A 2-Benzylmalonate Derivative as STAT3 Inhibitor Suppresses Tumor Growth in Hepatocellular Carcinoma by Upregulating β-TrCP E3 Ubiquitin Ligase
Abstract
1. Introduction
2. Results
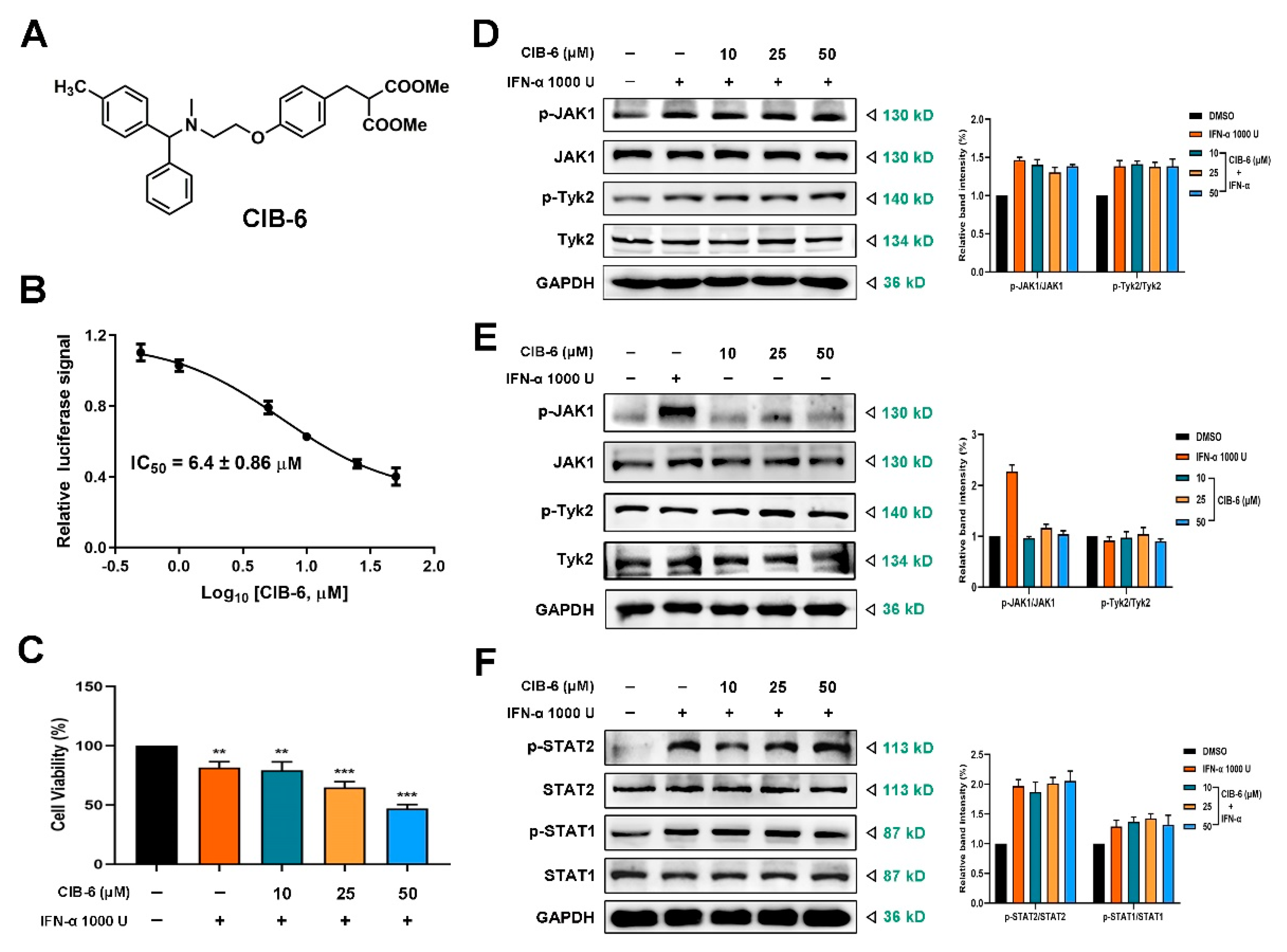
2.1. CIB-6 Inhibits the Interferon Stimulated Response Element (ISRE) Signal Induced by IFN-α without Upstream Kinase and STAT1/STAT2 Inhibition
2.2. CIB-6 Inhibits IFN-α Induced STAT3 Signaling
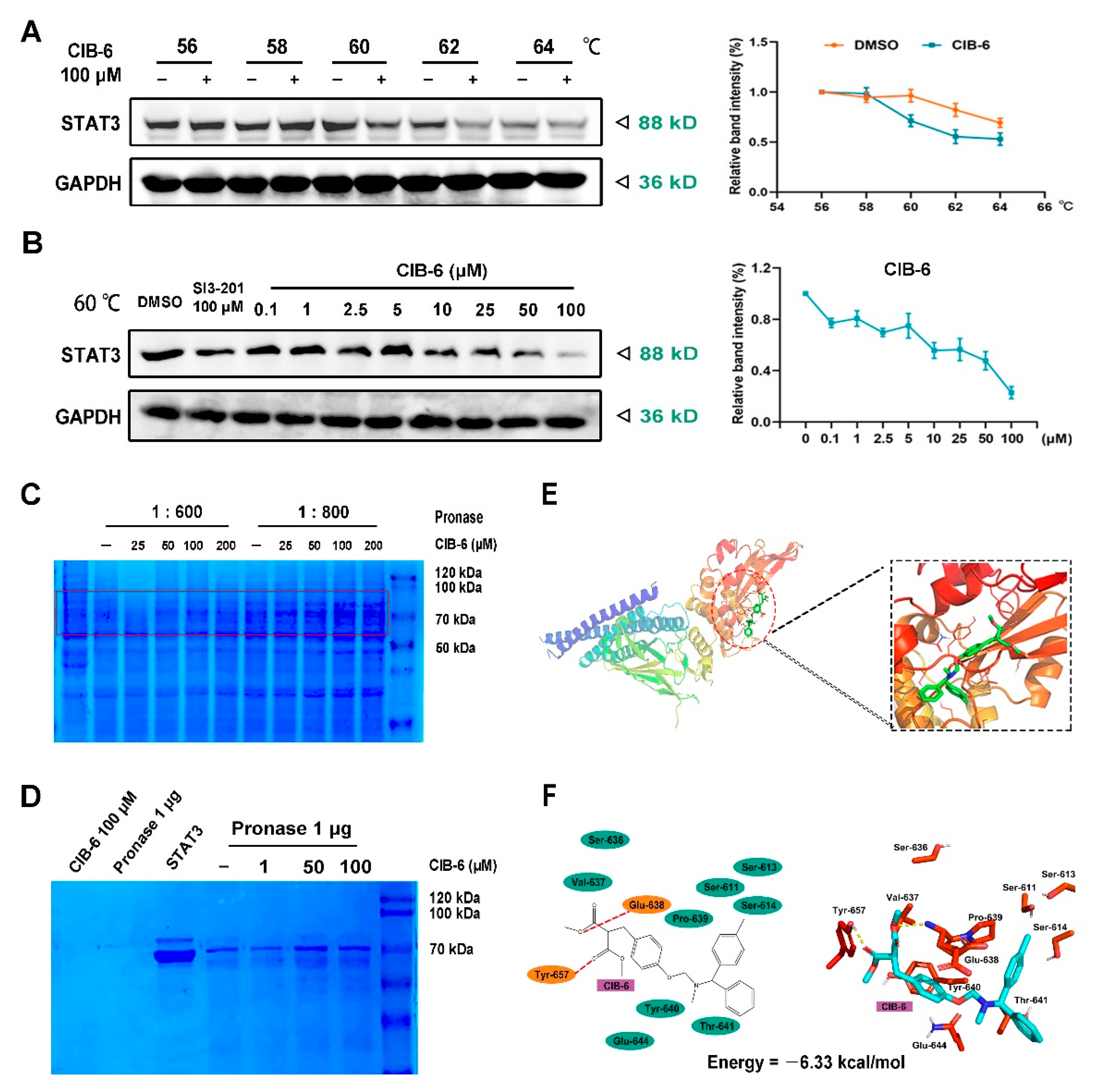
2.3. CIB-6 Directly Interacts with STAT3
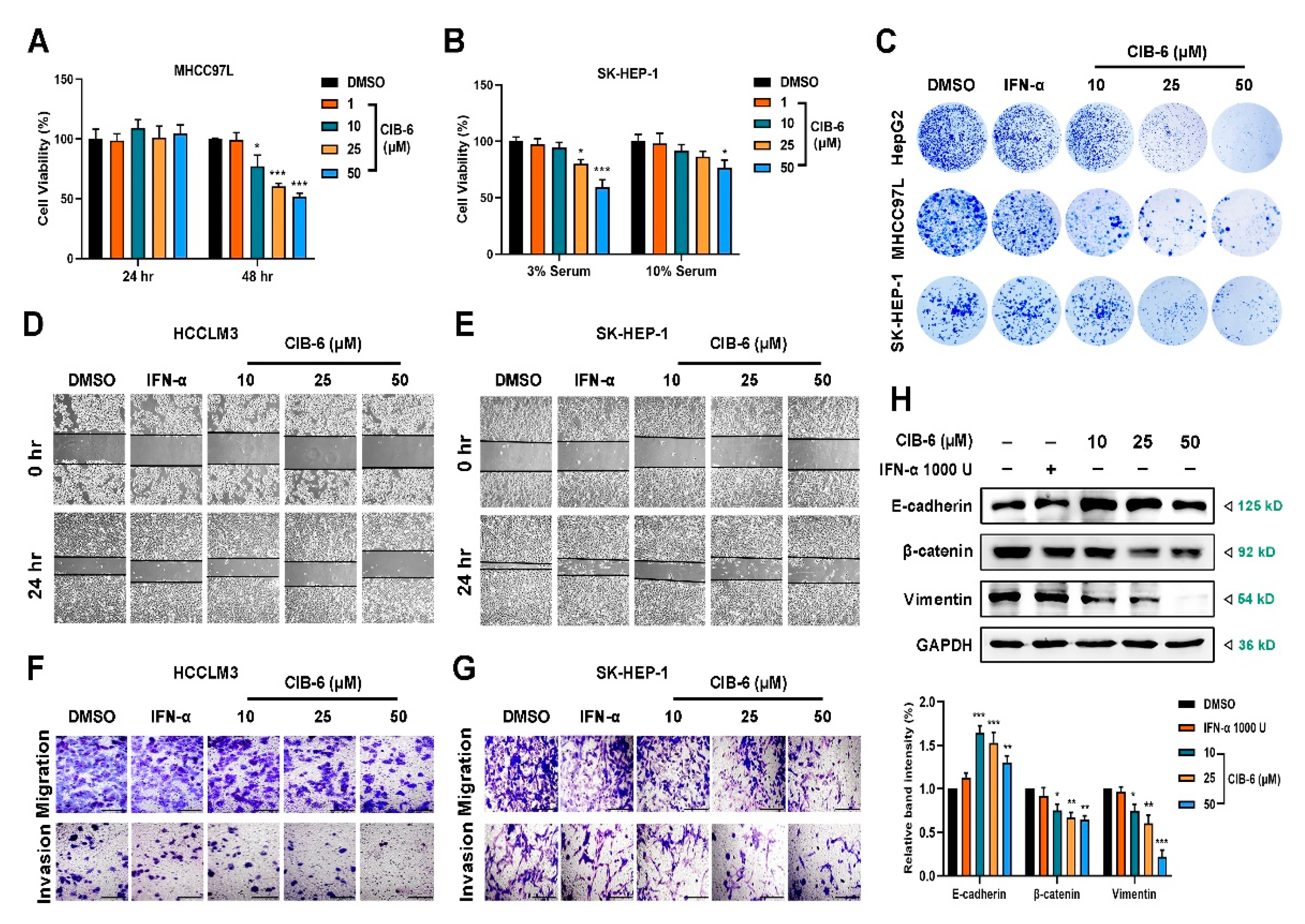
2.4. CIB-6 Inhibits the Viability, Migration, and Invasion of HCC Cells
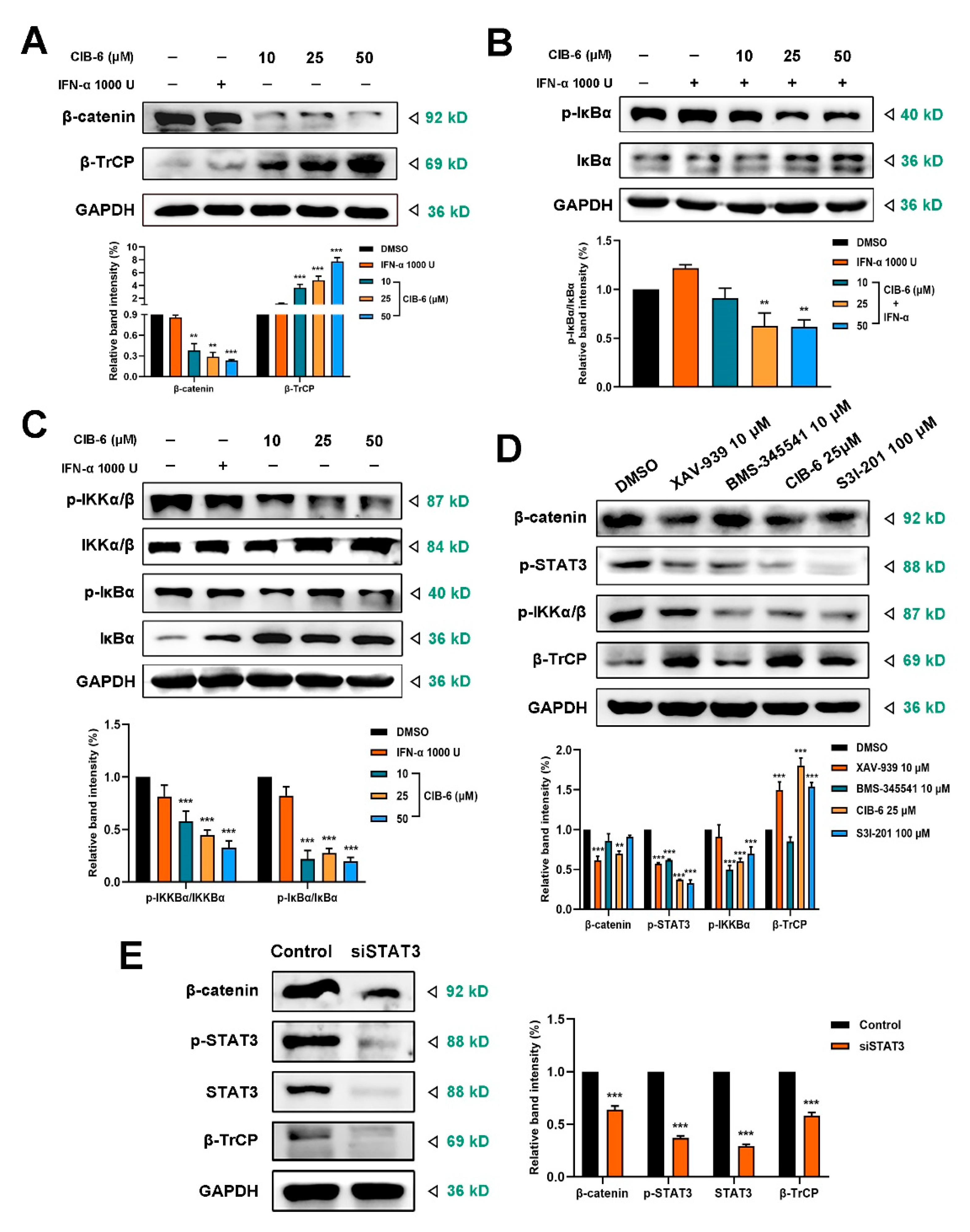
2.5. CIB-6 Suppresses β-TrCP/β-catenin/NF-κB axis by Inhibiting STAT3
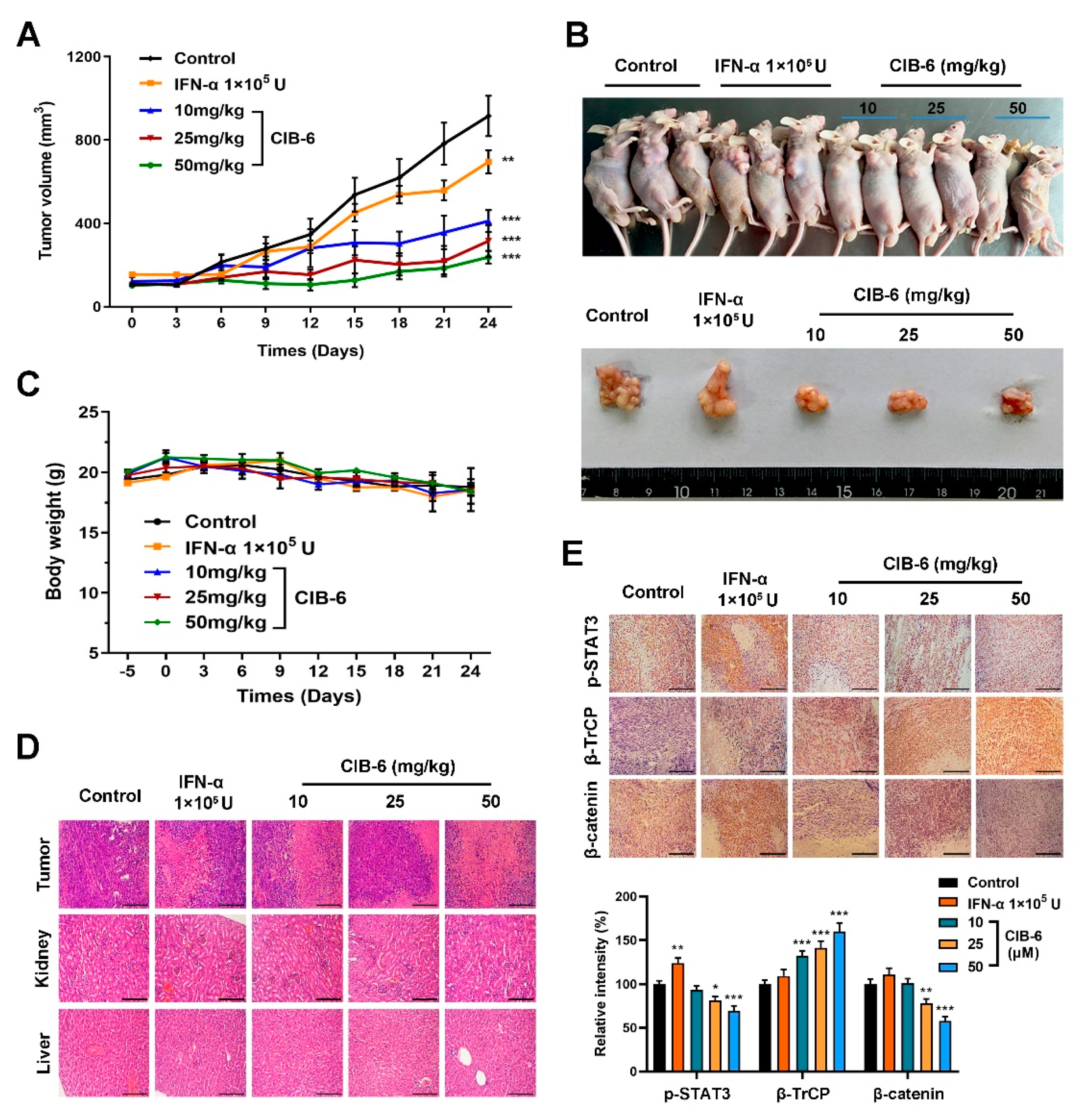
2.6. CIB-6 Exhibits Antitumor Activity in a Xenograft Mouse Model
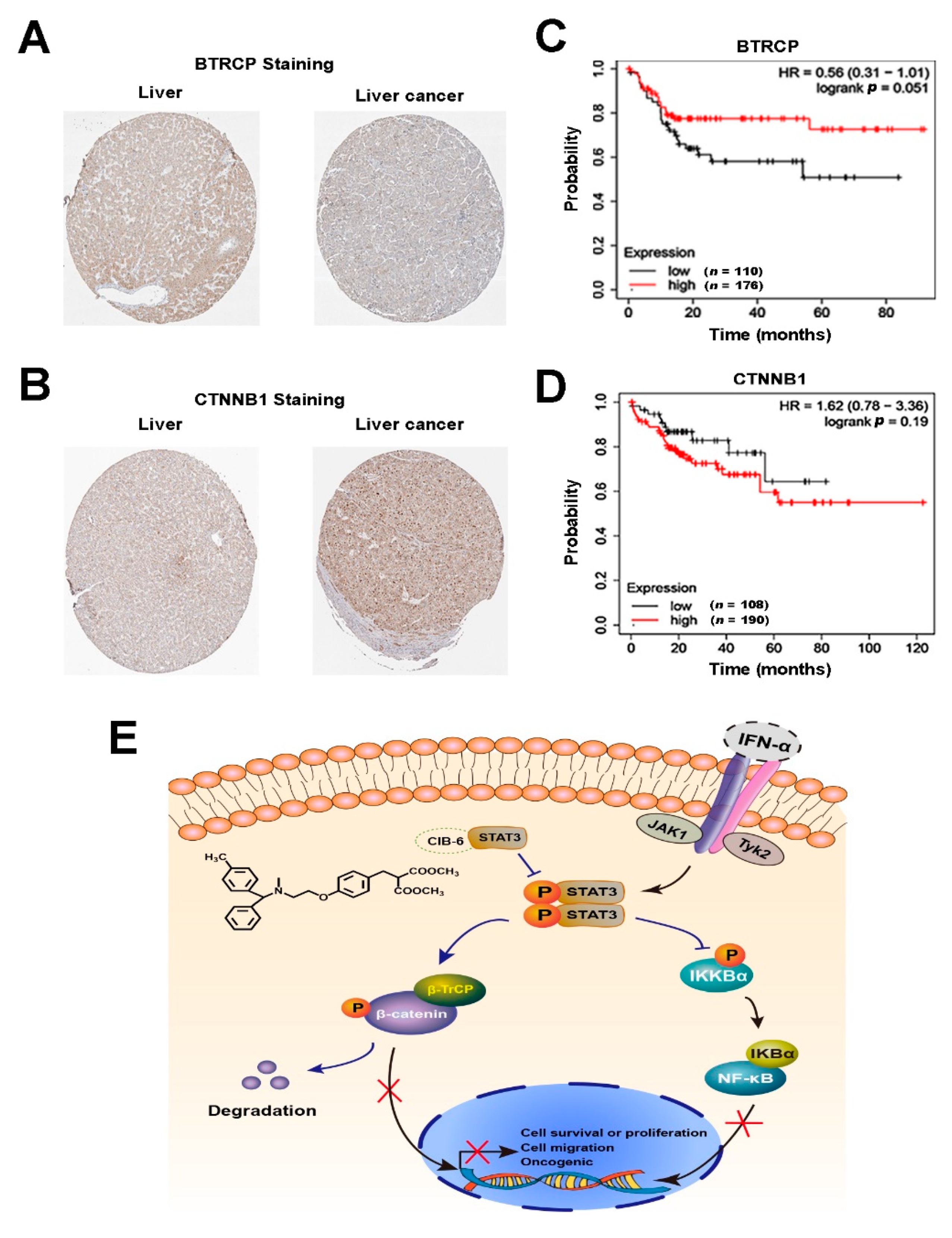
2.7. The Expression Levels of β-TrCP and β-catenin Correlate with the Development of HCC in Humans
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. Luciferase Reporter Assay
4.4. Cell Viability Assay
4.5. Western Blotting
4.6. Confocal Microscopy
4.7. STAT3-Responsive Reporter Expression
4.8. Cell Thermal Shift Assay (CETSA)
4.9. Protein Expression and Purification
4.10. Drug Affinity-Responsive Target Stability (DARTS)
4.11. Computer-Aided Virtual Calculation of the Molecular Properties
4.12. Colony Formation Assay
4.13. Wound Healing Assay
4.14. Cell Migration and Matrigel Invasion Assays
4.15. RNA Interference
4.16. Real-Time Quantitative PCR (RT-qPCR)
4.17. Animal Experiment
4.18. Hematoxylin–Eosin (H&E) Staining and Immunohistochemistry
