Osmolality Effects on CHO Cell Growth, Cell Volume, Antibody Productivity and Glycosylation
Abstract
:1. Introduction
2. Results and Discussion
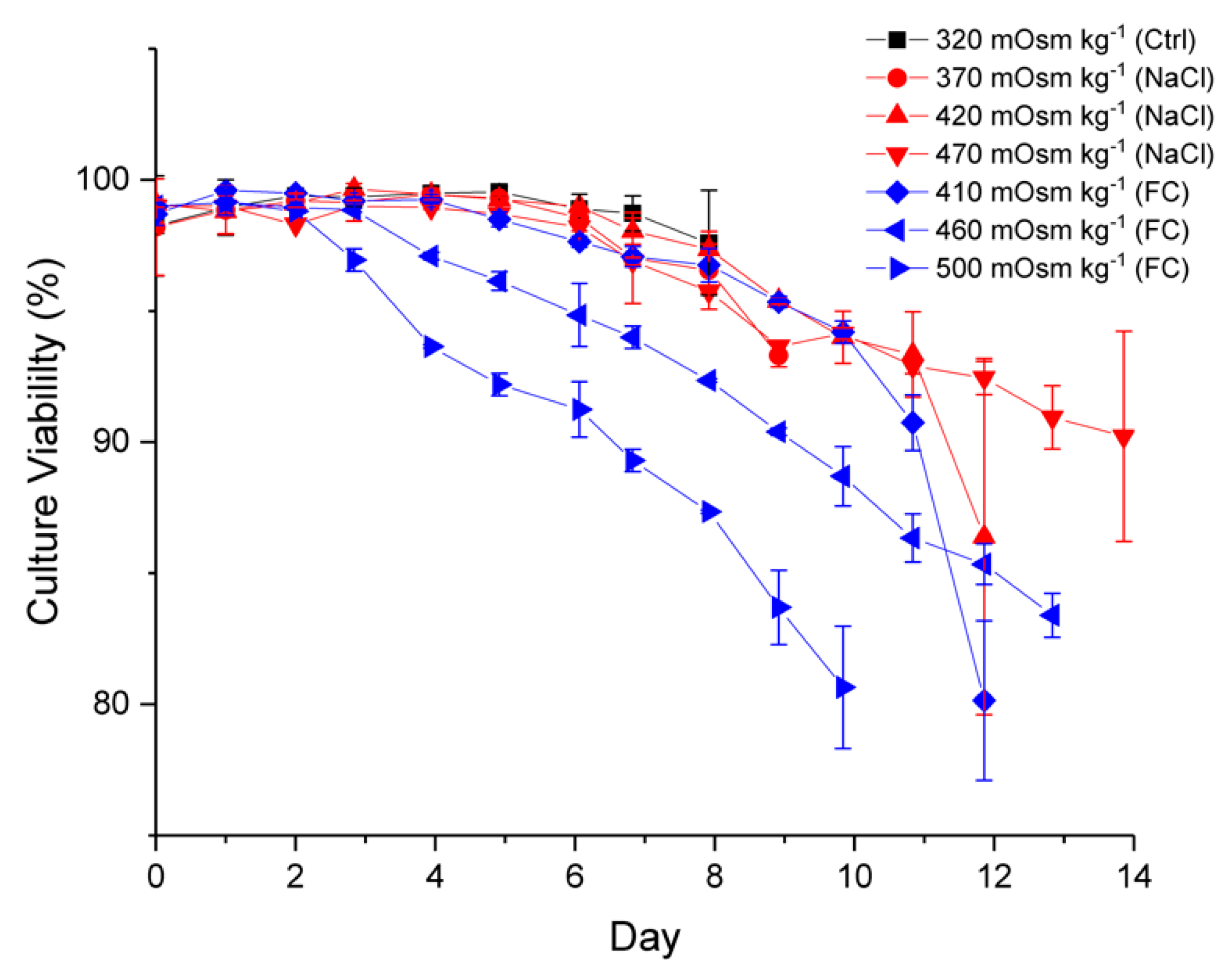
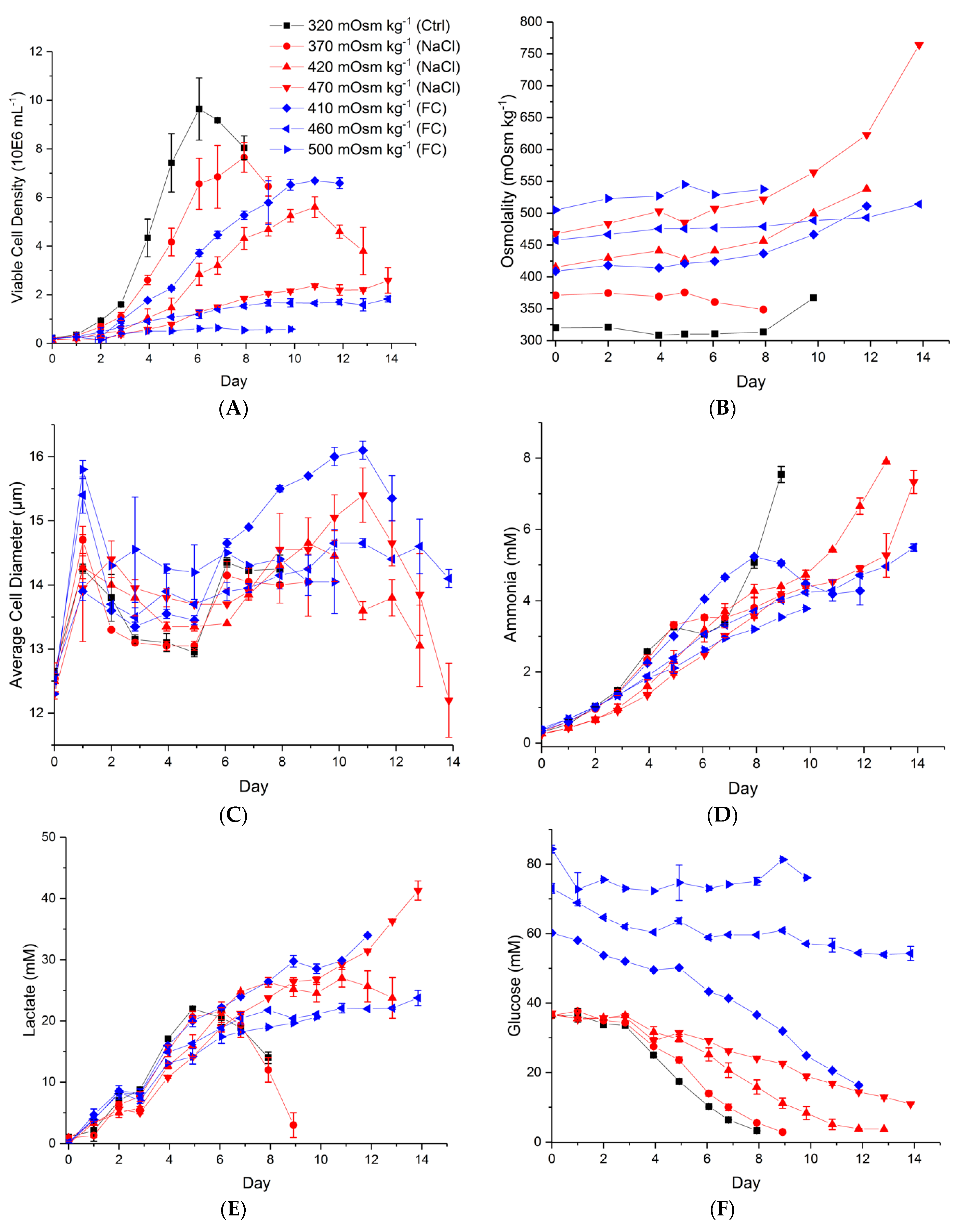
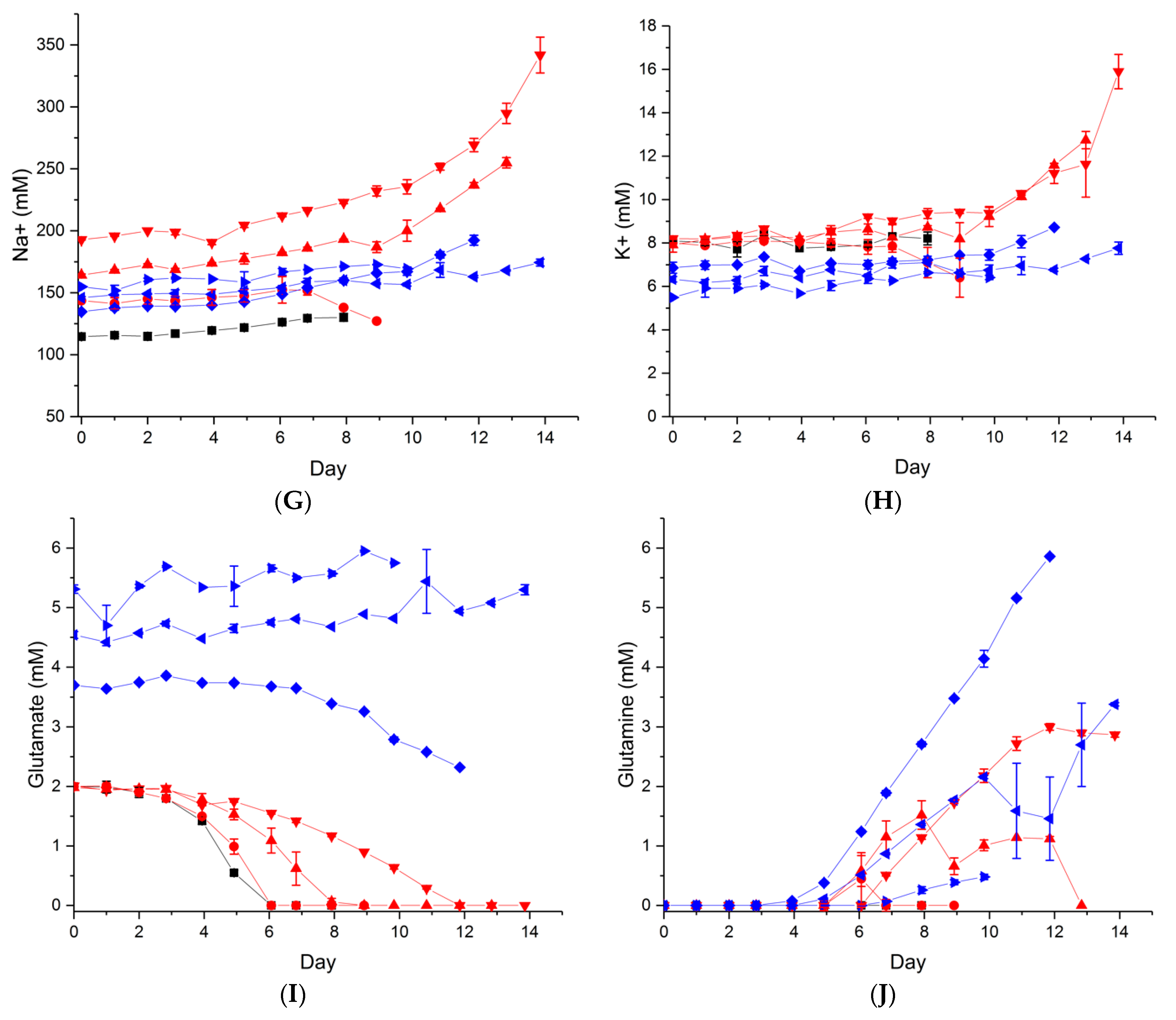
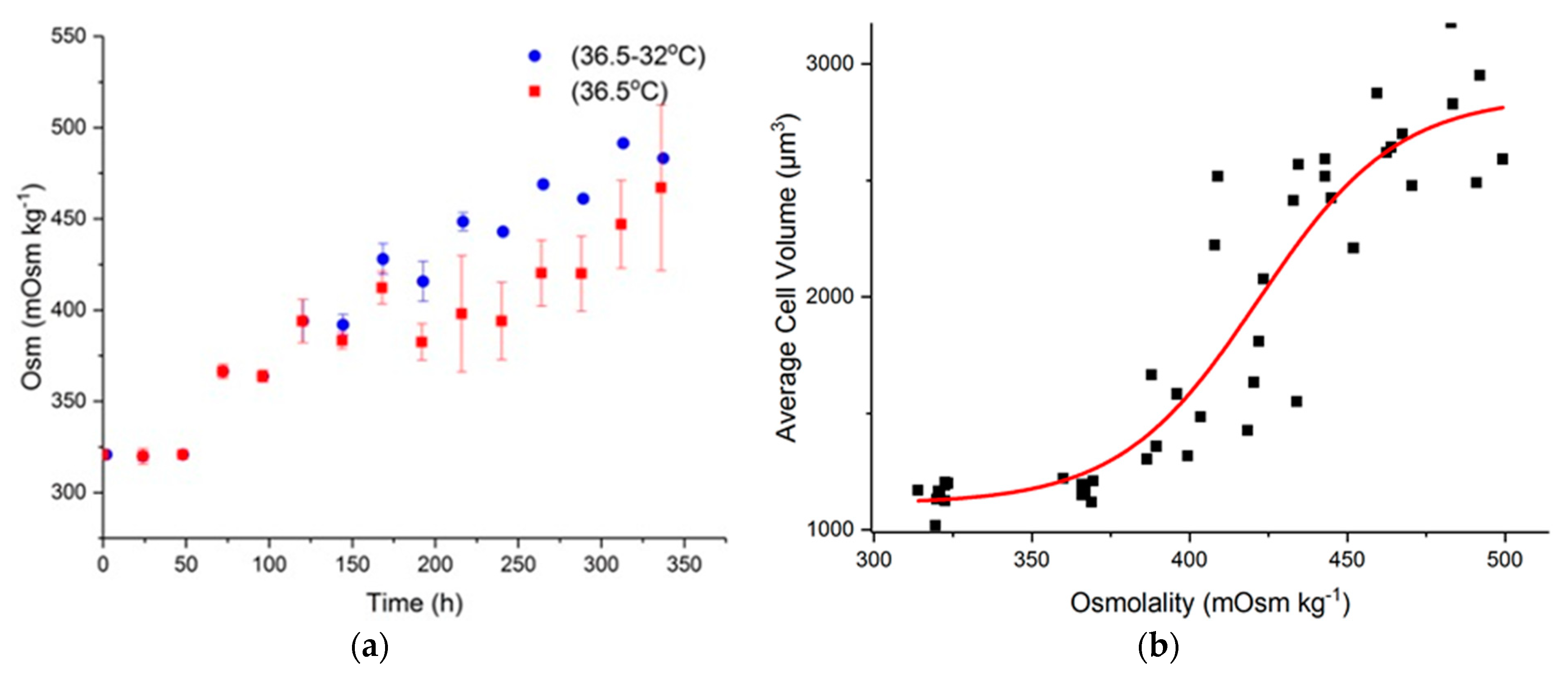
2.1. Effect of Hyperosmolality on Cell Growth and Metabolism
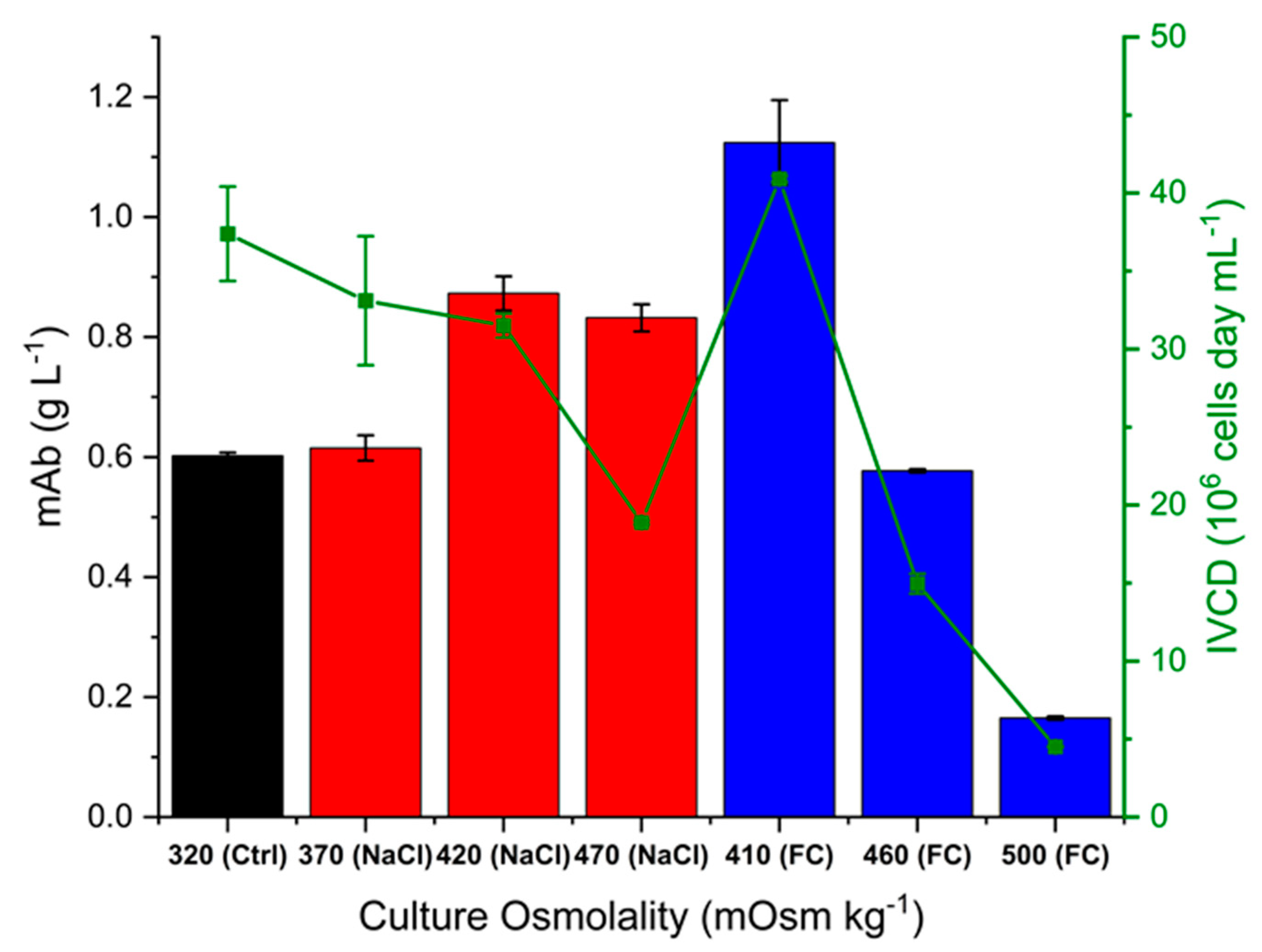
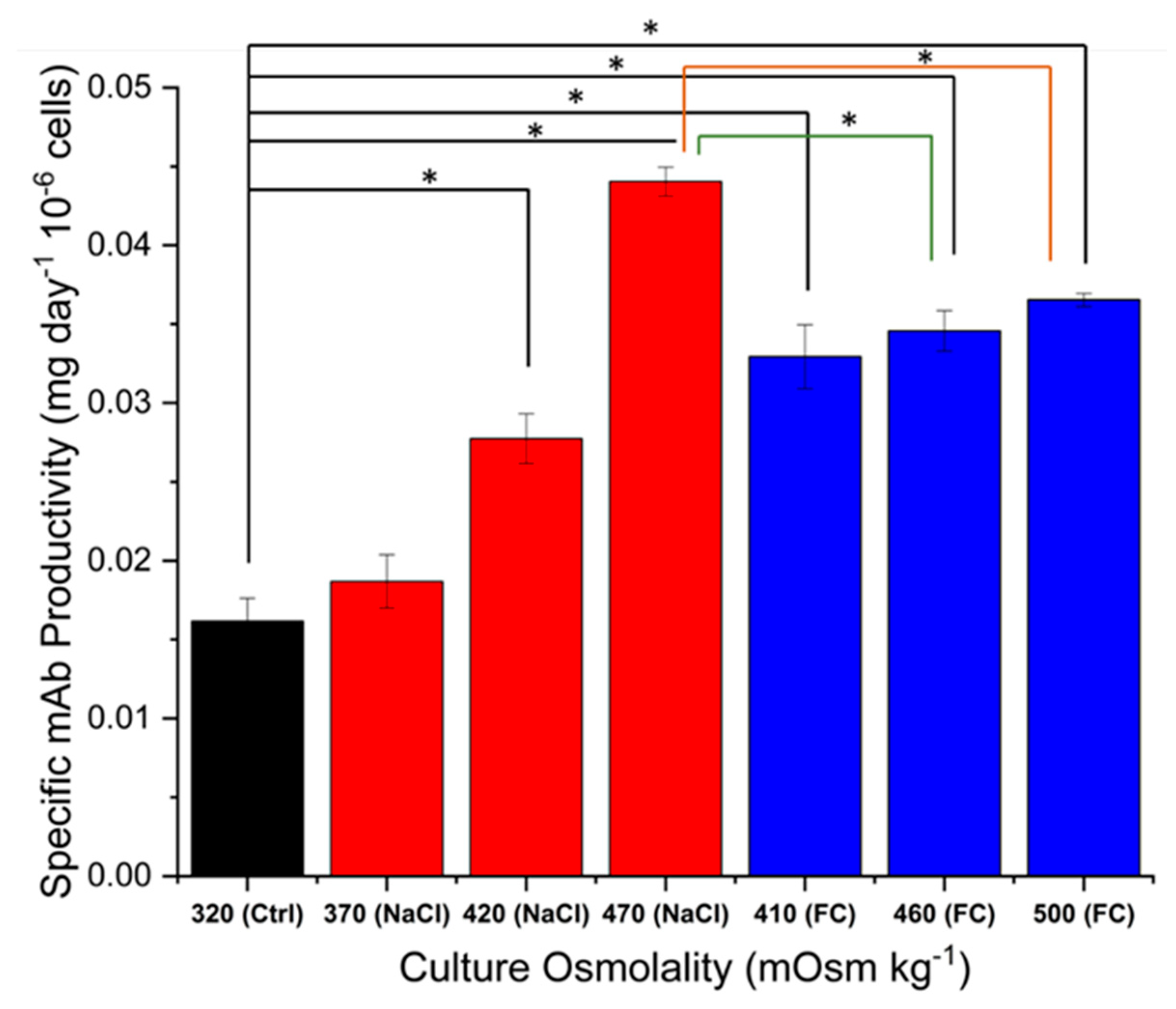
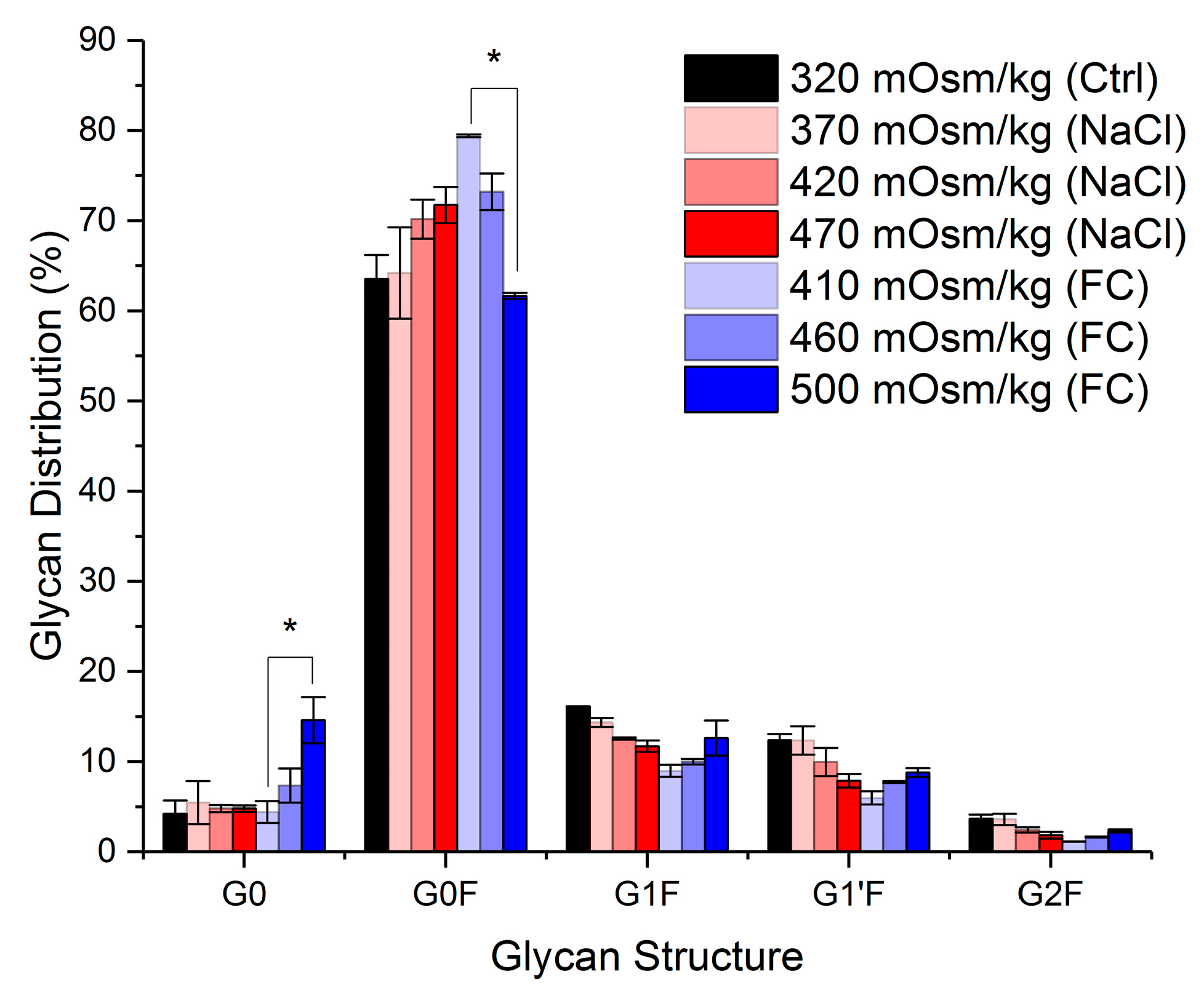
2.2. Effect of Hyperosmolality on Titre and Specific Antibody Productivity and Glycosylation
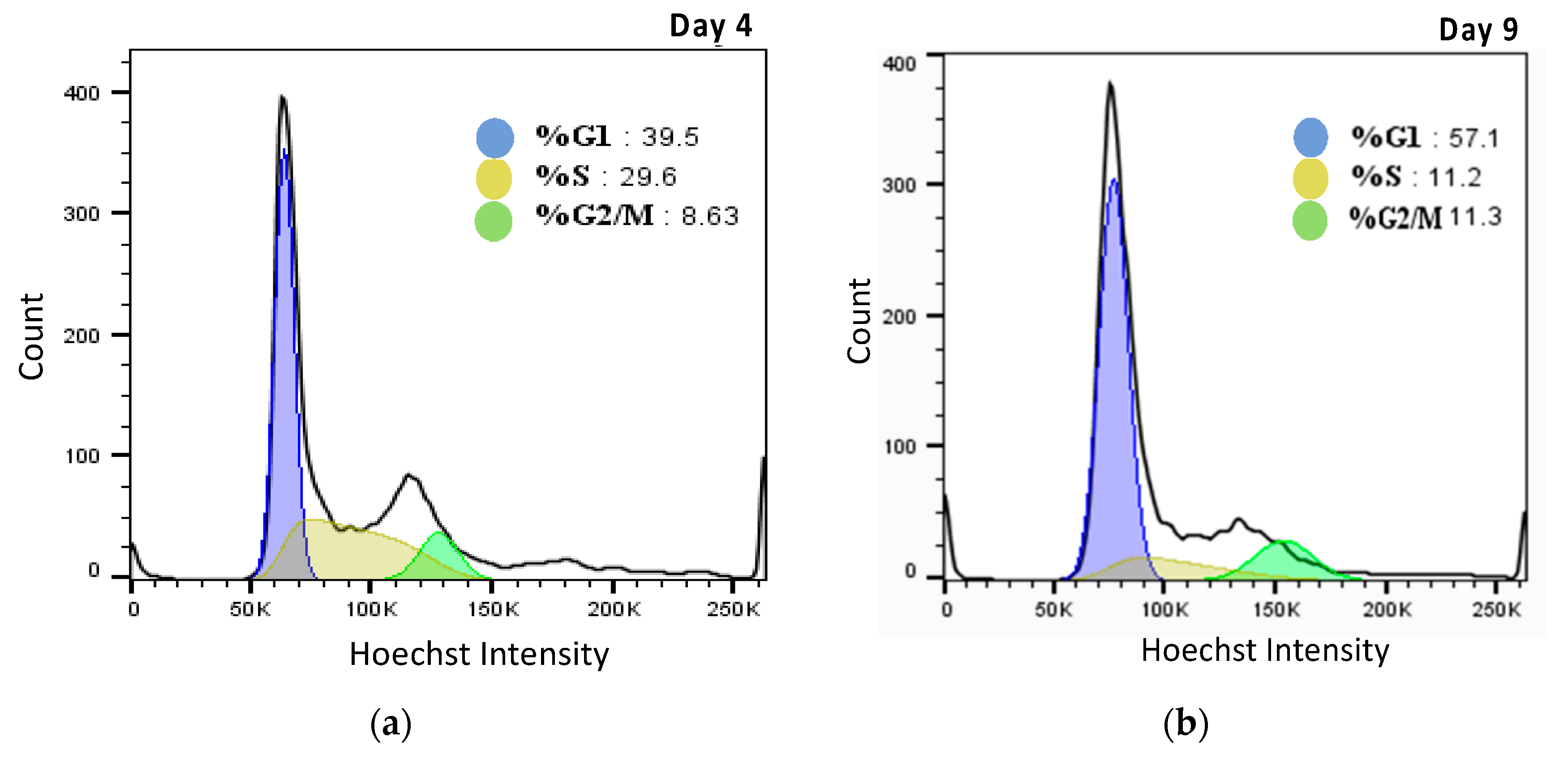
2.3. Effect of Hyperosmolality on Cell Volume
3. Materials and Methods
3.1. Cell Line Maintenance
3.2. Bioreactor Experiments
3.3. Flask-Based Osmolality Experiments
3.4. Analytical Methods
3.5. Flow Cytometry
3.6. Glycan Analysis
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
- (h−1) is growth rate in
- (106 cell mL−1) is viable cell density at time
- (μmol day−1 10−6 cell) is the metabolite (or mAb) uptake/secretion rate
- (mM) is the initial substrate (or mAb) concentration and (mM) the concentration at (day 6)
- (106 cell day mL−1) is the integral viable cell density at (day 6)


References
- Hendrick, V.; Winnepenninckx, P.; Abdelkafi, C.; Vandeputte, O.; Cherlet, M.; Marique, T.; Renemann, G.; Loa, A.; Kretzmer, G.; Werenne, J. Increased productivity of re-combinant tissular plasminogen activator (t-PA) by butyrate and shift of temperature: A cell cycle phases analysis. Cytotechnology 2001, 36, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.X.; Shuttleworth, J.; Al-Rubeai, M. Uncoupling of cell growth and proliferation results in enhancement of productivity in p21CIP1-arrested CHO cells. Biotechnol. Bioeng. 2004, 85, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Dalm, C.; Wijffels, R.H.; Martens, D.E. Metabolic characterization of a CHO cell size increase phase in fed-batch cultures. Appl. Microbiol. Biotechnol. 2017, 101, 8101–8113. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Alsayyari, A.A.; Dalm, C.; Hageman, J.A.; Wijffels, R.H.; Martens, D.E. Transcriptome Analysis of CHO Cell Size Increase During a Fed-Batch Process. Biotechnol. J. 2019, 14, 1800156. [Google Scholar] [CrossRef] [PubMed]
