Proteogenomic Characterization of the Cement and Adhesive Gland of the Pelagic Gooseneck Barnacle Lepas anatifera
Abstract
:1. Introduction
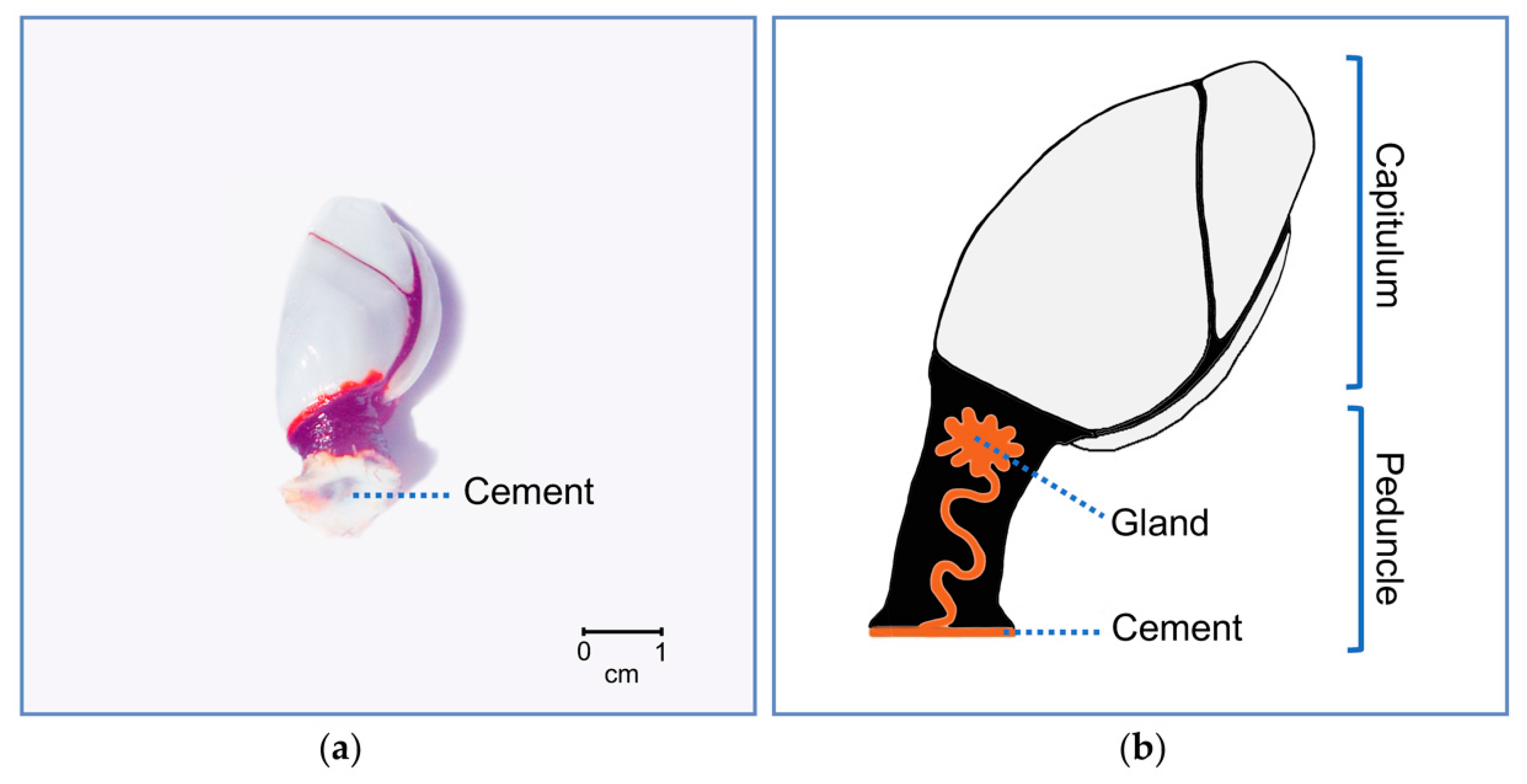
2. Results and Discussion
2.1. Protein Identification
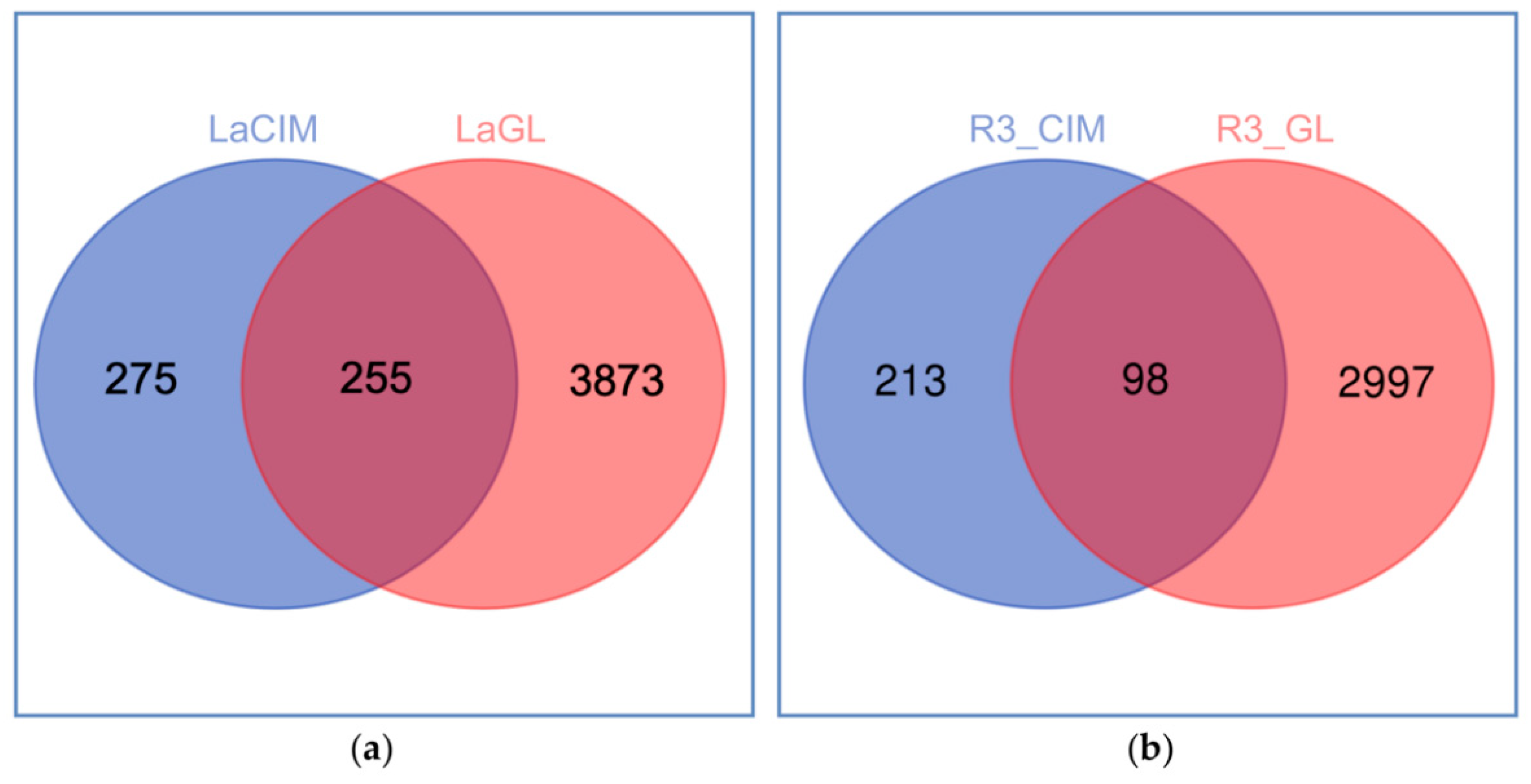
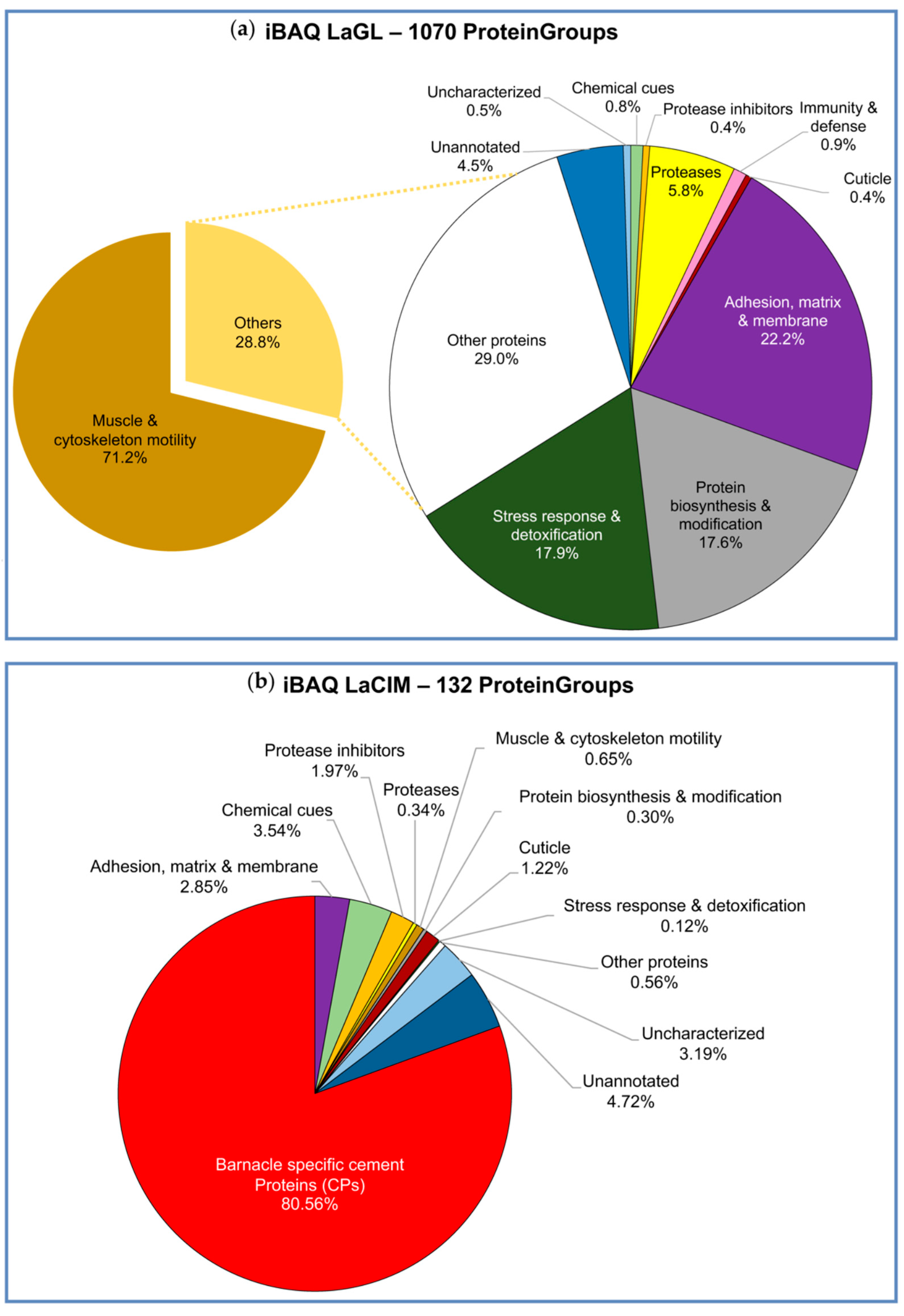
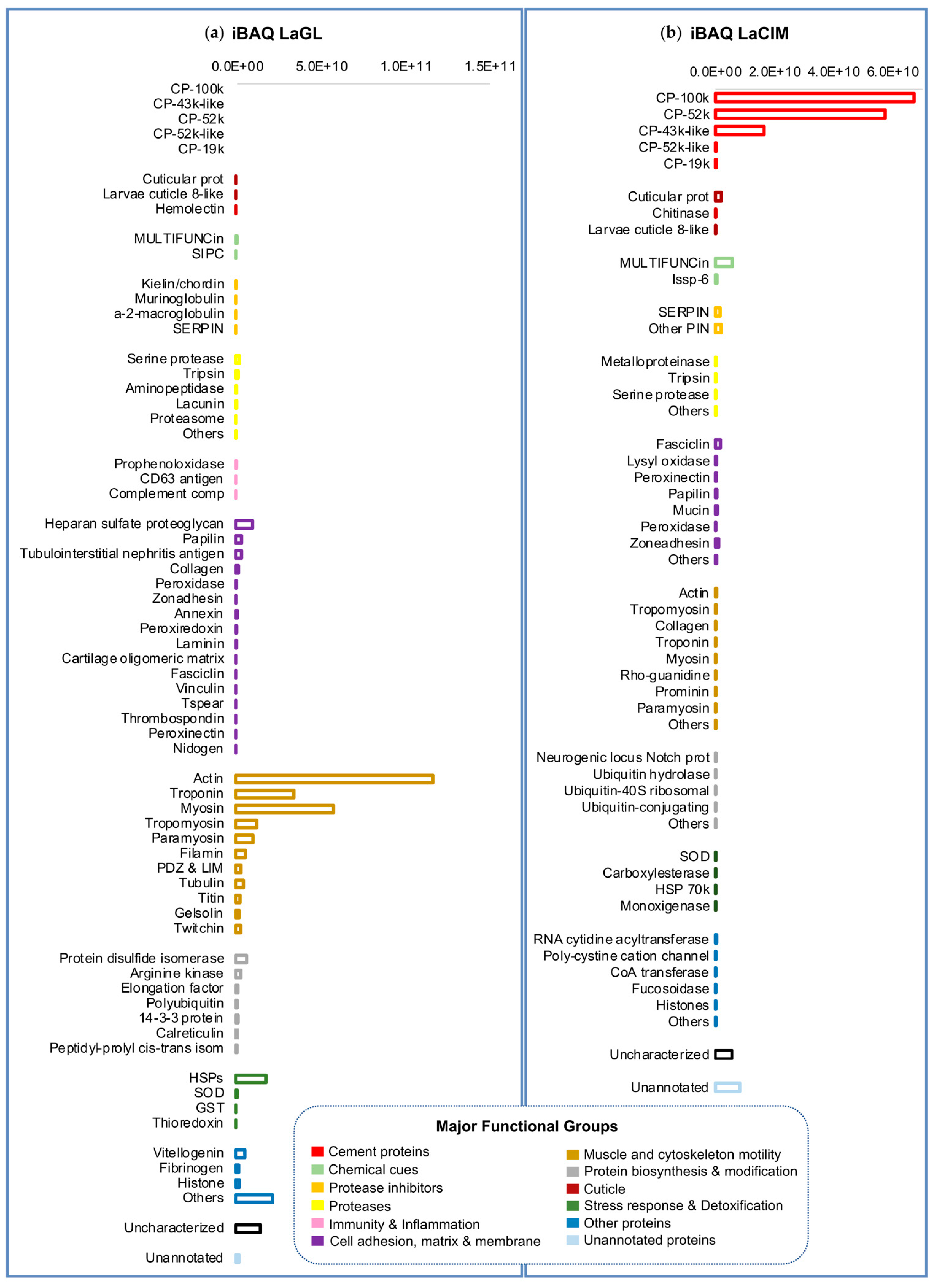
2.2. Quantitative Proteomic Analyses
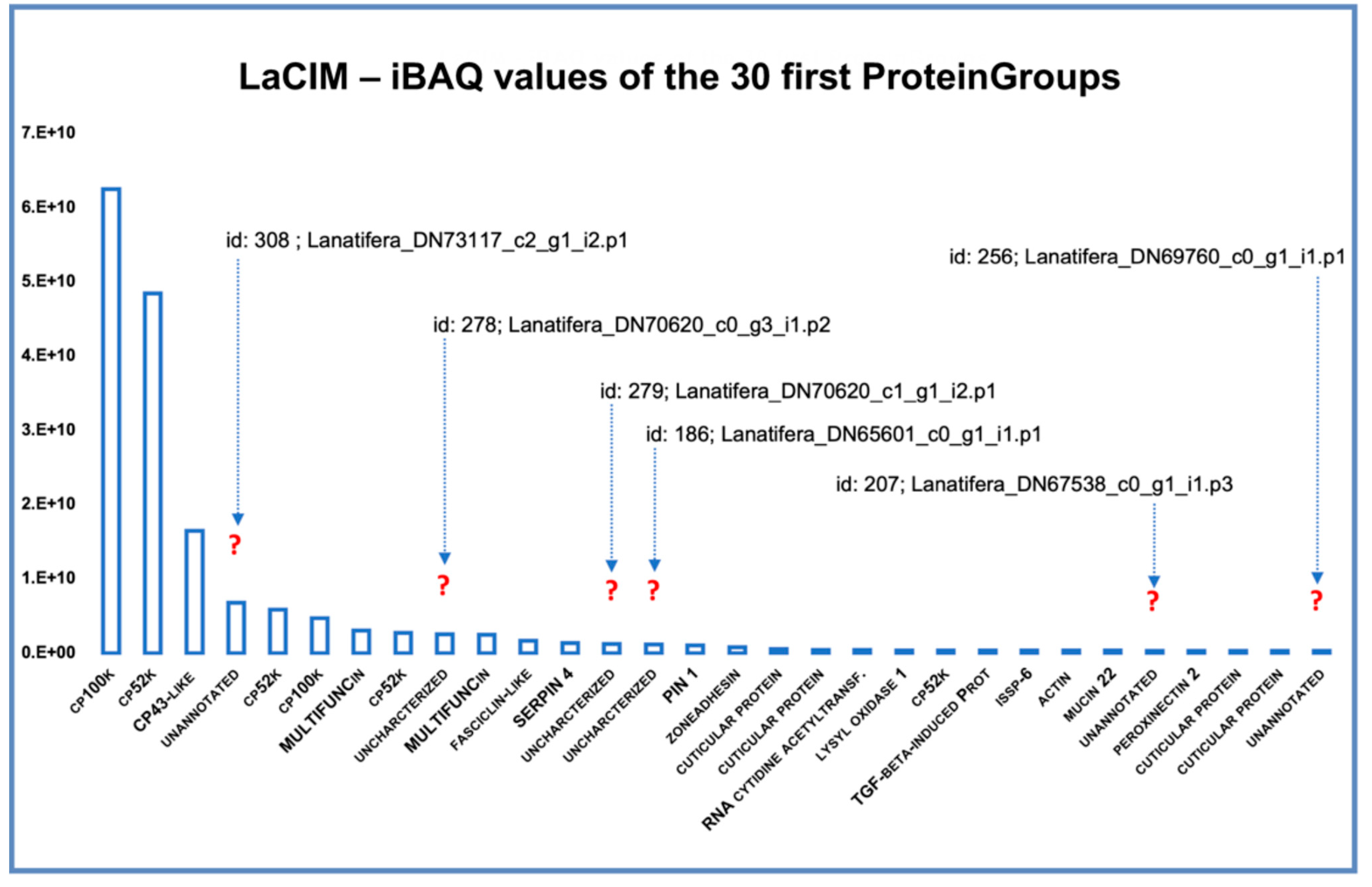
Unannotated Proteins of the Cement Proteome
3. Materials and Methods
3.1. Sampling, Protein Solubilization, and Extraction
3.2. LC-MS/MS Analyses
3.3. Protein Identification and Quantitative Proteomic Analyses
3.4. Data Filtration and Downstream Analyses
3.5. Characterization of Unannotated Cement Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lobo-da-Cunha, A.; Alves, Ï.; Oliveira, E.; Cunha, I. The cement apparatus of the stalked barnacle Pollicipes pollicipes. Mar. Biol. 2017, 164, 1–8. [Google Scholar] [CrossRef]
- Jonker, J.-L.; von Byern, J.; Flammang, P.; Klepal, W.; Power, A.M. Unusual adhesive production system in the barnacle Lepas anatifera: An ultrastructural and histochemical investigation. J. Morphol. 2012, 273, 1377–1391. [Google Scholar] [CrossRef]
