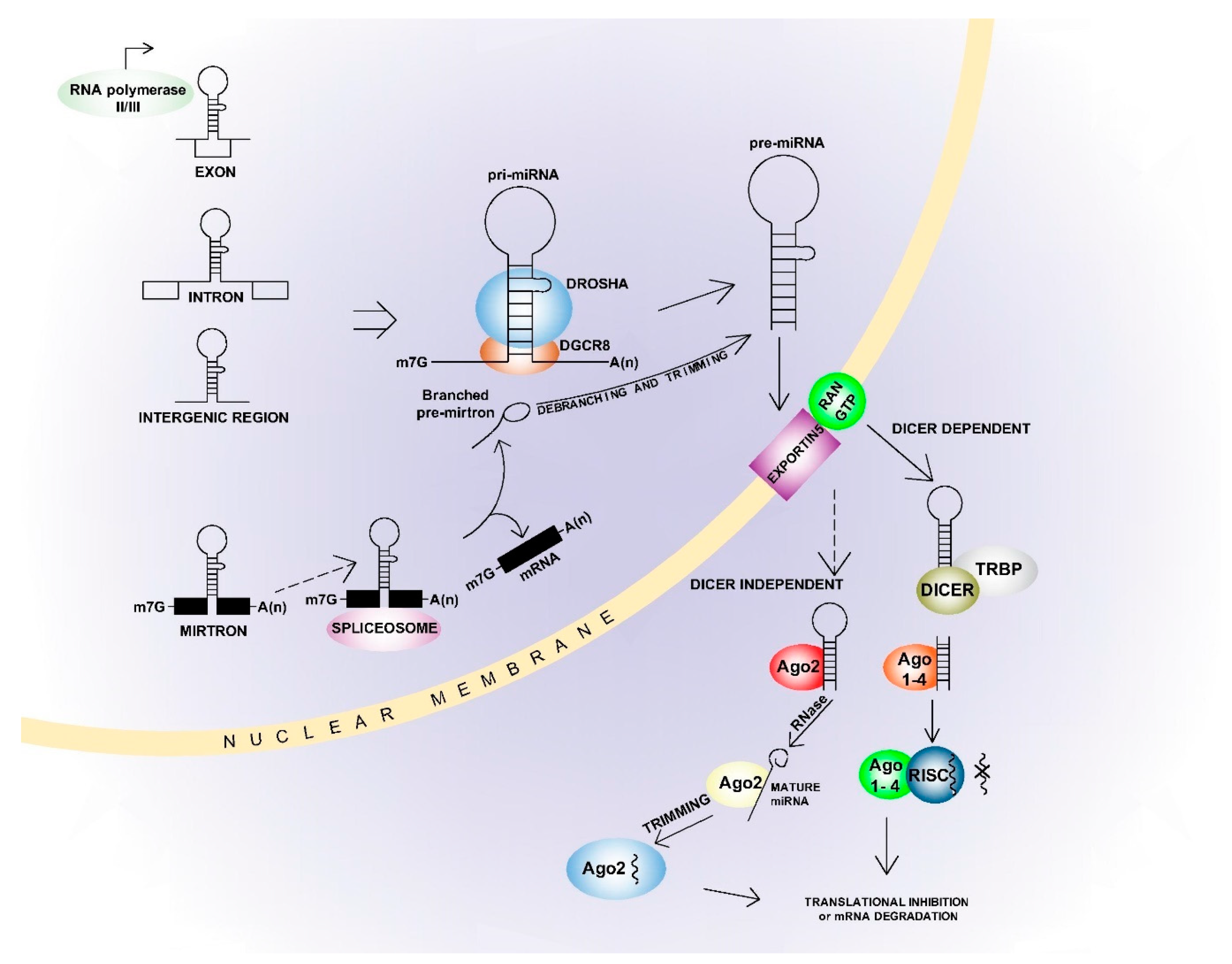
34]. The miRNA synthesis pathways can be canonical or non-canonical (
Figure 1) [
30,
31]. In the canonical pathway, miRNA genes are initially transcribed into the primary pri-miRNAs and then processed into the precursor pre-miRNAs. This process is mediated by a “micro-processor complex” comprising DGCR8, ribonuclease III and Drosha [
35]. The pre-miRNAs are then exported to the cytoplasm by Exportin 5 and processed to mature miRNA’s by the Dicer complex [
30,
31]. Double-stranded miRNAs are separated into guide strand and passenger strand, depending on orientation and sequence stability. Here, the strand with lower thermodynamic stability at the 5′ end usually features as the guide strand for incorporation into the Argonaute protein while the passenger strand is degraded [
30,
31]. Non-canonical biogenesis varies depending on the protein complexes involved, and is most often Drosha/DGCR-8 independent or Dicer independent [
30,
31]. While the miRNAs most often interact with the mRNA 3′UTR region, they can also interact with the 5′UTR region or directly with the gene promoter region [
36]. These interactions primarily result in repression of target gene expression, but some miRNAs also enhance gene expression [
30,
31]. This is usually seen in ‘starved’ cells, or under other non-physiological conditions [
37,
38]. In addition, one miRNA can inhibit several genes and one gene can be affected by multiple miRNAs [
30,
31]. This can result in complete cell signalling regulated by one or only a few miRNAs, and the miRNAs can have either a tumour-suppressive or oncogenic effect [
39].
3. Long Non-Coding RNAs
lncRNAs have a greater than 200 bp sequence [
26] and were initially considered redundant sequences following gene transcription. These lncRNAs undergo splicing and removal of intron-like sequences, similar to genes encoding for proteins. It is also currently estimated that over 270,000 lncRNAs are present in the human genome, although many have very low copy number [
40]. The lncRNAs are therefore very abundant and heterogeneous, and their sequences can be located close to the genes, in the space between genes or even overlapping them. Their binding is especially variable: in the 3′-5′direction, the 5′-3′direction, bi-directional or binding to intergenic regions, intron regions, and enhancer sequences [
41].
The lncRNAs division is as follows [
26]:
(a) mRNA-like transgenic transcripts, also known as lincRNAs. These are further spliced, capped and poly-adenylated
(b) Natural antisense transcripts (NAT) of protein coding genes
(c) primary RNA polymerase II transcript-derived unconventional lncRNAs. This group can be divided into the following subgroups based on the types of stabilisation and modifications they undergo:
(c1) MALAT and NEAT1 lncRNAs, processed by RNase P and stabilized by U-A-U triple helix structures at their 3′ ends; (c2) snoRNA-ended lncRNAs mainly derived from excised introns; (c3) 5′ snoRNA-ended and 3′-polyadenylated lncRNAs (SPA); (c4) Circular intronic RNAs (ciRNA) derived from excised introns; (c5) Circular RNAs (circRNA) produced by circular back-splicing of pre-mRNA exons. This division is depicted in
Figure 2.