- Moller, J.; Bhat, K.; Riecken, K.; Portner, R.; Zeng, A.P.; Jandt, U. Process-induced cell cycle oscillations in CHO cultures: Online monitoring and model-based investigation. Biotechnol. Bioeng. 2019, 116, 2931–2943. [Google Scholar] [CrossRef] [Green Version]
- Becerra, S.; Berrios, J.; Osses, N.; Altamirano, C. Exploring the effect of mild hypothermia on CHO cell productivity. Biochem. Eng. J. 2012, 60, 1–8. [Google Scholar] [CrossRef]
- Marshall, W.F.; Young, K.D.; Swaffer, M.; Wood, E.; Nurse, P.; Kimura, A.; Frankel, J.; Wallingford, J.; Walbot, V.; Qu, X. What determines cell size? BMC Biol. 2012, 10, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, J.S.; Lee, M.S.; Lee, G.M. Effects of cloned gene dosage on the response of recombinant CHO cells to hyperosmotic pressure in regard to cell growth and antibody production. Biotechnol. Prog. 2001, 17, 993–999. [Google Scholar] [CrossRef]
- Neurohr, G.E.; Terry, R.L.; Lengefeld, J.; Bonney, M.; Brittingham, G.P.; Moretto, F.; Miettinen, T.P.; Vaites, L.P.; Soares, L.M.; Paulo, J.A. Excessive Cell Growth Causes Cytoplasm Dilution And Contributes to Senescence. Cell 2019, 176, 1083–1097.e18. [Google Scholar] [CrossRef] [Green Version]
- Levy, D.L.; Heald, R. Mechanisms of Intracellular Scaling. Annu. Rev. Cell Dev. Biol. 2012, 28, 113–135. [Google Scholar] [CrossRef] [Green Version]
- Kafri, M.; Metzl-Raz, E.; Jona, G.; Barkai, N. The Cost of Protein Production. Cell Rep. 2016, 14, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, D.R.; Holmes, P.; Jackson, L.P.; Emery, A.N.; Al-Rubeai, M. Relationship between cell size, cell cycle and specific recom-binant protein productivity. Cytotechnology 2000, 34, 59–70. [Google Scholar] [CrossRef]
- Dreesen, I.A.; Fussenegger, M. Ectopic expression of human mTOR increases viability, robustness, cell size, proliferation, and antibody production of chinese hamster ovary cells. Biotechnol. Bioeng. 