- Foster, B.A. Barnacle ecology and adaptation. In Barnacle Biology; Southward, A.J., Crisp, D.J., Eds.; A.A. Balkema: Amsterdam, The Netherlands, 1987; ISBN 9061916283. [Google Scholar]
- Borja, A.; Liria, P.; Muxika, I.; Bald, J. Relationships between wave exposure and biomass of the goose barnacle (Pollicipes pollicipes, Gmelin, 1790) in the Gaztelugatxe Marine Reserve (Basque Country, northern Spain). ICES J. Mar. Sci. 2016, 63, 226–236. [Google Scholar] [CrossRef]
- Stewart, R.J. Protein-based underwater adhesives and the prospects for their biotechnological production. Appl. Microbiol. Biotechnol. 2011, 89, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.P.; Bendick, J.A.; Holm, E.R.; Hertel, W.M. Economic impact of biofouling on a naval surface ship. Biofouling 2011, 27, 87–98. [Google Scholar] [CrossRef]
- Kamino, K. Underwater Adhesive of Marine Organisms as the Vital Link Between Biological Science and Material Science. Mar. Biotechnol. 2008, 10, 111–121. [Google Scholar] [CrossRef]
- Bhagat, V.; Becker, M.L. Degradable Adhesives for Surgery and Tissue Engineering. Biomacromolecules 2017, 18, 3009–3039. [Google Scholar] [CrossRef] [PubMed]
- Meredith, H.J.; Jenkins, C.L.; Wilker, J.J. Enhancing the Adhesion of a Biomimetic Polymer Yields Performance Rivaling Commercial Glues. Adv. Funct. Mater. 2014, 24, 3259–3267. [Google Scholar] [CrossRef]
- Kamino, K. Barnacle underwater attachment. In Biological Adhesives; Springer Berlin Heidelberg: Berlin, Heidelberg, 2006; pp. 145–166. [Google Scholar]
- Kamino, K. Barnacle underwater attachment. In Biological Adhesives; Springer: Cham, Switzerland, 2016; pp. 153–176. [Google Scholar]
- So, C.R.; Fears, K.P.; Leary, D.H.; Scancella, J.M.; Wang, Z.; Liu, J.L.; Orihuela, B.; Rittschof, D.; Spillmann, C.M.; Wahl, K.J. Sequence basis of Barnacle Cement Nanostructure is Defined by Proteins with Silk Homology. Sci. Rep. 2016, 6, 36219. [Google Scholar] [CrossRef]
- Al-Aqeel, S.; Ryu, T.; Zhang, H.; Chandramouli, K.H.; Ravasi, T. Transcriptome and Proteome Studies Reveal Candidate Attachment Genes during the Development of the Barnacle Amphibalanus Amphitrite. Front. Mar. Sci. 2016, 3, 171. [Google Scholar] [CrossRef]
