Two Be or Not Two Be: The Nuclear Autoantigen La/SS-B Is Able to Form Dimers and Oligomers in a Redox Dependent Manner
Abstract
1. Introduction
2. Results
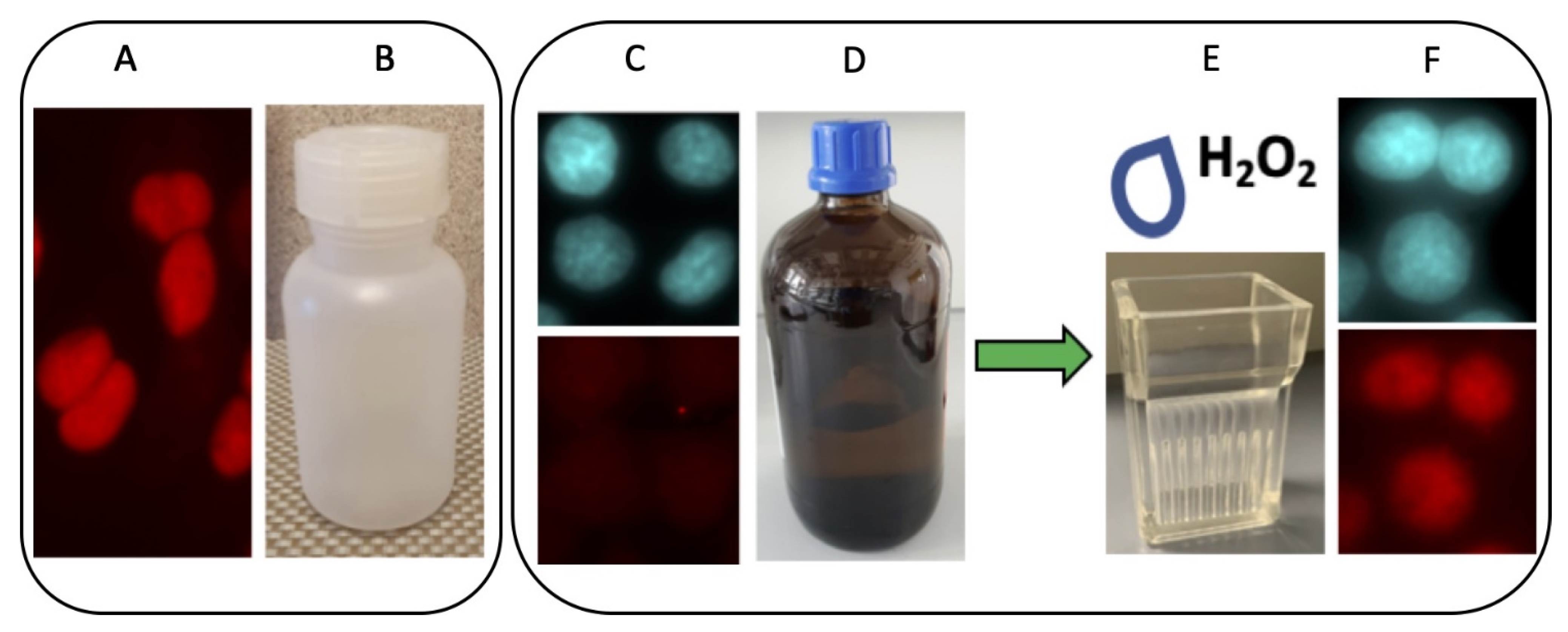
2.1. The Accessibility of the Epitope Recognized by the Anti-La mAb 7B6 