2011, 108, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Pilbrough, W.; Munro, T.P.; Gray, P. Intraclonal protein expression heterogeneity in recombinant CHO cells. PLoS ONE 2009, 4, e8432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd, D.R.; Leelavatcharamas, V.; Emery, A.N.; Al-Rubeai, M. The role of the cell cycle in determining gene expression and productivity in CHO cells. Cytotechnology 1999, 30, 49–57. [Google Scholar] [CrossRef]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd, A.C. The Regulation of Cell Size. Cell 2013, 154, 1194–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalonde, M.E.; Durocher, Y. Therapeutic glycoprotein production in mammalian cells. J. Biotechnol. 2017, 251, 128–140. [Google Scholar] [CrossRef]
- Kim, T.K.; Chung, J.Y.; Sung, Y.H.; Lee, G.M. Relationship between cell size and specific thrombopoietin productivity in Chinese hamster ovary cells during dihydrofolate reductase-mediated gene amplification. Biotechnol. Bioprocess Eng. 2001, 6, 332–336. [Google Scholar] [CrossRef]
- Duarte, T.M.; Carinhas, N.; Barreiro, L.C.; Carrondo, M.J.T.; Alves, P.M.; Teixeira, A.P. Metabolic Responses of CHO Cells to Limi-tation of Key Amino Acids. Biotechnol. Bioeng. 2014, 111, 2095–2106. [Google Scholar] [CrossRef]
- Suzuki, E.; Ollis, D.F. Cell-Cycle Model for Antibody-Production Kinetics. Biotechnol. Bioeng. 1989, 34, 1398–1402. [Google Scholar] [CrossRef]
- Zhang, X.W.; Garcia, I.F.; Baldi, L.; Hacker, D.L.; Wurm, F.M. Hyperosmolarity enhances transient recombinant protein yield in Chinese hamster ovary cells. Biotechnol. Lett. 2010, 32, 1587–1592. [Google Scholar] [CrossRef]
- Qin, J.Y.; Wu, X.; Xia, Z.G.; Huang, Z.; Zhang, Y.; Wang, Y.C.; Fu, Q.; Zheng, C. The effect of hyperosmolality application time on production, quality, and biopotency of monoclonal antibodies produced in CHO cell fed-batch and perfusion cultures. Appl. Microbiol. Biotechnol. 2019, 103, 1217–1229. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Kim, N.S.; Sung, Y.H.; Lee, G.M. Biphasic culture strategy based on hyperosmotic pressure for improved humanized antibody production in Chinese hamster ovary cell culture. Vitr. Cell Dev. Biol. Anim. 2002, 38, 314–319. [Google Scholar] [CrossRef]