- Chen, Z.-F.; Zhang, H.; Wang, H.; Matsumura, K.; Wong, Y.H.; Ravasi, T.; Qian, P.-Y. Quantitative Proteomics Study of Larval Settlement in the Barnacle Balanus amphitrite. PLoS ONE 2014, 9, e88744. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Perez, D.; Almeida, D.; Wissing, J.; Machado, A.M.; Jänsch, L.; Castro, L.F.; Antunes, A.; Vasconcelos, V.; Campos, A.; Cunha, I. The Quantitative Proteome of the Cement and Adhesive Gland of the Pedunculate Barnacle, Pollicipes pollicipes. Int. J. Mol. Sci. 2020, 21, 2524. [Google Scholar] [CrossRef]
- Liang, C.; Strickland, J.; Ye, Z.; Wu, W.; Hu, B.; Rittschof, D. Biochemistry of Barnacle Adhesion: An Updated Review. Front. Mar. Sci. 2019, 6, 565. [Google Scholar] [CrossRef]
- Rocha, M.; Antas, P.; Castro, L.F.C.; Campos, A.; Vasconcelos, V.; Pereira, F.; Cunha, I. Comparative Analysis of the Adhesive Proteins of the Adult Stalked Goose Barnacle Pollicipes pollicipes (Cirripedia: Pedunculata). Mar. Biotechnol. 2019, 21, 38–51. [Google Scholar] [CrossRef] [PubMed]