The lncRNAs also vary in their final effect. For example, they are preferentially located in the nucleus and can form a complex there with the HNRPNK nuclear matrix protein and participate in nuclear organisation [
26,
42,
43]. Xist lncRNA is involved in inactivating one of the X chromosomes [
44] and C0T-1 lncRNA affects chromosome decondensation [
45].
lncRNAs also act on chromatin remodelling by variable cis and trans mechanisms. Recruiting the PRC2 polycomb repressive complex is common, and this progresses to deposition of histone H3 lysine 27 trimethylation (H3K27me3) [
46]. Moreover, there is antagonistic effect of the MHRT (Myosin Heavy Chain Associated RNA Transcripts) cluster lncRNAs on the Brg1 catalytic subunit of the BAF chromatin remodelling complex [
47]. lncRNAs can interact with chromatin remodelling complexes, primarily the SWI/SNF complex [
48] and they can modify histones by altering their methylation [
49] and acetylation [
50], or affect DNA methylation levels [
51,
52]. Furthermore, they can either enhance or inhibit gene transcription as cis or trans regulators [
53]. The lncRNAs are also more tissue specific than miRNAs and although some conservation is observed in the formation of secondary and tertiary structures in closely related species, individual species’ evolutionary conservation is generally low [
54].
lncRNAs often function as competing endogenous RNAs (ceRNA) [
55]. These lncRNAs contain complementary binding sites to miRNA called microRNA response elements (MRE).This results in miRNA binding to this lncRNA instead of binding to the target mRNA sequence, and the latter is therefore not repressed. In addition, this effect can also be achieved with artificially prepared constructs called “miRNA sponges” [
40,
55]. These are often circular and contain variable numbers of copies of desired MREs. Their binding sites are usually specific for seed region and this enables blocking of entire miRNA families. Finally, some lncRNAs abundant in BC are precursors for various miRNAs [
26].
3.1. Long Non-Coding RNAs and BC
Description of H19, TINCR, MALAT and NEAT1 DANCR lncRNAs follows, because their abnormal expression is associated with BC development and metastasis. Additional lncRNAs reported to be abnormally expressed in at least one study and also lncRNAs associated with TNBC development are then summarised.
3.1.1. H19 lncRNA
The association of dysregulated lncRNA H19 (H19) expression and BC development has been described [
56,
57]. Here, the highly expressed H19 is apparent in over 70% of BC tumours, and this includes ER+ and ER-, and HER2+ and HER2- [
56,
57,
58]. Several typical mutational polymorphisms are also associated with higher expression of this lncRNA in BC [
58,
59]. H19 knockout can then suppress the Akt pathway, induce apoptosis, and restore paclitaxel sensitivity [
56,
57]. EZF1 also binds to the H19 promoter region, activates its expression [
60] and promotes cell cycle progression. EZF1 is over-expressed in BC tumours [
60].
In addition, lncRNA-H19 creates a precursor for miR-675 whose abnormal expression is demonstrably associated with BC development [
56,
61] and it also acts as a ceRNA in BC cells and interacts with miR-200b/c and let-7b. This increases the expression of
GIT2 and
CYTH3 genes, which are important contributors to rapid epithelial-to-mesenchymal transition (EMT) [
62]. In addition, the 200b/c and let-7b miRNAs are often abnormally expressed in metastatic BC tumours [
63]. The lncRNA-H19 can bind to miR-152 which targets DNA methyltransferase 1 (DNMT1), and this provides a further mechanism for altering BC methylation levels [
64]. Furthermore, the lncRNA-H19 binding to miR-9-5p depletes the latter’s levels and increases the expression of the important
STAT3 signal transducer gene in BC [
65].
Finally, lncRNA-H19 interacts with the Myc family of proto-oncogenes and these are among the most dysregulated transcription factors in various tumorous cells, including BC cells [
56]. c-Myc affects histone acetylation and H19 transcription initiation [
66], and n-Myc regulates DNA methyltransferase 3α2, thus altering H19 locus methylation status and expression [
67].
3.1.2. TINCR lncRNA
TINCR lncRNA (TINCR) affects the formation of primary BC tumours, and subsequent metastasis was discovered in 2018 [
68]. The qPCR methodology in that study of 24 patients determined higher TINCR BC expression than in non-BC participants. In addition, higher TINCR activity is triggered by SP1-zinc finger transcriptional factor which typically recognises the GC-rich sequences in promoter regions [
68]. TINCR also increased proliferation and inhibited apoptosis in the BC cell lines, and this occurred because TINCR competed with miR-7. This interaction between TINCR and miR-7 also resulted in modulation of
KLF4 gene expression. The miR-7 ability of supressing BC metastasis through
KLF4 activity has previously been demonstrated [
69]. Finally, experimental TINCR silencing suppressed proliferation in both in vivo and in vitro conditions [
68].
Further TINCR mechanisms were comprehensively elucidated by Dong et al. [
70]. These authors used SKBR-3-TR and BT474-TR trastuzumab-resistant cell lines, followed by mouse xenograft analysis. TINCR knockout decreased these cells’ chemoresistance, their EMT potential and EMT marker expression. The TINCR-lncRNA acted there as a ceRNA and inhibited activity of miR-125b which suppresses BC progression by regulating
HER2 expression [
71]. miR-125b was also found to target Snail-1 which is an important EMT and chemoresistance regulator. Thus, TINCR regulated the miR-125b-HER2/Snail-1 pathway and this resulted in trastuzumab resistance and EMT induction. The noted TINCR up-regulation was considered due to abnormal H3K27 acetylation of the TINCR promoter region. The final part of Dong et al.’s study [
70] concentrated on qPCR analysis of 60 HER2+ BC tumour tissues from 30 chemoresistant and 30 chemotherapy-sensitive patients, and results indicated higher TINCR activity in chemoresistant patients, who subsequently experienced worse overall survival rate.
That work was then complemented by Guo et al.’s recent research [
72], which revealed TINCR effect in MCF-7 and MDA-MB-231 BC cell lines. TINCR over-expression there inhibited miR-589-3p activity because TINCR acted as the miR-589’s ceRNA. This resulted in higher cancer cell proliferation, migration and invasion and also inhibition of cancer cell apoptosis. In addition, experimental miR-589-3p over-expression partly inhibited the TINCR effect on tumourigenesis.
The TINCR BC action can strongly participate in increasing IGFR/AKT pathway activity, because both of these components are miR-589-3p target genes [
73,
74]. IGFR is a well-known trans-membrane tyrosine kinase receptor important in cell mitosis, proliferation, differentiation, apoptosis and regulation of Akt pathway activity [
73,
74]. Finally, Guo et al. [
72] determined that higher TINCR expression was associated with poor survival rate in a cohort of 68 patients.
3.1.3. MALAT lncRNA
lncRNA MALAT (MALAT) is abnormally expressed in several BC types and this abnormal expression correlates with poor prognosis and metastasis [
75,
76]. It is further indicated that this lncRNA activity can be inhibited by high 17-β oestradiol concentration [
77]. One interesting study found that MALAT was elevated in patients with early post-BC-resection fever [
78] and this led to worse prognosis. In addition MALAT knockout in mouse 4T1 xenografts significantly decreased inflammation and reduced the lung metastases so often seen in BC [
78].
miR-1 was noted to directly target MALAT1 and small GTPase
CDC42. The MALAT binding with miR-1 resulted in over-expression of Cdc42 which then promoted BC cell line migration and invasion [
79]. MALAT1 therefore functioned as a ceRNA of
CDC42. Moreover, MALAT action also inhibits E-cadherin expression and induces vimentin expression, and this leads to higher epithelial mesenchymal transition ratio (EMT). This directly contrasts with miR-1 action [
79]. MALAT1 was also demonstrated to bind to miR-204, and there is reciprocal repression between miR-204 and MALAT1. In addition, E-box binding homeobox 2 (ZEB2) is a direct miR-204 target. ZEB2 is a very important factor in EMT promotion because it has been reported to inhibit E-cadherin and down-regulate a distinct set of constituents of desmosomes and adherens, gap and tight junctions [
80,
81].