Is Dependent on Oxidation
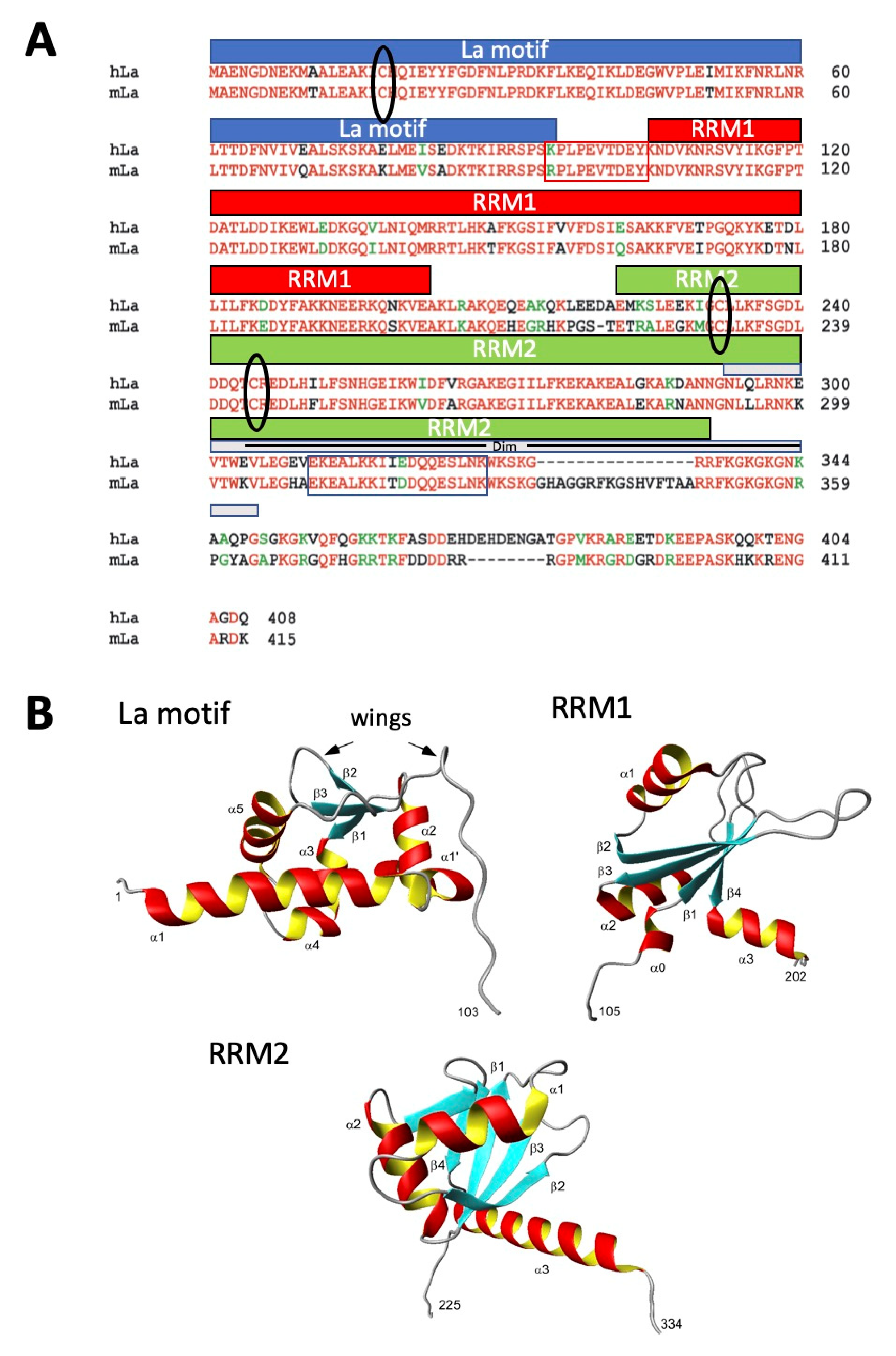
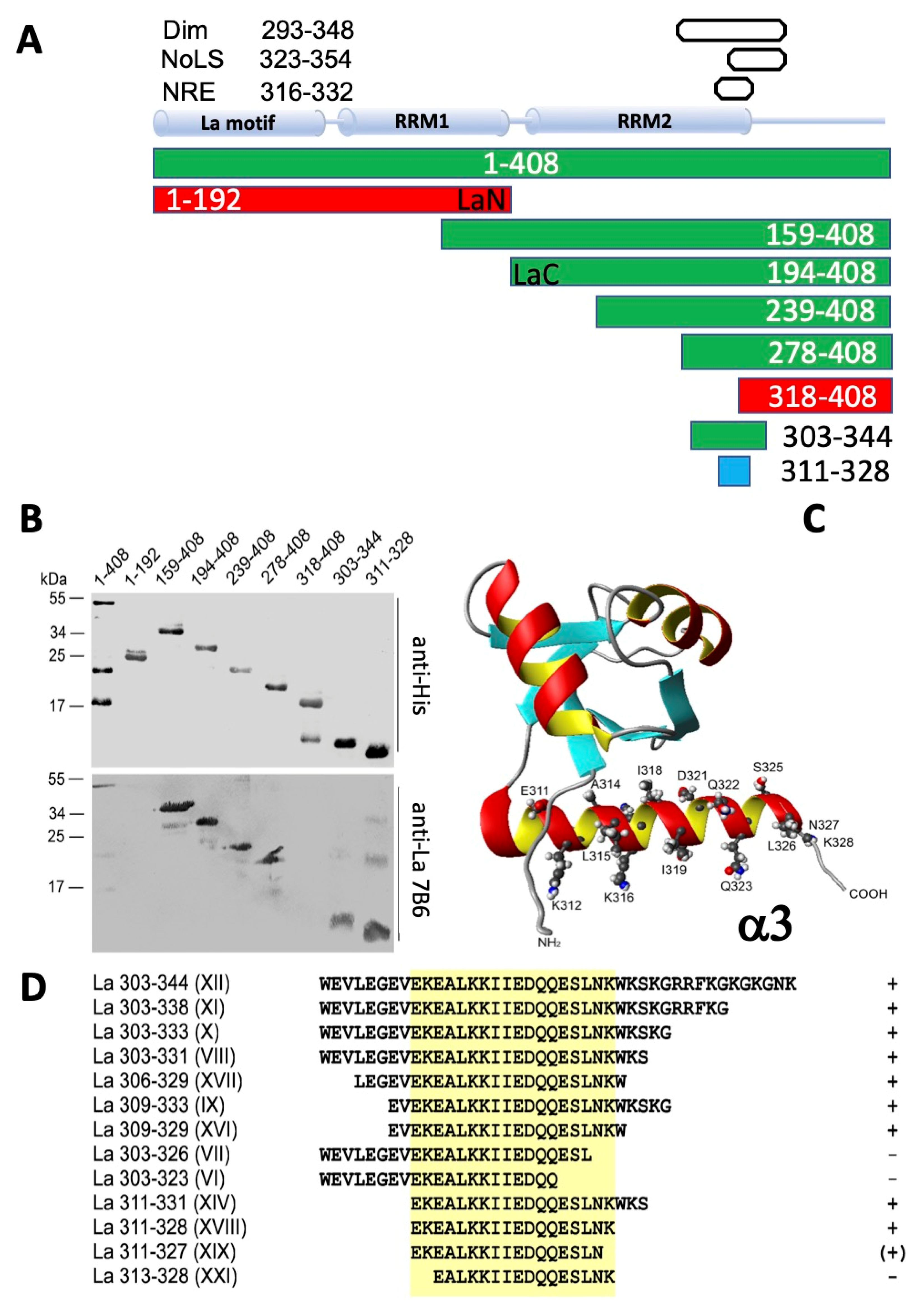
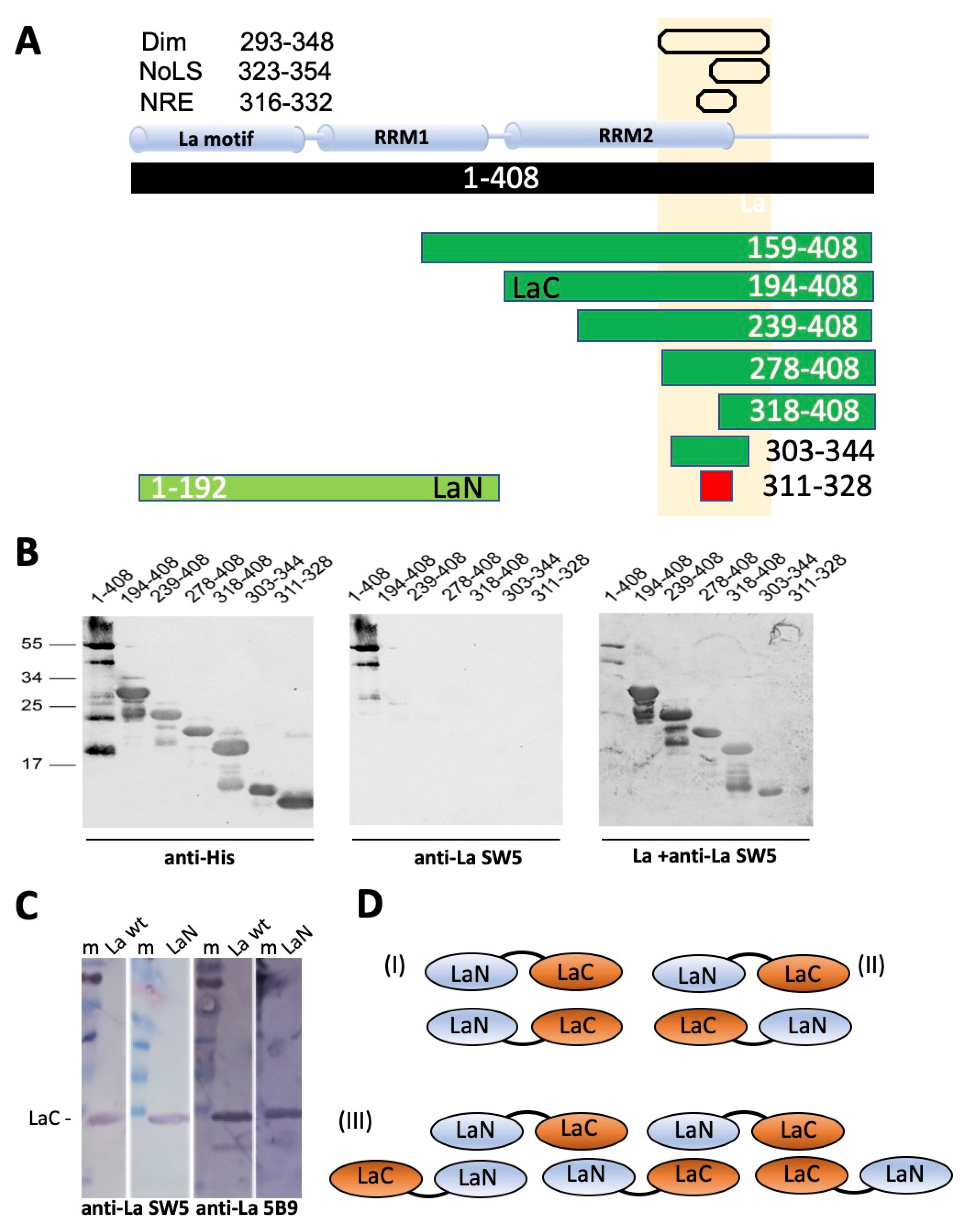