- So, C.R.; Scancella, J.M.; Fears, K.P.; Essock-Burns, T.; Haynes, S.E.; Leary, D.H.; Diana, Z.; Wang, C.; North, S.; Oh, C.S.; et al. Oxidase Activity of the Barnacle Adhesive Interface Involves Peroxide-Dependent Catechol Oxidase and Lysyl Oxidase Enzymes. Acs Appl. Mater. Interfaces 2017, 9, 11493–11505. [Google Scholar] [CrossRef] [PubMed]
- Fears, K.P.; Orihuela, B.; Rittschof, D.; Wahl, K.J. Acorn Barnacles Secrete Phase-Separating Fluid to Clear Surfaces Ahead of Cement Deposition. Adv. Sci. 2018, 5, 1–7. [Google Scholar] [CrossRef]
- Southward, A.J. Cirripedia—non-parasitic Thoracica. In European Register of Marine Species: A Check-List of the Marine Species in Europe and a Bibliography of Guides to Their Identification. Collection Patrimoines Naturels; Costello, M.J., Emblow, C., White, R.J., Eds.; Muséum National d’Histoire Naturelle: Paris, France, 2001; Volume 50, pp. 280–283. [Google Scholar]
- Worms Lepas Anatifera Linnaeus. 1758. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=106149 (accessed on 3 February 2021).