3.1.4. NEAT1 lncRNA
NEAT1 is an important oncogene in cancer and significantly affects EMT induction in BC [
82]. Abnormal NEAT1 activity affected chemoresistance and cancer cell stemness in a cohort of 179 BC patients and it was expressed 6.86 times more in BC patients than in 192 controls [
83]. NEAT1’s abnormally high expression was also instrumental in influencing tumour size, lymph node metastases and overall survival in a cohort of 40 BC patients [
84].
There were also changes in EMT marker levels following NEAT1 knockout in various cancer cell lines. This particularly applied to E-cadherin up-regulation and β-catenin and N-cadherin down-regulation following NEAT1 inhibition [
84]. Li et al. then indicated that NEAT1 activity in BC is largely oestrogen-inducible and NEAT1 mediated the interaction between FOXN3 and SIN3A in oestrogen-dependent BC [
85]. The FOXN3-NEAT1-SIN3A complex is considered to suppress genes which inhibits EMT, especially
GATA3 [
85], and connections between abrogated
GATA3 and increased EMT have previously been demonstrated [
86]. In addition,
NEAT1 and
FOXN3 over-expression results in decreased epithelial markers including E-cadherin, α-catenin and γ-catenin, and it corresponds with diminished GATA3 expression. This also induces mesenchymal markers such as fibronectin and vimentin. This study highlighted the FOXN3-NEAT1-SIN3A effect on EMT promotion, MCF-7 cell line invasion and higher dissemination and metastasis under in vivo conditions in BC mouse models [
85]. Finally, the authors also demonstrated higher NEAT1 and FOX3 expression and lower GATA3 expression in 24 BC patient tissue specimens [
85].
A further important BC feedback loop includes NEAT1, miR-124 and STAT3. The NEAT1 and STAT3 expression levels were elevated in BC tissues, and this correlated with decreased miR-124 expression levels. Moreover, NEAT1 over-expression markedly increased STAT3 protein expression levels, but this effect was reversed by miR-124 over-expression. The inhibitory effects of miR-124 over-expression were also abolished by STAT3 over-expression. Finally, STAT3 silencing inhibited NEAT1 transcription in BC cells. Therefore, NEAT1 and STAT3 form a feedback loop via sponging miR-124 to promote BC progression. NEAT1 modulation which regulates STAT3 appears very promising as a potential therapeutic approach, because STAT3 is a major BC marker with many upstream regulators and downstream targets which are known to promote BC malignancy and strong chemo-resistance [
87].
NEAT1 also acts as an ceRNA inhibiting function of miR-211 [
88] which is down-regulated in cancer and considered a tumour-suppressor [
89,
90]. This miRNA also binds to
HMGA2 which regulates a network of transcription factors that drive EMT [
88]. NEAT1 down-regulation ultimately decreases expression of the
HMGA2, and this NEAT1/HMGA2 activity is in direct proportion. NEAT1 activity has also been experimentally inhibited by sh-NEAT1, and this resulted in decreased 5-FU resistance. NEAT1 activity can therefore affect chemo-resistance, and further analysis of this interaction could prove beneficial.
3.1.5. DANCR lncRNA
DANCR lncRNA affects the activity of the EZH2 enzymatic subunit of the PRC2 polycomb repressive complex [
91]. The PRC2 complex changes histone methylation status and regulates EMT-inducers transcription [
92] and its abnormal activity is associated with various oncological diseases [
93]. Research suggests that DANCR most likely has tumour-suppressive function because of its low activity in BC cell lines and tissues [
91]. DANCR’s interaction with EZH2 is then thought to facilitate CD1 and EZH2 binding, and this causes EZH2 ubiquitination and degradation. DANCR therefore negatively regulates EZH2 action. While DANCR over-expression in mouse xenograft tumours formed from the injection of aggressive MDA-MB-231 cells decreased metastasis formation and spread, its knockout under in vitro conditions in MCF10A cells induced EMT, cell migration and invasion [
91].
Zhang et al., however, record different DANCR and EHZ2 interaction [
94]. Their study shows that DANCR lncRNA is more highly expressed and is considered to induce oncogenesis. DANCR did not initiate EZH2 degradation but promoted its binding to the
SOC3 gene promoter region, and this resulted in inhibited
SOC3 gene expression. This mechanism was observed under in vitro conditions in MCF-7, T47D, MDA-MB-231, and MDA-MB-468 cell lines. Finally, shDANCR knocked out DANCR in mouse xenografts with injected MDA-MB-231 and MDA-MB-468 cell lines, and this significantly reduced the number of metastatic BC nodules in mice lungs.
A further study demonstrated that DANCR was highly expressed in MCF-7 and MDA-MB-231 cell lines [
95]. However, a different action mechanism was noted in this study. DANCR was demonstrated as a ceRNA targeting miR-216-5p and its knock-out increased E-cadherin and decreased
Nanog,
OCT4, and
SOX2. The patients in this study were divided into DANCR expression groups, where patients with lower DANCR expression had slower disease development and better survival rates.
The DANCR-miR-758-3p-PAX6 molecular network is also affected in BC [
96]. DANCR was again observed to be more highly expressed in both BC cell lines and tissues, and miR-758-39 was significantly down-regulated. However, experimental DANCR down-regulation in BC cell lines inhibited cell malignancy and induced apoptosis by increasing typical apoptotic markers. In addition to DANCR inhibiting miR-758-3p activity, it also regulates the activity of
PAX6, which is a transcriptional factor abnormally expressed in BC, with subsequent poor prognosis [
97].
3.1.6. lncRNAs with Abnormal TNBC Expression
Recent studies have assessed the effect of lncRNA dysregulation in TNBC because this subtype is very aggressive and has poor prognosis [
98]. Aughoff et al. [
99] revealed that lncRNA LOC554202 is down-regulated in TNBC due to altered promoter region methylation, and this lncRNA includes a sequence encoding miR-31 which has a tumour-suppressive effect on BC formation and metastasis [
100].
Additional research by Koduru et al. [
101] employed freely available small RNA sequencing data from 24 TNBC patients and 14 healthy controls. They then used this in advanced data assembly and statistical analysis [
101]. Their results suggested that up to 258 lncRNAs can be abnormally expressed in TNBC tissues under the less stringent conditions of
p < 0.05. However, this decreased to 61 aberrantly expressed lncRNAs when
p < 0.01 was applied. It was further determined that SC5DL, PURA, EIF2C2 and ELP4 were the most important of the up-regulated lncRNAs, and PAPLN, FLT3LG, NEK8, FLOT2 and ZNF75D were the most significant of the 33 remaining down-regulated lncRNAs. Data for stage-wise lncRNA profiling assertion were then analysed. This confirmed that 160 lncRNAs were in BC Stage I, 155 in stage II, and 79 in stage III. There were 79 lncRNAs concurrently regulated in stages I and II, three in stages I and III, and only one in stages II and III. This indicates how individual lncRNA abnormal activity can affect TNBC progression, but further study is required to elucidate these relationships.
The small nucleolar RNA host gene 12 (SNHG12) lncRNA is abnormally expressed in several BC cell lines [
102] and this lncRNA’s pro-tumour effect was also found in TNBC tissues where higher expression correlated with tumour size and lymph node metastasis [
103]. Research has established that SNHG12 is a direct transcriptional target of c-MYC, and that c-MYC activation of SNHG12 resulted in higher
MMP13 activity. It is therefore highly likely that
MMP13 is also a target of SNHG12. The higher SNHG12 rate in TNBC tissue was also directly proportional to
MMP13 activity rate.