2.2. The 7B6 Epitope Is Part of the Dimerization Domain, the Nuclear Retention Element, and the Nucleolar Localization Signal
2.3. Further Characterization of the Epitope Sequence Recognized by the Anti-La mAb 7B6
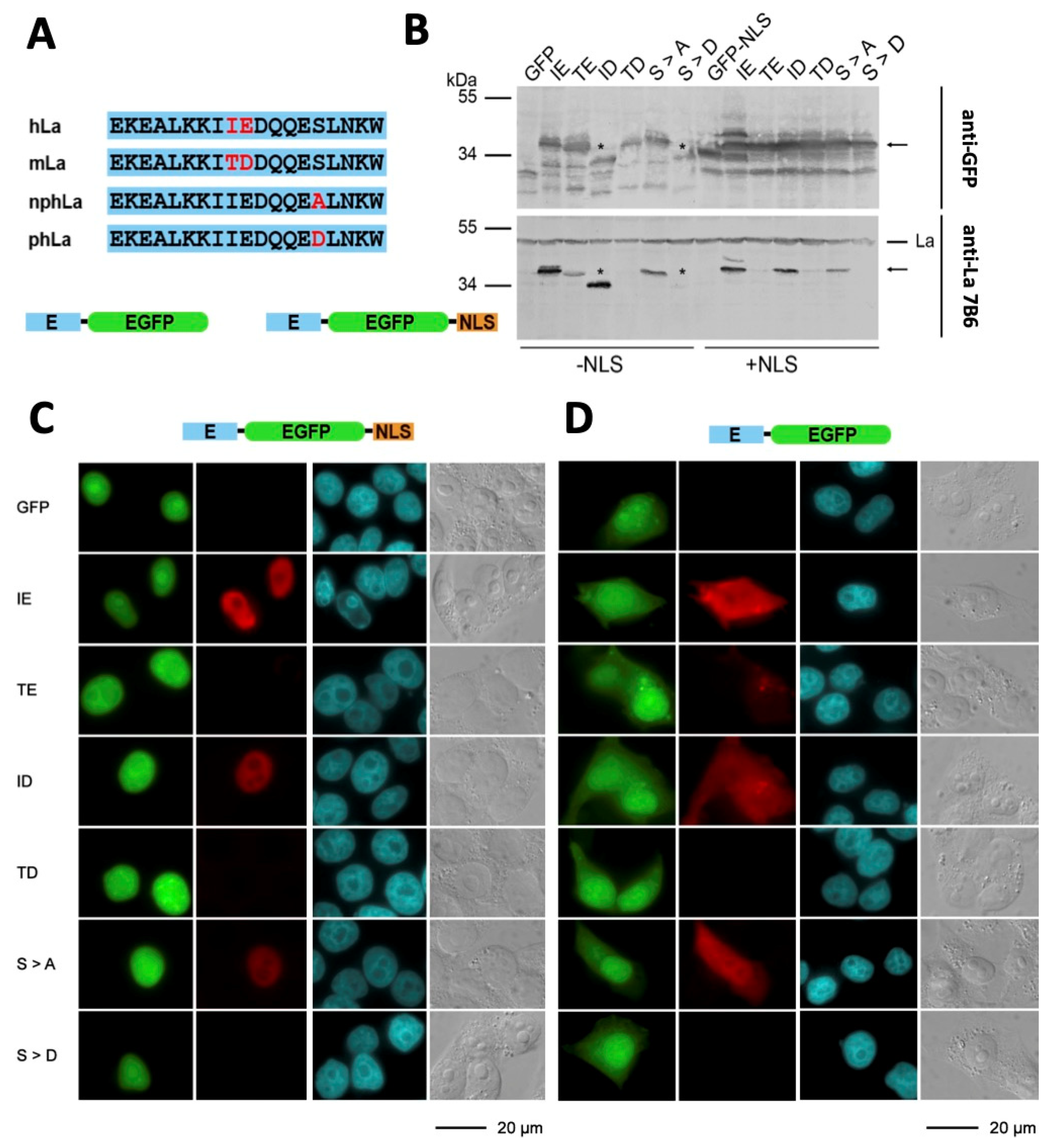