- Tsikhon-Lukanina, E.A.; Reznichenko, O.G.; Lukasheva, T.A. Feeding and Spawning of the Goose Barnacle Lepas anatifera (Cirripedia, Lepadidae) on Floating Substrates in the Open Northwestern Pacific Ocean. Entomol. Rev. C/C Entomol. Obozr. 2001, 81, S48–S54. [Google Scholar]
- Sousa, A.; Jacinto, D.; Penteado, N.; Martins, P.; Fernandes, J.; Silva, T.; Castro, J.J.; Cruz, T. Patterns of distribution and abundance of the stalked barnacle (Pollicipes pollicipes) in the central and southwest coast of continental Portugal. J. Sea Res. 2013, 83, 187–194. [Google Scholar] [CrossRef]
- Whitehead, T.O.; Biccard, A.; Griffiths, C.L. South African pelagic goose barnacles (Cirripedia, Thoracica): Substratum preferences and influence of plastic debris on abundance and distribution. Crustaceana 2011, 84, 635–649. [Google Scholar] [CrossRef]
- Lacombe, D.; Liguoru, V.R. Comparative Histological Studies of the Cement Apparatus of Lepas anatifera and Balanus tintinnabulum. Biol. Bull. 1969, 137, 170–180. [Google Scholar] [CrossRef]
- Gohad, N.V.; Aldred, N.; Hartshorn, C.M.; Lee, Y.J.; Cicerone, M.T.; Orihuela, B.; Clare, A.S.; Rittschof, D.; Mount, A.S. Synergistic roles for lipids and proteins in the permanent adhesive of barnacle larvae. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Kamino, K.; Nakano, M.; Kanai, S. Significance of the conformation of building blocks in curing of barnacle underwater adhesive. FEBS J. 2012, 279, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Anil, A.; Desai, D. Barnacles and Their Significance in Biofouling. In Operational and Environmental Consequences of Large Industrial Cooling Water Systems; Vayalam, J.P.V., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 65–93. [Google Scholar]
- Yap, F.C.; Wong, W.L.; Maule, A.G.; Brennan, G.P.; Chong, V.C.; Lim, L.H.S. First evidence for temporary and permanent adhesive systems in the stalked barnacle cyprid, Octolasmis angulata. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Kugele, M.; Yule, A.B. Active relocation in lepadomorph barnacles. J. Mar. Biol. Assoc. 2000, 80, 103–111. [Google Scholar] [CrossRef]
- Moriarty, J.E.; Sachs, J.A.; Jones, K. Directional locomotion in a turtle barnacle, Chelonibia testudinaria, on green turtles, Chelonia mydas. Mar. Turt. Newsl. 2008, 119, 1–4. [Google Scholar]
- Ferrier, G.A.; Kim, S.J.; Kaddis, C.S.; Loo, J.A.; Ann Zimmer, C.; Zimmer, R.K. Multifuncin: A Multifunctional Protein Cue Induces Habitat Selection by, and Predation on, Barnacles. In Proceedings of the Integrative and Comparative Biology, Portland, Oregon, 3–7 January 2016; Volume 56, pp. 901–913. [Google Scholar]
- Smith, P.A.; Clare, A.S.; Rees, H.H.; Prescott, M.C.; Wainwright, G.; Thorndyke, M.C. Identification of methyl farnesoate in the cypris larva of the barnacle, Balanus amphitrite, and its role as a juvenile hormone. Insect Biochem. Mol. Biol. 2000, 30, 885–890. [Google Scholar] [CrossRef]
- Naldrett, M.J.; Kaplan, D.L. Characterization of barnacle (Balanus eburneus and B. cenatus) adhesive proteins. Mar. Biol. 1997, 127, 629–635. [Google Scholar] [CrossRef]
- Mori, Y.; Urushida, Y.; Nakano, M.; Uchiyama, S.; Kamino, K. Calcite-specific coupling protein in barnacle underwater cement. FEBS J. 2007, 274, 6436–6446. [Google Scholar] [CrossRef]
- He, L.-S.; Zhang, G.; Qian, P.-Y. Characterization of Two 20kDa-Cement Protein (cp20k) Homologues in Amphibalanus amphitrite. PLoS ONE 2013, 8, e64130. [Google Scholar] [CrossRef]
- Lin, H.C.; Wong, Y.H.; Tsang, L.M.; Chu, K.H.; Qian, P.Y.; Chan, B.K.K. First study on gene expression of cement proteins and potential adhesion-related genes of a membranous-based barnacle as revealed from Next-Generation Sequencing technology. Biofouling 2014, 30, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [PubMed]