MMP13 is a metalloproteinase whose higher activity is typical in BC and its abnormal activity was initially identified in TNBC tumours [
104].
MMP13 degrades the extracellular matrix (ECM), but the process must be strictly regulated within physiological range. Moreover, an abnormal matrix is formed in tumour tissue when
MMP13 and other matrix metalloproteinase activity is too high, and this leads to greater invasion, metastasis and worse prognosis [
104]. However, SNHG12 expression in BT-549 and MDA-MB-231 cell lines was down-regulated by siRNA induced c-Myc depletion, with subsequent diminution of adhesion ability and cell proliferation [
103].
qPCR analysis determined that small nuclear NF90-associated (snaR) lncRNA has more than 15-fold expression in TNBC cell lines compared to controls [
105]. Consequently, Snar lncRNA knockdown significantly decreased cancer progression in triple negative MDA-MB-231 cell lines. Further research demonstrated that snaR activity is significantly influenced by NRON lncRNA, and the expression of these two lncRNAs is in indirect proportion. In particular, snaR expression increased from BC stage I to IV, while NRON expression decreased with increasing clinical staging [
106].
3.1.7. Additional lncRNAs
(1). The BCRT1 lncRNA is abnormally expressed in BC, where it functions as a ceRNA and competitively binds with miR-1303 to prevent degradation of its
PTBP3 target gene which is an important BC promoter [
107]. It is further considered that lncRNA BCRT1 can promote M2 polarisation, and that this lncRNA expression can be induced by direct hypoxia inducible factor-1a (HIF-1α) binding to hormone-responsive elements in its promoter region [
107].
(2). Chen et al. [
108] recorded the association of HIF-1α up-regulation with lncRNA action and BC formation. These researchers found that tumour associated macrophages (TAM) produce extracellular vesicles (EV) which contain HIF-1α-stabilising long-noncoding-RNA (HISLA). This HISLA lncRNA was naturally transmitted from EV to tumour cells where it blocked HIF-1α interaction with prolyl hydroxylase domain 2 (PHD2). This interaction prevented physiological HIF-1α degradation, and the subsequent rise in protein level increased cellular aerobic glycolysis [
109]. Experiments have also revealed that lactate released from glycolytic tumour cells up-regulates macrophage HISLA, and blocked transmission between EV and TAM results in glycolytic inhibition and decreased chemoresistance. Interaction between macrophages and tumour cells can therefore significantly influence tumour development, and modification of this interaction appears a worthy treatment target.
(3). The LINC0178 lncRNA can have an oncogenic effect in BC, because it influences tumour-suppressor miR-125 [
110] by inhibiting its binding to the Dicer complex and thus repressing miR-125 maturation.
(4). RP1-506.5 lncRNA is abnormally expressed in BC [
111]. This lncRNA represses the important p27Kip inhibitor of cyclin dependent kinase 1B due to formation of the RP1-p-4E-BP1/eIF4E complex. This complex precludes interaction of eIF4E and eIF4G eukaryotic translation initiation factors which normally activate p27Kip translation. The RP1-506.5 lncRNA also reduces
Snail1 gene activity, and it is further demonstrated that the
KLF5 gene affects this lncRNA’s activation. Finally, abnormal activity of the
KLF5 transcription factor and subsequent activation of its target genes have previously been associated with rapid BC growth and poor prognosis [
112].
(5). HOTAIR lncRNA is transcribed from the intergenic region of the
HOXC gene and this lncRNA activity dysregulation occurs in both ER positive and negative cancers with metastasis formation and worse disease course [
113,
114,
115]. HOTAIR is regulated by abundant oestrogen receptors and response elements in ER-positive BCs, and its activity is in direct proportion to oestradiol levels [
116]. Although the mechanisms of HOTAIR lncRNA’s increased activity in ER negative BCs is less understood, oestradiol involvement has been established also in this case [
117]. Oestradiol binds to the G-protein-coupled oestrogen receptors (GEDPR) in these tumours, and this triggers a cascade which culminates in inhibited miR-148 transcription [
118]. It is important that miR-148 is also a direct HOTAIR inhibitor. Therefore, the increased HOTAIR expression in ER- BC is not caused by direct initiation of transcription, as occurs in ER positive BC, but due to a loss of function of its inhibitor miR-148 [
117].
3.1.8. lncRNAs as Biomarkers in Liquid Biopsy
The important H19, NEAT1 and HOTAIR lncRNAs associated with BC development were analysed for detection of early-stage BC in patient plasma samples [
119]. While lncRNA H19 expression in plasma samples correlated with expression of its target miR-675 in both healthy and cancer-afflicted patients, the expression of NEAT1 and its target miR-204 correlated only in HER2+ patient plasma samples. In contrast, although HOTAIR could not be detected in plasma samples, its target miR-331 was detectable, and this was associated with nodal status. This miRNA was also differentially expressed in BC patient samples compared to samples from healthy subjects. Finally, all three lncRNAs were aberrantly expressed in MCF-7 cell lines, and their expression levels were in indirect proportion to their target miRNA levels.
Table 1 lists the abnormally expressed lncRNAs and their BC targets.
4. Piwi-Interacting RNAs
The piRNAs are 26–31 nucleotides long and associated with PIWI proteins which are a clade in the Argonaute protein family [
120,
121]. Piwi indicates “P-element–induced wimpy testes’, and their presence in eukaryotic cells was discovered in mouse sperm cells in 2006 [
122]. While their initial presence in human cells was demonstrated in germline and stem-cells [
123], they are also present in differentiated somatic cells, but in significantly smaller numbers [
120,
121]. However, over 30,000 piRNAs are currently described in the eukaryotic genome [
122].
The main piRNA role is silencing of transposable elements (TE) in the germline cells at both transcriptional and post-transcriptional levels. Although this function is conserved in most animal species, different proteins and mechanisms can be involved [
120,
121,
122]. Post transcriptional gene silencing (PTGS) of TE elements occurs in the cytoplasm, where the target transcript is sliced, and cleavage-competent Piwi-proteins are required for this processing. The best described PTGS mechanism is the “ping-pong-cycle” described later herein (
Figure 3). Transcriptional silencing (TGS) occurs in the nucleus where the piwi-piRNA complex recognises the nascent TE transcript by complementarity and this is followed by silencing thought the interaction with the heterochromatin silencing machinery. This results in H3K4me2 removal and H3K9me2/3 deposition, and this is followed by HP1a and histone H1 covering the target locus to maintain the repressed status [
124]. However, many of the molecular mechanisms of this process remain unknown and require investigation. Moreover, piwi-guided transcriptional silencing induces promoter DNA methylation changes in the mammalian germline [
125]. The piRNA-PIWI complex can activate or repress translation in early mouse spermatogenesis and affect mRNA degradation in a microRNA-like manner at later stages [
126]. The complex of piRNAs, Hsp90 and Hop protein are also involved in “canalisation” which ensures robust development [
127] and although piRNAs activity is mainly associated with germline cells, it also has the following functions in somatic cells; (1) piRNAs influence
CREB2 promoter methylation in nematodes and this affects long term memory formation [
128]; (2) involvement in dendritic spine development in the mouse hippocampus [
129]; (3) mediation of Nanos mRNA de-adenylation and degradation. The correct Nanos level is essential to form the Drosophila embryo anterior-posterior axis [
130]. In addition, it is presumed that piRNA function in somatic cells is ancestral and lost in some species [
120,
121].