4.19. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lemon, S.M.; McGivern, D.R. Is hepatitis C virus carcinogenic? Gastroenterology 2012, 142, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.C.; Llovet, J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015, 149, 1226–1239.e1224. [Google Scholar] [CrossRef]
- Goldstein, D.; Laszlo, J. The role of interferon in cancer therapy: A current perspective. CA Cancer J. Clin. 1988, 38, 258–277. [Google Scholar] [CrossRef] [PubMed]
- Bekisz, J.; Baron, S.; Balinsky, C.; Morrow, A.; Zoon, K.C. Antiproliferative properties of type I and type II interferon. Pharmaceuticals 2010, 3, 994–1015. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and distinct functions of type I and type III interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Snell, L.M.; McGaha, T.L.; Brooks, D.G. Type I interferon in chronic virus infection and cancer. Trends Immunol. 2017, 38, 542–557. [Google Scholar] [CrossRef]
- Huang, S.; Bucana, C.D.; Van Arsdall, M.; Fidler, I.J. Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene 2002, 21, 2504–2512. [Google Scholar] [CrossRef]
- Wang, W.B.; Levy, D.E.; Lee, C.K. STAT3 negatively regulates type I IFN-mediated antiviral response. J. Immunol. 2011, 187, 2578–2585. [Google Scholar] [CrossRef]
- Tsai, M.H.; Pai, L.M.; Lee, C.K. Fine-tuning of type I interferon response by STAT3. Front. Immunol. 2019, 10, 1448. [Google Scholar] [CrossRef]
- Tannapfel, A.; Anhalt, K.; Häusermann, P.; Sommerer, F.; Benicke, M.; Uhlmann, D.; Witzigmann, H.; Hauss, J.; Wittekind, C. Identification of novel proteins associated with hepatocellular carcinomas using protein microarrays. J. Pathol. 2003, 201, 238–249. [Google Scholar] [CrossRef]
- Li, N.; Grivennikov, S.I.; Karin, M. The unholy trinity: Inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell 2011, 19, 429–431. [Google Scholar] [CrossRef]