- Zhu, M.M.; Goyal, A.; Rank, D.L.; Gupta, S.K.; Vanden Boom, T.; Lee, S.S. Effects of elevated pCO2 and osmolality on growth of CHO cells and production of antibody-fusion protein B1: A case study. Biotechnol. Prog. 2005, 21, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Nasseri, S.S.; Ghaffari, N.; Braasch, K.; Jardon, M.A.; Butler, M.; Kennard, M.; Gopaluni, B.; Piret, J.M. Increased CHO cell fed-batch monoclonal an-tibody production using the autophagy inhibitor 3-MA or gradually increasing osmolality. Biochem. Eng. J. 2014, 91, 37–45. [Google Scholar] [CrossRef]
- Konno, Y.; Kobayashi, Y.; Takahashi, K.; Takahashi, E.; Sakae, S.; Wakitani, M.; Yamano, K.; Suzawa, T.; Yano, K.; Ohta, T.; et al. Fucose content of monoclonal antibodies can be controlled by culture medium osmolality for high antibody-dependent cellular cytotoxicity. Cytotechnology 2012, 64, 249–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Jeong, Y.R.; Kim, Y.G.; Lee, G.M. Understanding of decreased sialylation of Fc-fusion protein in hyperosmotic recom-binant Chinese hamster ovary cell culture: N-glycosylation gene expression and N-linked glycan antennary profile. Biotechnol. Bioeng. 2017, 114, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Pfizenmaier, J.; Matuszczyk, J.C.; Takors, R. Changes in intracellular ATP-content of CHO cells as response to hyperosmolality. Biotechnol. Prog. 2015, 31, 1212–1216. [Google Scholar] [CrossRef] [PubMed]
- Min Lee, G.; Koo, J. Osmolarity effects, Chinese hamster ovary cell culture. Encycl. Ind. Biotechnol. Bioprocess Biosep.Cell Technol. 2009, 3, 1–8. [Google Scholar]
- Takagi, M.; Hayashi, H.; Yoshida, T. The effect of osmolarity on metabolism and morphology in adhesion and suspension chinese hamster ovary cells producing tissue plasminogen activator. Cytotechnology 2000, 32, 171–179. [Google Scholar] [CrossRef]
- Perez-Rodriguez, S.; Ramirez-Lira, M.J.; Trujillo-Roldan, M.A.; Valdez-Cruz, N.A. Nutrient supplementation strategy improves cell concentration and longevity, monoclonal antibody production and lactate metabolism of Chinese hamster ovary cells. Bioengineered 2020, 11, 463–471. [Google Scholar] [CrossRef] [Green Version]
- Reinhart, D.; Damjanovic, L.; Castan, A.; Ernst, W.; Kunert, R. Differential gene expression of a feed-spiked super-producing CHO cell line. J. Biotechnol. 2018, 285, 23–37. [Google Scholar] [CrossRef]