piRNAs are transcribed from ‘piRNA clusters’ which are usually over 100 kb long and consist mostly of transposable elements, but also contain different repetitive regions accumulated during evolution [
120,
121]. A smaller number of piRNAs are transcribed from inter-genic non-coding regions and protein-coding 3′-UTR gene regions [
122,
123], and some studies suggest they can be transcribed from tRNA and snoRNA sequences [
122,
123]. The piRNA clusters can be uni-strand and dual-strand. Uni-strand clusters produce piRNA precursors from only one genomic DNA strand. They harbour H3K4me2 marks, and the piRNA precursors transcribed from uni-strand clusters are both 5′-capped and 3′-polyadenylated. In contrast, the dual-strand clusters produce piRNA precursors mapping to both genomic strands. They do not have similar marking, and their transcribed precursors are only 5′capped [
122,
123]. The piRNA precursors are transferred from the nucleus after transcription and undergo primary biogenesis. Although the precursors are predominantly anti-sense (5′-3′) to the target TE sequence, this is not an absolute rule. For example, the mouse piRNA precursors have sense polarity (3′-5′).
These precursors are then shortened and bind to specific PIWIL1/HIWI, PIWIL2/HILI, PIWIL3 and PIWIL4/HIWI2 Piwi proteins or Aubergine (Aub). Here, the HIWI indicates human protein, in order to distinguish it from non-human protein. Multi-step processing then occurs to give the final PIWI/AUB-piRNA complex, followed by methylation of the piRNA 3′ end by HEN1 methyltransferase. Finally, the entire complex enters the nucleus for transcriptional silencing or, alternatively, it is involved in secondary biogenesis (ping-pong cycle) [
122,
123,
131,
132] (
Figure 3).
A summary of the entire process is as follows; an initial pool of piRNAs is created in primary biogenesis and this targets multiple TEs. This is followed by amplification of RNA sequences that target active transposons in secondary biogenesis and these then repress transposons via slicer-dependent PTGS (
Figure 3).
The Aub-piwi complex from primary biogenesis recognises and cleaves the cognate transcript of transposon mRNAs with opposite orientation, and the cleaved product is then converted into a new sense piRNA associated with Ago3.
Ago3 with incorporated piRNA is further modified and subsequently recognises and cleaves the cluster transcripts. The products of this slicing then re-initiate the cycle. This process ensures amplification of particular piwiRNAs and the degradation of both sense and anti-sense transposon transcripts [
122,
123]. Although the finer details of piRNA biogenesis have not been elucidated and most of our understanding comes from
Drosophila germline cells, it is anticipated that the mechanisms in human cells are the same, or very similar, to those documented in drosophila [
122,
123].
Martinez et al.’s comprehensive study [
123] highlights that although only approximately 1% of piRNAs is expressed in healthy somatic cells compared to germline cells, almost two-fold piRNAs over-expression occurs in cancer [
123]. Their expression level there differentiates between healthy and tumor tissue [
123], and level changes are associated with cancer cell migration and invasion [
133,
134]. It is therefore presumed that the normal physiological mechanisms of piRNA-induced target regulation in germlines become abnormal in cancer cells and contribute to their formation and development. This includes physiological abnormality in DNA methylation, histone modificiation, translation initiation and inhibition and mRNA degradation. The connection between abnormal piRNA activity and the development of various cancers has previously been documented [
122,
135,
136].
4.1. Piwi-Interacting RNAs and BC
The presence of piRNAs and their altered function in tumour cells was first demonstrated in 2010 in the well-known HeLa cells [
137]. The piRNA effect on tumorigenesis by binding to the target mRNA or altering epigenetic status was then demonstrated in several tumours, especially in lung cancer [
138,
139] and colon cancer [
140].
The initial association between abnormal piRNAs activity and BC tumorigenesis was demonstrated in 2011 in incidental research by Cheng et al. [
141]. Although their work focused on the abnormal activity of piRNA-651 in gastric cancer tissue, the authors then recognised that this piRNA is also up-regulated in FFPE samples from BC. The piRNA-651 up-regulation in tumorigenesis has since been repeatedly demonstrated in vitro, and it especially contributes to increased proliferation and migration and inhibited apoptosis [
142,
143,
144]. The piRNA-651 is also considered to up-regulate cyclin D1 and CDK, but the precise mechanisms require elucidation [
143].
Huang et al. [
145] produced the first study focused exclusively on multiple piRNAs presence in BC tissues. These researchers performed deep sequencing of 4 tumour samples and five controls, and results were subsequently verified by qPCR analysis of 50 samples. Deep sequencing revealed piRNA-4897, piRNA-19825, piRNA-20365, piRNA-20485, and piRNA-20582 were up-regulated, and that piRNA-17485 was down-regulated. Similar results were demonstrated by qPCR except for piRNA-19825. Finally, the piRNA-4897 abnormal expression there was associated with lymph-node positivity.
The piRNA expression in MCF-7, SKBR3 and ZR-75.1 BC cell lines is affected by cell cycle phase and oestrogen receptor activity [
146]. Research results documented that 39 piRNAs were differentially expressed in exponentially growing cells compared to senescence cells, and 25 piRNAs had different expression-levels in ERβ+ and ERβ- cells [
146]. The authors subsequently performed RNA sequencing analyses on four paired BC tissues. The abnormal expression of almost 150 piRNAs was revealed under less strict
p < 0.05 statistical conditions, and subsequent
p < 0.01 statistical analysis determined that the eight PiRNA-34736, PiRNA-36249, PiRNA-35407, PiRNA-34377, PiRNA-36318, PiRNA-36026, PiRNA-31106 and PiRNA-36743 piRNAs have statistically significantly higher activity. This study also determined that active piRNAs in somatic cells made up only 1% of those active in germline cells [
146].
4.1.1. piRNA Effects on Expression of Other Genes
There was significantly higher piRNA-932 activity in cells with high metastatic potential, and especially in those with induced EMT [
147]. The authors consider that piRNA-932 forms immune complexes through precipitation with piwil2, and that the complexes will subsequently affect latexin gene expression by hyper-methylation of its promoter region. Latexin is a tumour-suppressor and negative stem cell regulator which induces removal of old stem cells and prevents their eventual transformation into tumour cells [
147].
Further piRNA effects indicate that piRNA-021285 significantly influences
ARHGAP11A gene promoter methylation level [
148]. This gene encodes the Rho GTPase-activating protein which is considered oncogenic [
148], and the study demonstrated that MCF-7 cell lines mimic-transfected with piRNA-021285 mutational variant have significantly lower
ARHGAP11A gene promoter region methylation levels compared to piRNA-021285 wild type mimic-transfected MCF7 cell. This was subsequently connected with higher expression of this gene and increased cell invasion. Importantly, the SNV rs1326306 G>T in piR-021285 was identified in a Connecticut-US sub-population and was strongly associated with increased likelihood of BC [
148]. Finally, the
ARHGAP11A gene was abnormally expressed in basal-like BC and it stimulated BC development in in vitro studies [
149].
piRNA-36712 also has important functions in BC development [
150]. In contrast to piRNAs with higher BC tissue expression, this piRNA’s activity was down-regulated in resected cancerous tissues and in MCF-7 and ZR75-1 cancer cell lines. The piRNA-36712 tumour-suppressive effect is therefore expected, and its expression rate in BC patients correlates with outcome. Those with lower expression levels have lower progression-free survival rates. This piRNA’s effect is related to its interaction with
SEPWP1 gene mRNA whose levels were increased following piRNA-36712 knockout.