2.4. The 7B6 Epitope Undergoes Posttranslational Modifications
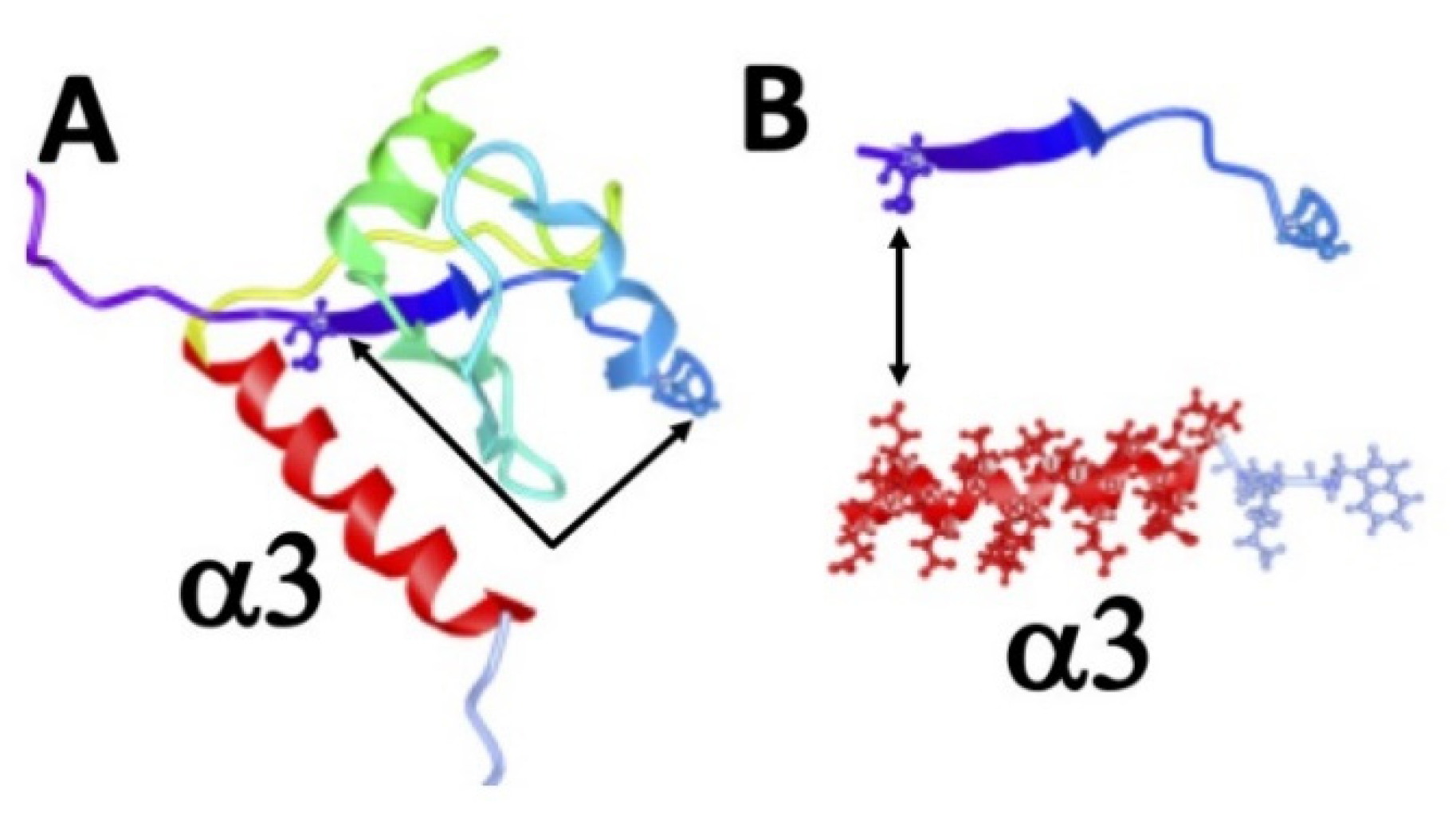
2.5. The 7B6 Epitope Representing