- Pan, X.; Streefland, M.; Dalm, C.; Wijffels, R.H.; Martens, D.E. Selection of chemically defined media for CHO cell fed-batch culture processes. Cytotechnology 2017, 69, 39–56. [Google Scholar] [CrossRef] [Green Version]
- Okada, Y.; Maeno, E.; Shimizu, T.; Dezaki, K.; Wang, J.; Morishima, S. Receptor-mediated control of regulatory volume decrease (RVD) and apoptotic volume decrease (AVD). J. Physiol. Lond. 2001, 532, 3–16. [Google Scholar] [CrossRef]
- Kyriakopoulos, S.; Kontoravdi, C. A framework for the systematic design of fed-batch strategies in mammalian cell culture. Biotechnol. Bioeng. 2014, 111, 2466–2476. [Google Scholar] [CrossRef] [PubMed]
- Hartley, F.; Walker, T.; Chung, V.; Morten, K. Mechanisms driving the lactate switch in Chinese hamster ovary cells. Biotechnol. Bioeng. 2018, 115, 1890–1903. [Google Scholar] [CrossRef] [PubMed]
- Bortner, C.D.; Cidlowski, J.A. Cell shrinkage and monovalent cation fluxes: Role in apoptosis. ell shrinkage and monovalent cation fluxes: Role in apoptosis. Arch. Biochem. Biophys. 2007, 462, 176–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- deZengotita, V.M.; Schmelzer, A.E.; Miller, W.M. Characterization of hybridoma cell responses to elevated pCO(2) and osmo-lality: Intracellular pH, cell size, apoptosis, and metabolism. Biotechnol. Bioeng. 2002, 77, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.O.; Lee, G.M. Nutrient deprivation induces autophagy as well as apoptosis in Chinese hamster ovary cell culture. Biotechnol. Bioeng. 2008, 99, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Schmelzer, A.E.; Miller, W.M. Effects of osmoprotectant compounds on NCAM polysialylation under hyperosmotic stress and elevated pCO(2). Biotechnol. Bioeng. 2002, 77, 359–368. [Google Scholar] [CrossRef]
- Zeng, A.P.; Hu, W.S.; Deckwer, W.D. Variation of stoichiometric ratios and their correlation for monitoring and control of animal cell cultures. Biotechnol. Prog. 1998, 14, 434–441. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.J.; Guo, L.Y.; Li, P.P.; Zhao, Z.Q.; Zhou, H.S.; Di, L.-J. Molecular link between glucose and glutamine consumption in cancer cells mediated by CtBP and SIRT4. Oncogenesis 2018, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, P.M.; James, D.C. Systems biotechnology of mammalian cell factories. Brief. Funct. Genom. Proteom. 