SEPW1 is an important cell cycle regulator and its depletion increases p53 and p21 activity by suppressing their degradation by ubiquitination and this leads to G1 cell-cycle arrest [
151,
152]. Moreover, authors found that up-regulation of p53 reduced Slug and increased E-cadherin expression. This is supported by research that p53 can suppress cancer invasion by inducing MDM2-mediated Slug degradation. Slug is a transcription repressor that regulates E-cadherin expression [
153,
154].
In addition piR-sno 75, which is a snoRNA-derived piRNA has quite complex function in balancing BC epigenetic status [
155]. piR-sno 75 has significantly lower expression in BC and experimental over-expression leads to TRAIL up-regulation. Here, piR-sno75 was shown to guide the recruitment of MLL3/COMPASS-like complex to the TRAIL promoter and this induced H3K4 methylation and H3K27 de-methylation and led to increased
TRAIL expression and subsequent apoptosis induction.
Table 2 summarises the abnormally expressed piRNAs and piwi proteins in BC.
4.1.2. Abnormalities in PIWI Proteins in BC
The increased expression of Piwi proteins required for general piRNA function has also been reported in BC tissues. Piwil2 and Piwil4 are highly expressed in SKBR3 cell lines [
146], and altered Piwil2 expression was demonstrated in 30.8% CD44+/CD24- cells cultured from resected tumour tissues. These cells form a fraction of the stem cells typically present in BC tumorous masses [
156]. The Piwil2 expression was also significantly higher in BC than in para-cancerous tissues and hyperplasias, and its protein expression rate was associated with age, histological type, tumour stage and size, and lymph node metastasis. Moreover, metastasis occurred in over 50% of patients with abnormally expressed Piwil2. This was evident even after operative removal of the primary tumour, and this strongly contrasts with the metastasis noted in only 13% of patients with lower expressed Piwil2 [
147].
5. Small Nucleolar RNAs
snoRNAs are short non-coding RNAs from 60 to 300 bp long, and most are encoded in intronic or other non-coding regions of genes encoding proteins involved in ribosome synthesis. The final snoRNA transcription is also intimately associated with host-gene expression [
157]. The snoRNA processing commences in the nucleus. It continues in the cytoplasm, and it is necessary to incorporate them into the snorP protein complex. This process improves their stability so that they can be incorporated in the nucleus, usually in cajal bodies. Cajal bodies provide the major sites for splicing and final modification [
158].
The two major snoRNA groups are C/D box and H/ACA box [
157,
159]. Their major function is ribosomal RNA modification and processing, with the following functional division; the C/D BOX snoRNAs act primarily as sequence-specific guides which direct rRNA modification by 2′-O-ribose methylation and the H/ACA group are especially involved in pseudo-uridylation of selected regions essential for ribosomal function [
157,
159]. However, further research established a broad spectrum of snoRNA functions; snoRNAs influence the modification of various cellular RNAs such as snRNAs and miRNAs [
157], by affecting pre-RNA cleavage [
9,
160]. They are also considered to directly affect mRNA splicing and influence alternative splicing in serotonin receptor subtype 2C [
161], and
DPM2,
TAF1,
RALGPS1,
PBRM1, and
CRHR1 pre-mRNAs [
162].
Moreover, the expression of more than 200 genes in HEK 293T cells is altered in vitro by SNORD15 and SNORD16 RNA over-expression, and researchers presume direct association of these events [
163]. The snoRNAs are also expected to regulate E2F7 transcriptional factor splicing [
163], and abnormal snoRNA activity has been observed under oxidative stress from palmitate and hydrogen peroxide treatment [
164]. The snoRNA U60 is involved in plasma-membrane cholesterol trafficking and reduces cholesterol synthesis [
165]. Jinn et al. reported that snoRNA U17 influence on the hypoxia up-regulated mitochondrial movement regulator mRNA (HUMMR) also leads to modification of cholesterol synthesis and trafficking [
166]. Finally, there is a reportedly important snoRNA involvement in the phosphoinositide 3-kinase (PI3K)/ACT, p53, and Wnt/β-catenin cell-signalling pathways [
157].
In summary, although snoRNAs have diverse cell functions and mechanisms, these are not adequately elucidated. For example, there are also ‘orphan snoRNAs’ without rRNA modification function, which possess no sequence complementary to any known rRNAs and have no involvement in ribosomal biogenesis [
167]. Elucidation of the precise functions of these orphan snoRNAs will add to existing knowledge of snoRNA mechanisms in healthy and tumorous cells.
5.1. SnoRNAs and BC
Increased ribosome biogenesis and nucleolar activity are associated with cancer development [
168,
169] and enlarged nucleoli are markers of tumour aggression [
170]. However, the impact of snoRNA abnormalities in tumourigenesis was largely overlooked until recent research focused on their effects in this development.
5.1.1. SnoRNAs Affect p53 Response
Su et al. [
170] revealed the strong association between abnormally high expression of several snoRNAs and BC development in animal models and human tumours. The U15a, U15b, U22, MBI-43 and U87 SnoRNAs were abnormally expressed in animal models and HBII, U22, U3, U8, U15b, U94 and U97 in human tissues. These authors also reported the in vitro association of abnormal snoRNAs, p53, and fibrillarin (FBL) activity. Fibrillarin is an important enzymatic component which affects snoRNAs accumulation and higher snoRNA and FBL expressions correlate in direct proportion [
171]. Fibrillarin mRNA and protein levels were up-regulated in 60% of human BC tumour tissues and its inhibition decreased the frequency of tumour formation and volume in mouse models. There was also reciprocal FBL interaction in MCF-7 cell lines [
172], where FBL deletion increased p53 activity and its over-expression reduced p53 response. The experimental suppression of snoRNAs biogenesis also induced p53 dependent cycle arrest, and the authors therefore considered that p53 responds to snoRNA pathway inhibition. A summary of these multiple interaction indicates ‘vast cross-talk’ between p53, FBL and snoRNAs and modulation of this entire interaction system can be beneficial in improving BC treatment
Langhendries et al. demonstrated abnormally expressed C/D box snoRNAs U3 and U8 in BC cells [
173]. Depletion of both these snoRNAs in MCF-7 cells resulted in a strong anti-tumour p53 stress response which led to higher p53 stability, cell cycle arrest and apoptosis. This was observed in MCF-7 BC cell lines and H1944 lung cancer cell lines. The tumourigenic potential of H1944 cells in mouse xenografts was significantly reduced after U3 suppression, and completely by U8 suppression. However, this experiment was not performed with BC cell lines, and outcomes are therefore doubtful, so follow-up analysis on BC cell lines would be efficacious.
5.1.2. Association between Abnormal snoRNA and miRNA Expression
The RNU44, RNU48, RNU43 and RNU6B snoRNAs were variably expressed in various BC cancer types in a cohort of 219 patient [
174]. This was associated with expression of miR-21, miR-210, and miR-10b, and this is important because miR-21 is a significant promoter of BC progression, proliferation and metastasis [
175]. Altered snoRNAs expression was also associated with greater disease aggression. Moreover, RNU44 snoRNA is transcribed from the
GAS5 gene intronic region. This gene regulates cell cycle arrest in stress conditions and low RNU44 levels therefore mark poor prognosis. In addition, the RNU48 and RNU43 snoRNAs are transcribed from the RPL3 and C6orf48 cancer-associated genes [
175].