- Schlessinger, A.; Rost, B. Protein flexibility and rigidity predicted from sequence. Proteins Struct. Funct. Bioinforma. 2005, 61, 115–126. [Google Scholar] [CrossRef]
- Campos, A.; Danielsson, G.; Farinha, A.P.; Kuruvilla, J.; Warholm, P.; Cristobal, S. Shotgun proteomics to unravel marine mussel (Mytilus edulis) response to long-term exposure to low salinity and propranolol in a Baltic Sea microcosm. J. Proteom. 2016, 137, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Scheerlinck, E.; Dhaenens, M.; Van Soom, A.; Peelman, L.; De Sutter, P.; Van Steendam, K.; Deforce, D. Minimizing technical variation during sample preparation prior to label-free quantitative mass spectrometry. Anal. Biochem. 2015, 490, 14–19. [Google Scholar] [CrossRef]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef]
- Sielaff, M.; Kuharev, J.; Bohn, T.; Hahlbrock, J.; Bopp, T.; Tenzer, S.; Distler, U. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. 2017, 16, 4060–4072. [Google Scholar] [CrossRef]
- Machado, A.M.; Sarropoulou, E.; Castro, L.F.C.; Vasconcelos, V.; Cunha, I. An important resource for understanding bio-adhesion mechanisms: Cement gland transcriptomes of two goose barnacles, Pollicipes pollicipes and Lepas anatifera (Cirripedia, Thoracica). Mar. Genom. 2019, 45. [Google Scholar] [CrossRef]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Hönigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. PredictProtein-an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, 337–343. [Google Scholar] [CrossRef]
- Lengerer, B.; Algrain, M.; Lefevre, M.; Delroisse, J.; Hennebert, E.; Flammang, P. Interspecies comparison of sea star adhesive proteins. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Davey, P.A.; Power, A.M.; Santos, R.; Bertemes, P.; Ladurner, P.; Palmowski, P.; Clarke, J.; Flammang, P.; Lengerer, B.; Hennebert, E.; et al. Omics-based molecular analyses of adhesion by aquatic invertebrates. Biol. Rev. 2021, brv.12691. [Google Scholar] [CrossRef] [PubMed]

| iBAQ Rank | Barnacle Specific Leading Cement Proteins | No. Res. | MM (×1000) | pI | Instability Index a | Hydropathy (GRAVY) b | % Neg. Res. | % Pos. Res. | Aliphatic Index c | Aromatic Res. (%) | Secondary Structure Composition (%) | Solvent Accessibility (%) | Protein Disorder d (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loop | α-Helix | β-Sheet | Exposed | Interm. | Buried | ||||||||||||
| 4 | DN73117_c2_g1_i2.p1 | 341 | 39.35 | 11.37 | 80.43 | −1.603 | 17.0 | 24.6 | 15.43 | 13.2 | 96.19 | 0.00 | 3.81 | 91.2 | 0.59 | 8.21 | 70.01 |
| 9 | DN70620_c0_g3_i1.p2 | 134 | 14.93 | 11.78 | 34.87 | −0.26 | 4.5 | 21.6 | 98.28 | 5.2 | 35.82 | 43.28 | 20.9 | 51.49 | 12.69 | 35.82 | 62.69 |
| 13 | DN70620_c1_g1_i2.p1 | 163 | 17.71 | 9.91 | 25.09 | 0.082 | 6.8 | 12.9 | 101.66 | 4.3 | 43.56 | 34.97 | 21.47 | 46.63 | 14.11 | 39.26 | 28.22 |
| 14 | DN65601_c0_g1_i1.p1 | 538 | 57.15 | 5.14 | 37.24 | −0.463 | 12.4 | 11.0 | 71.12 | 4.4 | 63.94 | 9.85 | 26.21 | 48.33 | 5.02 | 46.65 | 17.66 |
| 23 | DN64372_c0_g1_i2.p1 | 161 | 17.39 | 6.29 | 24.32 | 0.181 | 11.8 | 11.2 | 107.83 | 8.7 | 48.45 | 23.60 | 27.95 | 45.96 | 6.21 | 47.83 | 16.8 |
| 26 | DN67538_c0_g1_i1.p3 | 108 | 12.42 | 9.05 | 67.8 | −0.936 | 5.6 | 13.9 | 45.41 | 7.4 | 72.22 | 15.74 | 12.04 | 64.81 | 6.48 | 28.7 | 81.48 |
| 30 | DN69760_c0_g1_i1.p1 | 128 | 13.38 | 8.15 | 41.51 | −0.045 | 9.4 | 10.2 | 89.38 | 3.9 | 51.59 | 42.97 | 5.47 | 54.69 | 3.91 | 41.41 | 78.9 |
| 42 | DN61611_c0_g1_i1.p1 | 413 | 42.31 | 6.07 | 35.18 | −0.289 | 9.7 | 7.8 | 71.67 | 2.2 | 79.18 | 11.62 | 9.2 | 61.5 | 3.39 | 35.11 | 55.2 |
| 44 | DN63562_c0_g1_i1.p1 | 420 | 43.24 | 10.18 | 66.90 | −0.979 | 10.9 | 15.0 | 42.71 | 4.7 | 97.86 | 0.00 | 2.14 | 92.62 | 1.19 | 6.19 | 97.6 |
| 46 | DN64031_c0_g1_i1.p1 | 368 | 40.99 | 9.75 | 60.52 | −0.791 | 12.0 | 15.5 | 67.04 | 4.9 | 71.47 | 12.50 | 16.03 | 56.52 | 2.72 | 40.76 | 30.16 |