- Perugorria, M.J.; Olaizola, P.; Labiano, I.; Esparza-Baquer, A.; Marzioni, M.; Marin, J.J.G.; Bujanda, L.; Banales, J.M. Wnt-β-catenin signalling in liver development, health and disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Herrmann, A.; Deng, J.H.; Kujawski, M.; Niu, G.; Li, Z.; Forman, S.; Jove, R.; Pardoll, D.M.; Yu, H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 2009, 15, 283–293. [Google Scholar] [CrossRef]
- Varshavsky, A. The ubiquitin system, autophagy, and regulated protein degradation. Annu. Rev. Biochem. 2017, 86, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Peng, Y.; Zhang, J.; Long, J.; Liu, J.; Wei, W. Targeting SCF E3 ligases for cancer therapies. Adv. Exp. Med. Biol. 2020, 1217, 123–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Lai, W.C.; Chuang, K.A.; Shen, Y.J.; Hu, W.S.; Ho, C.H.; Chen, Y.B.; Hsu, M.F.; Hsu, H.C.; Lieu, C.H. Blockade of JAK2 activity suppressed accumulation of β-catenin in leukemic cells. J. Cell. Biochem. 2010, 111, 402–411. [Google Scholar] [CrossRef]
- Park, I.H.; Li, C. Characterization of molecular recognition of STAT3 SH2 domain inhibitors through molecular simulation. J. Mol. Recognit. 2011, 24, 254–265. [Google Scholar] [CrossRef]
- You, L.; Wang, Z.; Li, H.; Shou, J.; Jing, Z.; Xie, J.; Sui, X.; Pan, H.; Han, W. The role of STAT3 in autophagy. Autophagy 2015, 11, 729–739. [Google Scholar] [CrossRef]
- Zhao, W.; Jaganathan, S.; Turkson, J. A cell-permeable Stat3 SH2 domain mimetic inhibits Stat3 activation and induces antitumor cell effects in vitro. J. Biol. Chem. 2010, 285, 35855–35865. [Google Scholar] [CrossRef]