5.1.3. The Effect in BC of Micro-RNA-Like Fragments Arising from snoRNAs
Some snoRNAs can proceed to short and stable micro-RNA-like fragments. These are synonymously named sno-miRNA and sdRNA (‘small nucleolar RNAs-derived microRNAs’). sdRNA-93 is markedly over-expressed in MCF-7 and MDA-MB-231 BC cell lines and while sdRNA93 induces cell invasion, its inhibition prevents it [
176]. Although the authors reported increased sdRNA-93 expression in Luminal B Her2+ tumour tissue, its expression was only minimally increased in healthy tissues and BC subtypes other than luminal B Her2+. In addition, sdRNA93 is involved in regulation of the
Pipox gene whose protein product regulates sarcosine metabolism and alterations in this protein product are associated with BC metastasis formation [
177].
Abnormal sno-miR-28 expression has been detected in BC [
172] and this targets
TAF9B which stabilises p53 in physiological conditions. A brief explanation of the processes involved here is that the, the interaction between p53, NHG1, sno-miR-28, and TAF9B results in a signalling cascade, which significantly affects p53 and modifies its down-stream gene network.
Table 3 below lists the snoRNA actions.
6. Small Nuclear RNAs
snRNA are small non-coding RNAs in the Cajal bodies and splicing speckles in the nucleus [
178,
179]. The snRNAs are approximately 150 nt, and have the primary function of pre-mRNA post-transcriptional modification [
178]. The following two main classes differ in common sequences and interacting proteins.
(1) the Sm-class comprise U1, U2, U4, U4atac, U5, U7, U11, and U12 snRNAs transcribed by RNA polymerase II and their processing involves transfer to the cytoplasm and return to the nucleus.
(2) the Lsm-class contains only U6 and U6atac. These are transcribed by polymerase III and remains in the cell nucleus [
180,
181].
The snRNAs unite with an approximately 150 proteins to form a major or minor spliceosome complex dependent on the snRNAs involved [
182,
183]; a major spliceosome complex which removes 99.5% of introns [
178] is formed with U1, U2, U4, U5, and U6 snRNAs, and a minor complex with U4atac, U5, U6atac, U11, and U12. It is clear here that only snRNA U5 is present in both complexes [
181]. Spliceosomal abnormality is evident in the following pathologies; the congenital abnormalities that occur in dwarfism [
181] and neurodegenerative diseases such as Alzheimer [
184] Somatic spliceosome abnormalities are typical in the snRNA component mutations responsible for pre-cancerous and cancerous lesions [
180,
181]. An example of this is highlighted in in vitro experiments which reveal that snRNA U1 over-expression causes altered expression levels in over 900 genes, and 73 of these are directly involved in 12 important cell signalling pathways directly associated with various cancer development [
184].
SnRNAs and BC
Although aberrant mRNA splicing occurs in many cancers [
185], oncology research usually addresses only altered function and activity of the “regulatory” factors which control splicing, and there has been minimal analysis of snRNAs as “basal factors” required for catalysing the splicing process. However, recent research by Dvinge et al. [
186] content that varied snRNA abundance in different BC types is not random and most BC samples have subtype-specific snRNA expression patterns.
These researchers also considered that TNBC could be divided into two subtypes based solely on snRNA levels, and that splicing changes predominantly affected only one exon after snRNA knock-down in MCF-7 cells. They subsequently performed deep sequencing of 136 invasive ductal carcinomas to compare naturally occurring splicing anomalies with those associated with snRNA knock-down in MCF-7 cells. The results confirmed that splicing anomalies accorded with snRNA expression, and that the observed abnormal splicing was mostly snRNA related. There was also confirmed relative and absolute snRNA expression variability in a wide range of biological conditions and in healthyand tumorous tissues.
The U1 snRNA exerts significant impact on migration and invasion in BC cell lines [
187]. While shortening of the mRNA 3-UTR region occurs from altered proximal polyadenylation signals (PAS) in introns and the last exon [
188,
189,
190], PAS activity can be silenced by snRNP U1 [
191]. These shorter mRNA isoforms are typical in immune cells, neurons, and some cultivated cell lines, and they are also present in various cancers including BC [
188,
189,
190]. Research has highlighted that U1 inhibition results in premature transcription termination and mRNA shortening in BC [
187], but U1 over-expression not only negates these effects but significantly decreases cell line ability to migrate and spread. This U1 snRNP therefore presents a suitable target for inhibiting BC spread.
Although snRNAs are typically situated in the nucleus, they can be detected by liquid biopsy and potentially used for early non-invasive cancer detection [
192]. For example, higher snRNA U6 levels have been found in BC patient plasma samples [
193]. This is typical in both active and inactive disease and in ER+ and ER- BC subtypes, but not in healthy women. The permanently elevated U6 levels also indicate increased polymerase III activity in BC, regardless of disease progression [
193].
Table 4 lists snRNA actions in BC.
7. Small-Interfering and Short Hairpin RNAs
The double-stranded small interfering RNAs (siRNAs) with 20–25 nt were first discovered in
Caenorhabditis elegans [
194,
195] and then in plants and invertebrates [
196]. The siRNAs’ function in plant, fungal, unicellular and invertebrate cells is regulation of post transcriptional expression and protection against viral elements [
197]. The siRNAs in these taxa can also originate as exogenous transcripts derived from viral elements [
194]. It remains debatable whether siRNAs can be naturally present in higher eukaryotes, but some studies also admit this possibility [
198]. However, the presence of dsRNAs in higher eukaryotes cells leads to interferon response [
199].
Artificially prepared siRNAs are often used studies in the expectation of modulating individual gene activity. This would be therapeutic in cancer and in diseases where conventional treatment is insufficiently effective [
194,
200]. However siRNAs cannot passively diffuse through cellular membranes [
201,
202,
203] and their application is therefore limited by delivery to the cell target area. This remains a challenge because siRNAs are rapidly degraded by ubiquitous endo- and exonucleases, and administrative, vascular, cellular and immune barriers must be overcome for effective siRNA delivery. Additional requirements are that siRNAs must be chemically modified for increased stability and their length must be limited to 30 nt [
204].
Finally, the siRNA delivery system can be viral or non-viral [
201], where the non-viral delivery systems are predominantly lipid-based, polymer-based or utilise nucleotide-derived nanoparticles [
194,
201,
202,
203,
204].
Following delivery of artificially prepared siRNA to the cytoplasm, the siRNAs are either loaded directly into the RISC complex or processed by the Dicer complex before loading. The strands are subsequently separated, and the RISC complex is directed to a specific mRNA depending on the guide-strand. Ago-2 then cleaves the target mRNA between bases 10 and 11 relative to the 5′ end of the siRNA antisense strand and mRNA is degraded. There can also be alternative action by different Ago proteins which do not have catalytic activity, and this leads to translation repression through mRNA sequestration in processing bodies (P-bodies) [
194,
205] (
Figure 4).
In addition, intact siRNAs have been reported present in the nucleus of transfected cells, and also to remain there for at least 15 min [
206]. Moreover, while the Ago/RISC complex loaded with siRNA shuffled between nucleus and cytoplasm [
207], the mechanism for this process has not been elucidated. Recent research indicates that Ago2 loaded with small RNA and localised in the nucleus can regulate both miRNA and lncRNA transcription levels [
208].