| 52 | DN61926_c0_g1_i3.p1 | 112 | 12.10 | 9.25 | 30.56 | 0.004 | 4.5 | 16.1 | 68.04 | 8.1 | 66.07 | 25.00 | 8.93 | 52.68 | 5.36 | 41.96 | 60.71 |
| 53 | DN56671_c0_g2_i1.p1 | 116 | 12.63 | 6.26 | 44.18 | 0.199 | 9.5 | 8.6 | 95.95 | 6.9 | 49.14 | 44.83 | 6.03 | 49.14 | 6.03 | 44.83 | 47.41 |
| 54 | DN72668_c0_g1_i2.p1 | 1120 | 124.85 | 9.27 | 49.24 | −0.584 | 10.5 | 14.1 | 66.96 | 7.4 | 63.48 | 17.32 | 19.2 | 51.07 | 5.09 | 43.84 | 28.75 |
| 61 | DN69827_c0_g1_i5.p1 | 285 | 30.58 | 5.39 | 81.17 | −0.672 | 11.9 | 9.5 | 57.19 | 6.4 | 94.74 | 2.81 | 2.46 | 77.89 | 2.81 | 19.3 | 90.18 |
| 66 | DN53050_c0_g1_i2.p1 | 112 | 12.56 | 5.26 | 43.07 | −0.848 | 17.9 | 16.1 | 52.95 | 8.1 | 66.96 | 14.29 | 18.75 | 71.43 | 6.25 | 22.32 | 97.54 |
| 89 | DN67416_c2_g1_i13.p1 | 122 | 12.47 | 9.93 | 42.42 | −0.301 | 8.2 | 9.8 | 70.33 | 4.9 | 66.39 | 22.13 | 11.48 | 68.03 | 2.46 | 29.51 | 77.87 |
| 93 | DN62666_c1_g1_i3.p1 | 312 | 34.10 | 5.46 | 59.45 | −0.508 | 13.5 | 10.9 | 84.1 | 3.5 | 75.32 | 12.50 | 12.18 | 56.41 | 3.85 | 39.74 | 17.63 |
| 116 | DN62022_c1_g1_i2.p1 | 344 | 37.47 | 10.03 | 48.67 | −0.394 | 9.6 | 16.6 | 83.63 | 4.4 | 57.56 | 22.67 | 19.77 | 49.13 | 9.59 | 41.28 | 23.84 |
| 118 | DN58739_c0_g1_i1.p1 | 171 | 18.71 | 9.38 | 75.75 | −0.309 | 7.0 | 14.6 | 76.9 | 4.6 | 76.61 | 10.53 | 12.87 | 52.05 | 8.19 | 39.77 | 87.72 |
| 1 | DN69987_c0_g1_i2.p1 (LA100k) | 924 | 99.8 | 9.81 | 10.07 | 0.205 | 5.8 | 8.1 | 107.6 | 8.60 | 54.11 | 35.61 | 10.28 | 37.01 | 9.52 | 53.46 | 0.32 |
| 2 | DN63945_c0_g1_i4.p1 (LA52k) | 380 | 43.3 | 10.38 | 50.11 | −0.104 | 2.4 | 12.1 | 89.6 | 15.8 | 55.79 | 42.37 | 1.84 | 38.84 | 12.37 | 50.79 | 0.8 |
| 3 | DN40455_c0_g1_i1.p1 (LA43k-L) | 484 | 46.9 | 4.42 | 38.05 | −0.323 | 8.9 | 5.4 | 67.9 | 1.40 | 92.98 | 3.10 | 3.93 | 91.32 | 0.62 | 8.06 | 90.70 |
| 5 | DN67731_c0_g1_i1.p1 (LA52k) | 775 | 85.6 | 10.06 | 41.77 | 0.033 | 3.0 | 10.1 | 101.5 | 12.70 | 65.55 | 26.06 | 8.39 | 35.23 | 11.61 | 53.16 | 0.00 |
| 6 | DN69987_c0_g2_i1.p1 (LA100k) | 155 | 16.5 | 6.08 | 39.81 | 0.228 | 6.5 | 5.8 | 113.3 | 5.70 | 57.42 | 40.00 | 2.58 | 60.00 | 4.52 | 35.48 | 0.0 |
| 8 | DN63945_c0_g1_i2.p1 (LA52k) | 380 | 43.2 | 10.43 | 49.74 | −0.113 | 2.4 | 11.8 | 89.4 | 15.60 | 56.32 | 41.84 | 1.84 | 36.84 | 12.11 | 51.05 | 2.37 |
| 21 | DN62610_c0_g1_i4.p1 (LA52k) | 1253 | 134.1 | 10.11 | 54.20 | 0.126 | 5.9 | 9.3 | 102.7 | 6.10 | 73.18 | 22.91 | 3.91 | 52.19 | 6.70 | 41.10 | 29.93 |
| 31 | DN66462_c0_g1_i1.p1 (LA52k-L3) ¥ | 267 | 26.7 | 4.58 | 30.19 | −0.061 | 12.4 | 7.5 | 83.5 | 2.20 | 73.78 | 16.85 | 9.36 | 73.03 | 0.37 | 26.59 | 83.52 |
| 33 | DN68159_c0_g1_i3.p1 (LA19k) | 218 | 21.7 | 4.50 | 50.61 | −0.172 | 13.3 | 6.9 | 78.0 | 1.80 | 68.81 | 6.88 | 24.31 | 68.81 | 3.21 | 27.98 | 88.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domínguez-Pérez, D.; Almeida, D.; Wissing, J.; Machado, A.M.; Jänsch, L.; Antunes, A.; Castro, L.F.; Vasconcelos, V.; Campos, A.; Cunha, I. Proteogenomic Characterization of the Cement and Adhesive Gland of the Pelagic Gooseneck Barnacle Lepas anatifera. Int. J. Mol. Sci. 2021, 22, 3370. https://doi.org/10.3390/ijms22073370
Domínguez-Pérez D, Almeida D, Wissing J, Machado AM, Jänsch L, Antunes A, Castro LF, Vasconcelos V, Campos A, Cunha I. Proteogenomic Characterization of the Cement and Adhesive Gland of the Pelagic Gooseneck Barnacle Lepas anatifera. International Journal of Molecular Sciences. 2021; 22(7):3370. https://doi.org/10.3390/ijms22073370
Chicago/Turabian StyleDomínguez-Pérez, Dany, Daniela Almeida, Josef Wissing, André M. Machado, Lothar Jänsch, Agostinho Antunes, Luís Filipe Castro, Vitor Vasconcelos, Alexandre Campos, and Isabel Cunha. 2021. "Proteogenomic Characterization of the Cement and Adhesive Gland of the Pelagic Gooseneck Barnacle Lepas anatifera" International Journal of Molecular Sciences 22, no. 7: 3370. https://doi.org/10.3390/ijms22073370