- Tai, Z.F.; Zhang, G.L.; Wang, F. Identification of small molecule activators of the janus kinase/signal transducer and activator of transcription pathway using a cell-based screen. Biol. Pharm. Bull. 2012, 35, 65–71. [Google Scholar] [CrossRef]
- Chen, W.; Shen, X.; Xia, X.; Xu, G.; Ma, T.; Bai, X.; Liang, T. NSC 74859-mediated inhibition of STAT3 enhances the anti-proliferative activity of cetuximab in hepatocellular carcinoma. Liver Int. 2012, 32, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Al Zaid Siddiquee, K.; Turkson, J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008, 18, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial-mesenchymal transition. Cells 2020, 9, 217. [Google Scholar] [CrossRef]
- Kanarek, N.; Ben-Neriah, Y. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol. Rev. 2012, 246, 77–94. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Roca Suarez, A.A.; Van Renne, N.; Baumert, T.F.; Lupberger, J. Viral manipulation of STAT3: Evade, exploit, and injure. PLoS Pathog. 2018, 14, e1006839. [Google Scholar] [CrossRef]
- Waziry, R.; Hajarizadeh, B.; Grebely, J.; Amin, J.; Law, M.; Danta, M.; George, J.; Dore, G.J. Hepatocellular carcinoma risk following direct-acting antiviral HCV therapy: A systematic review, meta-analyses, and meta-regression. J. Hepatol. 2017, 67, 1204–1212. [Google Scholar] [CrossRef]
- Cai, X.H.; Xie, B.; Guo, H. Synthesis and evaluation of methyl 2-methoxycarbonyl-3-phenylproplonate derivatives as a new type of angiotensin converting enzyme inhibitors. Can. J. Chem. 2006, 84, 1110–1113. [Google Scholar] [CrossRef]
- Egami, K.; Murohara, T.; Shimada, T.; Sasaki, K.; Shintani, S.; Sugaya, T.; Ishii, M.; Akagi, T.; Ikeda, H.; Matsuishi, T.; et al. Role of host angiotensin II type 1 receptor in tumor angiogenesis and growth. J. Clin. Investig. 2003, 112, 67–75. [Google Scholar] [CrossRef]