2008, 7, 95–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai-Nishiya, H.; Mori, K.; Inoue, M.; Wakitani, M.; Iida, S.; Shitara, K.; Satoh, M. Double knockdown of α1,6-fucosyltransferase (FUT8) and GDP-mannose 4,6-dehydratase (GMD) in antibody-producing cells: A new strategy for generating fully non-fucosylated therapeutic antibodies with enhanced ADCC. BMC Biotechnol. 2007, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Wells, E.; Song, L.; Greer, M.; Luo, Y.; Kurian, V.; Ogunnaike, B.; Robinson, S.A. Media supplementation for targeted manipulation of monoclonal antibody galactosylation and fucosylation. Biotechnol. Bioeng. 2020, 117, 3310–3321. [Google Scholar] [CrossRef] [PubMed]
- Gramer, M.J.; Eckblad, J.J.; Donahue, R.; Brown, J.; Shultz, C.; Vickerman, K.; Priem, P.; van den Bremer, E.T.J.; Gerritsen, J.; van Berkel, P.H.C. Modulation of antibody galactosylation through feeding of uridine, manganese chloride, and galactose. Biotechnol. Bioeng. 2011, 108, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Kotidis, P.; Jedrzejewski, P.; Sou, S.N.; Sellick, C.; Polizzi, K.; del Val, I.J.; Kontoravdi, C. Model-based optimization of antibody galactosylation in CHO cell culture. Biotechnol. Bioeng. 2019, 116, 1612–1626. [Google Scholar] [CrossRef]
- Thomann, M.; Reckermann, K.; Reusch, D.; Prasser, J.; Tejada, M.L. Fc-galactosylation modulates antibody-dependent cellular cytotoxicity of therapeutic antibodies. Mol. Immunol. 2016, 73, 69–75. [Google Scholar] [CrossRef]
- Jimenez del Val, I.; Fan, Y.; Weilguny, D. Dynamics of immature mAb glycoform secretion during CHO cell culture: An in-tegrated modelling framework. Biotechnol. J. 2016, 11, 610–623. [Google Scholar] [CrossRef] [Green Version]
- Aghamohseni, H.; Ohadi, K.; Spearman, M.; Krahn, N.; Moo-Young, M.; Scharer, J.M.; Butler, M.; Budman, H.M. Effects of nutrient levels and average culture pH on the glycosylation pattern of camelid-humanized monoclonal antibody. J. Biotechnol. 2014, 186, 98–109. [Google Scholar] [CrossRef]