The 20–25 nt short hairpin RNAs (shRNA) are artificially prepared, and these can also be used to alter gene expression. However, in contrast to siRNAs which must be delivered to the cytoplasm, shRNAs must be delivered by appropriate plasmid or viral vector to the nucleus for transcription, and the primary transcript (pri-shRNA) is then processed by the Drosha complex. Precursor shRNAs (pre-shRNAs) are formed as a result of this processing, and these are incorporated in the Dicer complex after Exportin-5-mediated transport from the nucleus. The shRNA hairpin loop is excised by the Dicer complex and this then forms an active double-stranded siRNA with 2nt 3′ overhangs which is subsequently incorporated in the Ago/RISC complex. The action mechanisms which then lead to mRNA cleavage or translation repression are then the same as in siRNAs [
209] (
Figure 5).
7.1. SiRNAs and BC
7.1.1. siRNA Based Inhibition of BC Associated Pathways
Researchers have endeavoured to decrease BC growth using siRNAs which inhibit abnormally active and typically dysregulated signalling pathways such as NF-κB [
210]. The application of micelleplex containing siRNA-p65 and cisplatin-prodrug inhibited this pathway in 4T1 BC murine cells [
211]. P65 is an essential protein, involved in NF-κB heterodimer formation, nuclear translocation and activation [
212]. The application of micelleplexes containing siRNA-p65 was first successfully applied in cell lines and has subsequently proven efficacious in in vivo T4 mice xenograft tumours. This application inhibited tumour growth-rate compared to controls. In addition, the siRNA-p65 mediated silencing of p65 protein decreased in 4T1 cell lines
MMP9 expression by 40% and cyclin D1 expression by 70%.
Frequent abnormalities in the Wnt/β-catenine/EMT pathway were also noted in BC tissues [
213]. The DANCR lncRNA is known to have significant impact on this pathway, and Vaidya et al. therefore examined siRNA DANCR inhibition [
214]. Their application of RGD-PEG-ECO/siDANCR nanoparticles produced up to 90% DANCR knockdown in MDA-MB-231 and BT549 cancer lines within seven days. At the molecular level, decreased PRC2-mediated H3K27-trimethylation and altered phosphorylation profiles of several kinases occurred after DANCR inhibition. In addition, application of the above nanoparticles in TNBC mouse xenografts suppressed tumours without side effects.
Abnormalities in the VEGF pathway cause rapid angiogenesis, and this can hasten cancer development, including BC [
215]. VEGF pathway abnormality in BC is also connected with increased lymphangiogenesis, which can result in aggressive spread to the lymph nodes and even metastases in the lungs [
216]. Feng et al. demonstrated that vapreotide-modified core-shell type nanoparticles which co-encapsulated VEGF targeted siRNA and paclitaxel can be effectively transported into BC cells via samostatin receptors. This resulted in siRNA interference which reduced VEGF activity in MCF-7 cell lines [
217], and the nanoparticle delivery to mouse xenografts resulted in significantly decreased tumour vascularity and growth.
7.1.2. siRNA-Based Inhibition of Cell Cycle Regulators and Transcriptional Factors in BC
Polo-like kinase 1 (PLK1) is a serine-threonine kinase, a trigger of G2/M transition involved in centrosome maturation [
218].
PLK1 is considered a proto-oncogene which induces centrosome abnormalities and cell-cycle progression defects [
218]. Further, PLK1 phosphorylates BRCA1 thus impairing its involvement in homologous recombination [
219], and reported
PLK1 expression is higher in TNBC than in both healthy cells and benign tumours [
220]. Moreover, PLK1 inhibition by BI-2536 siRNA leads to G2/M cell-cycle arrest and apoptosis initiation in the MDA-MB-231, Hs578T, MDA-MB-436, MDA-MB- 468 and HCC1937 BC cell lines [
220].
The PLK-inhibiting siRNA-siPLK1 delivered to TNBC cell lines in meso-porous-silica nanoparticles reduced all cell line viability [
221] and decreased
PLK1 mRNA levels by 69 to 87% and protein levels by 64–91%, depending on cell lines. In addition, meso-porous-nanoparticles lacking incorporated siRNA should have ROS scavenging ability, and empty particle application consequently resulted in reduced NOX4 expression and an overall decrease in cellular ROS. Finally, BC metastasis to lungs has been reported in animal models injected with LM2-4lucþ/H2N cells, and the application of the meso-porous-silica nanoparticles with siPLK1 resulted in 80% mRNA knockout and approximately 90% decrease in tumour size [
221].
Nachreiner et al. [
222]. investigated siRNA-inhibition of
PLK1 and the
EEF2,
GRK4 and
SKIP5 genes. Here, siRNAs were applied to MCF-7 cells using the highly specific HER3 aptamers A30. These characteristically bind to the human epidermal growth factor receptor-3 extracellular domain, and this domain is routinely over-expressed on the BC cell surfaces [
222]. This siRNA application reduced cell viability and also the expression of all analysed genes at the mRNA level. In contrast, there was no loss of cell viability in controls.
Further research showed that RNase-resistant nanoparticles suppress
XBP1 gene activity in mouse HER2+ BC [
223]. Silencing this gene by 3WJ-HER2apt-siXBP1 nanoparticles inhibited cell proliferation in mouse xenografts, significantly suppressed BC growth and promoted chemotherapeutic sensitivity. It is most important that these nanoparticles bind strongly to tumour tissues, but not to healthy tissues.
7.1.3. siRNA Based Inhibition of BC Metastases
Metastasis formation presents a problem for RNA interference and BC metastases often migrate to the lungs [
224]. The exosome-coated biomimetic nanoparticles can target these metastases in animal models, and this is possible because the exosome surfaces have specific integrins binding to α6β4 and α6β1 laminin receptors on the lung surface, and the organism does not recognise these as foreign elements [
224]. In particular, exosome membrane coated bio-mimetic nanoparticles have been used to inhibit
S100A4 gene. These comprise a cationine bovine serum albumin, a siS100A4 core and an exosome membrane shell. Abnormal S100A4 expression is positively associated with lung cancer tumour progression [
225] and importantly its function demonstrably contributes to BC metastases migration to the lungs [
226]. The nanoparticles were intravenously injected into BALB/c mice with established post-operative BC lung metastases on a daily basis for four days. This effected significant decrease in S100A4 at the protein level, accompanied by significant tumour nodule decrease after the 30-day study. It was thus confirmed that the exosome nanoparticles had great lung-targeting ability.
7.1.4. Possibility of siRNA Use as a Potential Method of Improving Adoptive Transfer Therapy
Tumour-infiltrating lymphocytes (TIL) migrate from the bloodstream around and inside tumorous tissues. They are tumour-antigen specific and have a relatively high ability for solid-tumour destruction [
227,
228]. TIL have become important in the last twenty years in ‘adoptive transfer therapy’ where T-cells are removed from resected tissue, cultivated and re-infused in patients [
229]. Their action in tumour tissues can be inhibited by immunosuppressive molecules such as PD-L1 which inhibits TIL anti-tumour activity by binding to the PD-1 receptor. The PD-1 and PD-L1 expression is rapidly elevated in later BC stages and this is associated with poor prognosis [
230]. Wu et al. [
231] used lipid-coated calcium phosphate nanoparticles in their inhibition of PD-1 and PD-L1. This enabled efficient siRNA entry into MCF-7 BC cell lines and subsequent inhibition of both the PD 1 receptor and ligands. The authors further reported that the elevated TIL cancer cytotoxicity was connected with increased secretion of pro-inflammatory cytokines such as IFN-γ and TNF-α.
Table 5 summarises the RNA interferences performed in BC.