- Ager, E.I.; Neo, J.; Christophi, C. The renin-angiotensin system and malignancy. Carcinogenesis 2008, 29, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Chen, Z.; Wang, Q.; Guo, Q.; Xu, J.; Wu, X. XJP-1, a novel ACEI, with anti-inflammatory properties in HUVECs. Atherosclerosis 2011, 219, 40–48. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Fan, X.; Yang, W.; Yu, B.; Kou, J.; Li, F. Captopril attenuates TAC-induced heart failure via inhibiting Wnt3a/β-catenin and Jak2/Stat3 pathways. Biomed. Pharmacother. 2019, 113, 108780. [Google Scholar] [CrossRef]
- Furqan, M.; Akinleye, A.; Mukhi, N.; Mittal, V.; Chen, Y.; Liu, D. STAT inhibitors for cancer therapy. J. Hematol. Oncol. 2013, 6, 90. [Google Scholar] [CrossRef]
- Xu, X.; Kasembeli, M.M.; Jiang, X.; Tweardy, B.J.; Tweardy, D.J. Chemical probes that competitively and selectively inhibit Stat3 activation. PLoS ONE 2009, 4, e4783. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Buettner, R.; Yuan, Y.C.; Yip, R.; Horne, D.; Jove, R.; Vaidehi, N. Molecular dynamics simulations of the conformational changes in signal transducers and activators of transcription, Stat1 and Stat3. J. Mol. Graph. Model. 2009, 28, 347–356. [Google Scholar] [CrossRef]
- Yesylevskyy, S.O.; Ramseyer, C.; Pudlo, M.; Pallandre, J.R.; Borg, C. Selective inhibition of STAT3 with respect to STAT1: Insights from molecular dynamics and ensemble docking simulations. J. Chem. Inf. Model. 2016, 56, 1588–1596. [Google Scholar] [CrossRef]
- Fagard, R.; Metelev, V.; Souissi, I.; Baran-Marszak, F. STAT3 inhibitors for cancer therapy: Have all roads been explored? JAKSTAT 2013, 2, e22882. [Google Scholar] [CrossRef]