- Sou, S.N.; Sellick, C.; Lee, K.; Mason, A.; Kyriakopoulos, S.; Polizzi, K.M.; Kontoravd, C. How does mild hypothermia affect monoclonal antibody glycosylation? Biotechnol. Bioeng. 2015, 112, 1165–1176. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Butler, M. Effects of ammonia on CHO cell growth, erythropoietin production, and glycosylation. Biotechnol. Bioeng. 2000, 68, 370–380. [Google Scholar] [CrossRef]
- Ehret, J.; Zimmermann, M.; Eichhorn, T.; Zimmer, A. Impact of cell culture media additives on IgG glycosylation produced in Chinese hamster ovary cells. Biotechnol. Bioeng. 2019, 116, 816–830. [Google Scholar]
- Villiger, T.K.; Roulet, A.; Perilleux, A.; Stettler, M.; Broly, H.; Morbidelli, M.; Soos, M. Controlling the time evolution of mAb N-linked glycosylation, Part I: Microbioreactor experiments. Biotechnol. Prog. 2016, 32, 1123–1134. [Google Scholar] [CrossRef]
- Gawlitzek, M.; Ryll, T.; Lofgren, J.; Sliwkowski, M.B. Ammonium alters N-glycan structures of recombinant TNFR-IgG: Deg-radative versus biosynthetic mechanisms. Biotechnol. Bioeng. 2000, 68, 637–646. [Google Scholar] [CrossRef]
- McCracken, N.A.; Kowle, R.; Ouyang, A. Control of galactosylated glycoforms distribution in cell culture system. Biotechnol. Prog. 2014, 30, 547–553. [Google Scholar] [CrossRef]
- Chen, P.; Harcum, S.W. Effects of elevated ammonium on glycosylation gene expression in CHO cells. Metab. Eng. 2006, 8, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Sumit, M.; Dolatshahi, S.; Chu, A.-H.A.; Cote, K.; Scarcelli, J.J.; Marshall, J.K.; Cornell, R.J.; Weiss, R.; Lauffenburger, D.A.; Mulukutla, B.C.; et al. Dissecting N-Glycosylation Dynamics in Chinese Hamster Ovary Cells Fed-batch Cultures using Time Course Omics Analyses. iScience 2019, 12, 102–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhart, D.; Damjanovic, L.; Kaisermayer, C.; Sommeregger, W.; Gili, A.; Gasselhuber, B.; Castan, A.; Mayrhofer, P.; Grünwald-Gruber, C.; Kunert, R. Bioprocessing of Recombinant CHO-K1, CHO-DG44, and CHO-S: CHO Expression Hosts Favor Either mAb Production or Biomass Synthesis. Biotechnol. J. 2019, 14, 1700686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freund, N.W.; Croughan, M.S. A Simple Method to Reduce both Lactic Acid and Ammonium Production in Industrial Animal Cell Culture. Int. J. Mol. Sci. 2018, 19, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goey, C.H.; Tsang, J.M.H.; Bell, D.; Kontoravdi, C. Cascading effect in bioprocessing-The impact of mild hypothermia on CHO cell behavior and host cell protein composition. Biotechnol. Bioeng. 2017, 114, 2771–2781. [Google Scholar] [CrossRef] [PubMed]