- Zhang, X.; Yue, P.; Fletcher, S.; Zhao, W.; Gunning, P.T.; Turkson, J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem. Pharmacol. 2010, 79, 1398–1409. [Google Scholar] [CrossRef]
- Lachenmayer, A.; Alsinet, C.; Savic, R.; Cabellos, L.; Toffanin, S.; Hoshida, Y.; Villanueva, A.; Minguez, B.; Newell, P.; Tsai, H.W.; et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin. Cancer. Res. 2012, 18, 4997–5007. [Google Scholar] [CrossRef] [PubMed]
- Yuzugullu, H.; Benhaj, K.; Ozturk, N.; Senturk, S.; Celik, E.; Toylu, A.; Tasdemir, N.; Yilmaz, M.; Erdal, E.; Akcali, K.C.; et al. Canonical Wnt signaling is antagonized by noncanonical Wnt5a in hepatocellular carcinoma cells. Mol. Cancer 2009, 8, 90. [Google Scholar] [CrossRef]
- Kawada, M.; Seno, H.; Uenoyama, Y.; Sawabu, T.; Kanda, N.; Fukui, H.; Shimahara, Y.; Chiba, T. Signal transducers and activators of transcription 3 activation is involved in nuclear accumulation of beta-catenin in colorectal cancer. Cancer Res. 2006, 66, 2913–2917. [Google Scholar] [CrossRef]
- Fuchs, S.Y.; Spiegelman, V.S.; Kumar, K.G. The many faces of beta-TrCP E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene 2004, 23, 2028–2036. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, V.S.; Slaga, T.J.; Pagano, M.; Minamoto, T.; Ronai, Z.; Fuchs, S.Y. Wnt/beta-catenin signaling induces the expression and activity of betaTrCP ubiquitin ligase receptor. Mol. Cell 2000, 5, 877–882. [Google Scholar] [CrossRef]
- Caruso, S.; O’Brien, D.R.; Cleary, S.P.; Roberts, L.R.; Zucman-Rossi, J. Genetics of HCC: Novel approaches to explore molecular diversity. Hepatology 2020, 73, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Pez, F.; Lopez, A.; Kim, M.; Wands, J.R.; Caron de Fromentel, C.; Merle, P. Wnt signaling and hepatocarcinogenesis: Molecular targets for the development of innovative anticancer drugs. J. Hepatol. 2013, 59, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007, 6, 975–990. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Murti, A.; Pfeffer, S.R.; Kim, J.G.; Donner, D.B.; Pfeffer, L.M. Interferon alpha/beta promotes cell survival by activating nuclear factor kappa B through phosphatidylinositol 3-kinase and Akt. J. Biol. Chem. 2001, 276, 13756–13761. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Luedde, T.; Schwabe, R.F. NF-κB in the liver-linking injury, fibrosis and hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Gao, F.; Lu, Z.; Ren, Z.; Ramesh, R.; Birtwistle, J.S.; Kaluarachchi, K.K.; Chen, X.; Bast, R.C., Jr.; Liao, W.S.; et al. Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3. J. Med. Chem. 2011, 54, 3549–3563. [Google Scholar] [CrossRef]
- Schmidt, M.L.; Donninger, H.; Clark, G.J. Ras regulates SCF(β-TrCP) protein activity and specificity via its effector protein NORE1A. J. Biol. Chem. 2014, 289, 31102–31110. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, Y.; Xu, J.; Xiao, K.; Xu, Y.; Guo, T.; Zhang, L.; Wang, J.; Zheng, H. β-TrCP restricts lipopolysaccharide (LPS)-induced activation of TRAF6-IKK pathway upstream of IκBα signaling. Front. Immunol. 2018, 9, 2930. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Iwasaki, A. Type I and type III interferons-induction, signaling, evasion and application to combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- National Institutes of Health. ImageJ-Image Processing and Analysis in Java, Version 1.52a; National Institutes of Health: Bethesda, MD, USA, 1997.