- Kiehl, T.R.; Shen, D.; Khattak, S.F.; Jian Li, Z.; Sharfstein, S.T. Observations of cell size dynamics under osmotic stress. Cytom. Part A 2011, 79, 560–569. [Google Scholar] [CrossRef]
- Cooper, S. Reappraisal of serum starvation, the restriction point, G0, and G1 phase arrest points. FASEB J. 2003, 17, 333–340. [Google Scholar] [CrossRef]
- Martens, D.E.; Degooijer, C.D.; Vanderveldendegroot, C.A.M.; Beuvery, E.C.; Tramper, J. Effect of Dilution Rate on Growth, Productivity, Cell-Cycle and Size, and Shear Sensitivity of a Hybridoma Cell in a Continuous Culture. Biotechnol. Bioeng. 1993, 41, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Yao, G. Modelling mammalian cellular quiescence. Interface Focus 2014, 4, 20130074. [Google Scholar] [CrossRef] [Green Version]
- Pasantes-Morales, H. Channels and Volume Changes in the Life and Death of the Cell. Mol. Pharmacol. 2016, 90, 358–370. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhuthali, S.; Kotidis, P.; Kontoravdi, C. Osmolality Effects on CHO Cell Growth, Cell Volume, Antibody Productivity and Glycosylation. Int. J. Mol. Sci. 2021, 22, 3290. https://doi.org/10.3390/ijms22073290
Alhuthali S, Kotidis P, Kontoravdi C. Osmolality Effects on CHO Cell Growth, Cell Volume, Antibody Productivity and Glycosylation. International Journal of Molecular Sciences. 2021; 22(7):3290. https://doi.org/10.3390/ijms22073290
Chicago/Turabian StyleAlhuthali, Sakhr, Pavlos Kotidis, and Cleo Kontoravdi. 2021. "Osmolality Effects on CHO Cell Growth, Cell Volume, Antibody Productivity and Glycosylation" International Journal of Molecular Sciences 22, no. 7: 3290. https://doi.org/10.3390/ijms22073290
APA StyleAlhuthali, S., Kotidis, P., & Kontoravdi, C. (2021). Osmolality Effects on CHO Cell Growth, Cell Volume, Antibody Productivity and Glycosylation. International Journal of Molecular Sciences, 22(7), 3290. https://doi.org/10.3390/ijms22073290