- Martinez Molina, D.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- The Scripps Research Institute. Autodock, version 4.2.6; The Scripps Research Institute: La Jolla, CA, USA, 2014. [Google Scholar]
- GraphPad Software Inc. GraphPad Prism, version 8.0.0; GraphPad Software Inc.: San Diego, CA, USA, 2018. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, T.; Wonganan, O.; Zhang, Z.; Yu, J.; Xi, R.; Cao, Y.; Suksamrarn, A.; Zhang, G.; Wang, F. A 2-Benzylmalonate Derivative as STAT3 Inhibitor Suppresses Tumor Growth in Hepatocellular Carcinoma by Upregulating β-TrCP E3 Ubiquitin Ligase. Int. J. Mol. Sci. 2021, 22, 3354. https://doi.org/10.3390/ijms22073354
Peng T, Wonganan O, Zhang Z, Yu J, Xi R, Cao Y, Suksamrarn A, Zhang G, Wang F. A 2-Benzylmalonate Derivative as STAT3 Inhibitor Suppresses Tumor Growth in Hepatocellular Carcinoma by Upregulating β-TrCP E3 Ubiquitin Ligase. International Journal of Molecular Sciences. 2021; 22(7):3354. https://doi.org/10.3390/ijms22073354
Chicago/Turabian StylePeng, Ting, Orawan Wonganan, Zhonghui Zhang, Jialing Yu, Ruiying Xi, Yu Cao, Apichart Suksamrarn, Guolin Zhang, and Fei Wang. 2021. "A 2-Benzylmalonate Derivative as STAT3 Inhibitor Suppresses Tumor Growth in Hepatocellular Carcinoma by Upregulating β-TrCP E3 Ubiquitin Ligase" International Journal of Molecular Sciences 22, no. 7: 3354. https://doi.org/10.3390/ijms22073354
APA StylePeng, T., Wonganan, O., Zhang, Z., Yu, J., Xi, R., Cao, Y., Suksamrarn, A., Zhang, G., & Wang, F. (2021). A 2-Benzylmalonate Derivative as STAT3 Inhibitor Suppresses Tumor Growth in Hepatocellular Carcinoma by Upregulating β-TrCP E3 Ubiquitin Ligase. International Journal of Molecular Sciences, 22(7), 3354. https://doi.org/10.3390/ijms22073354