the α3−Helix of the RRM2 Domain of La Protein Alone Is Not Sufficient for Dimerization
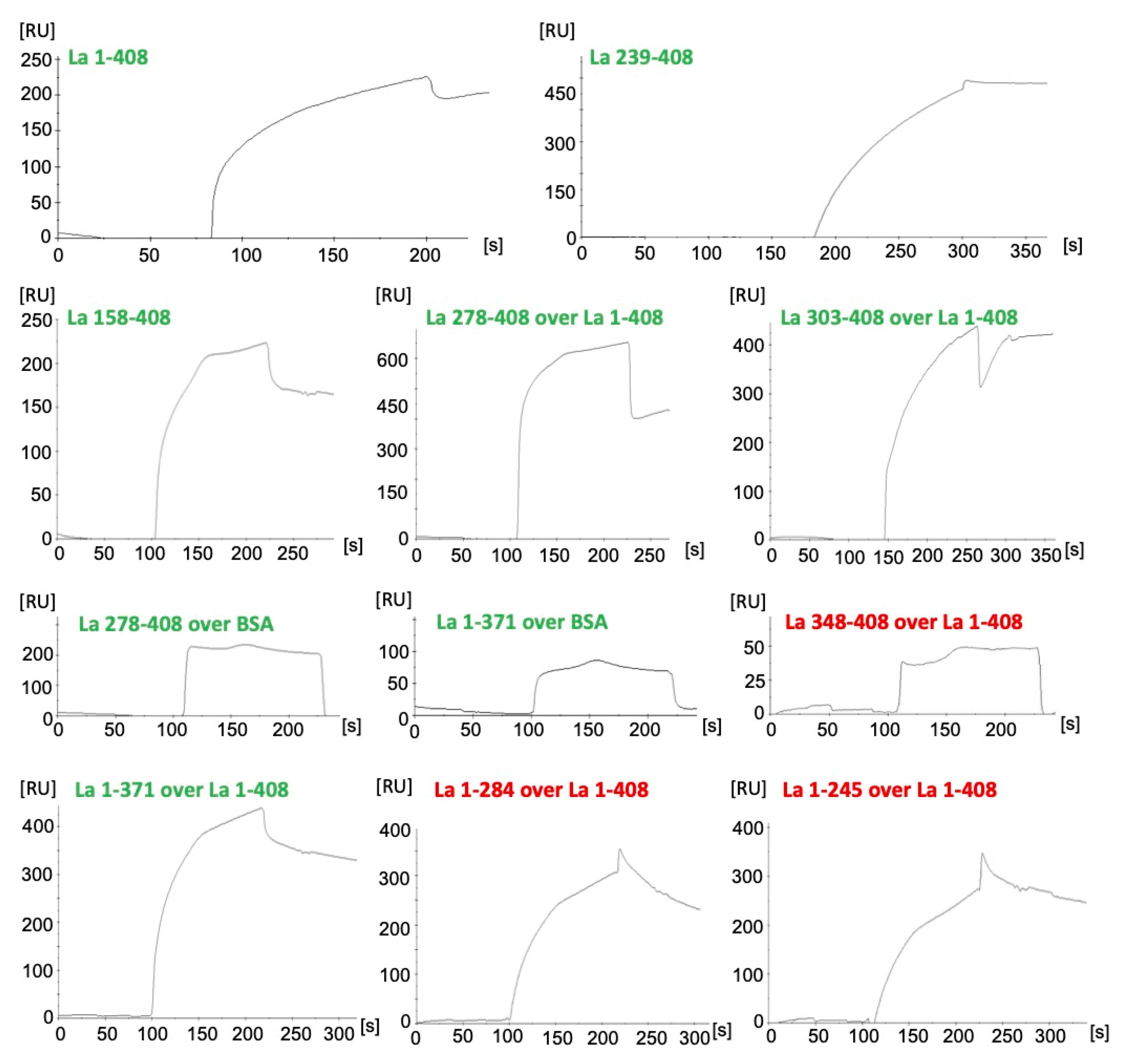
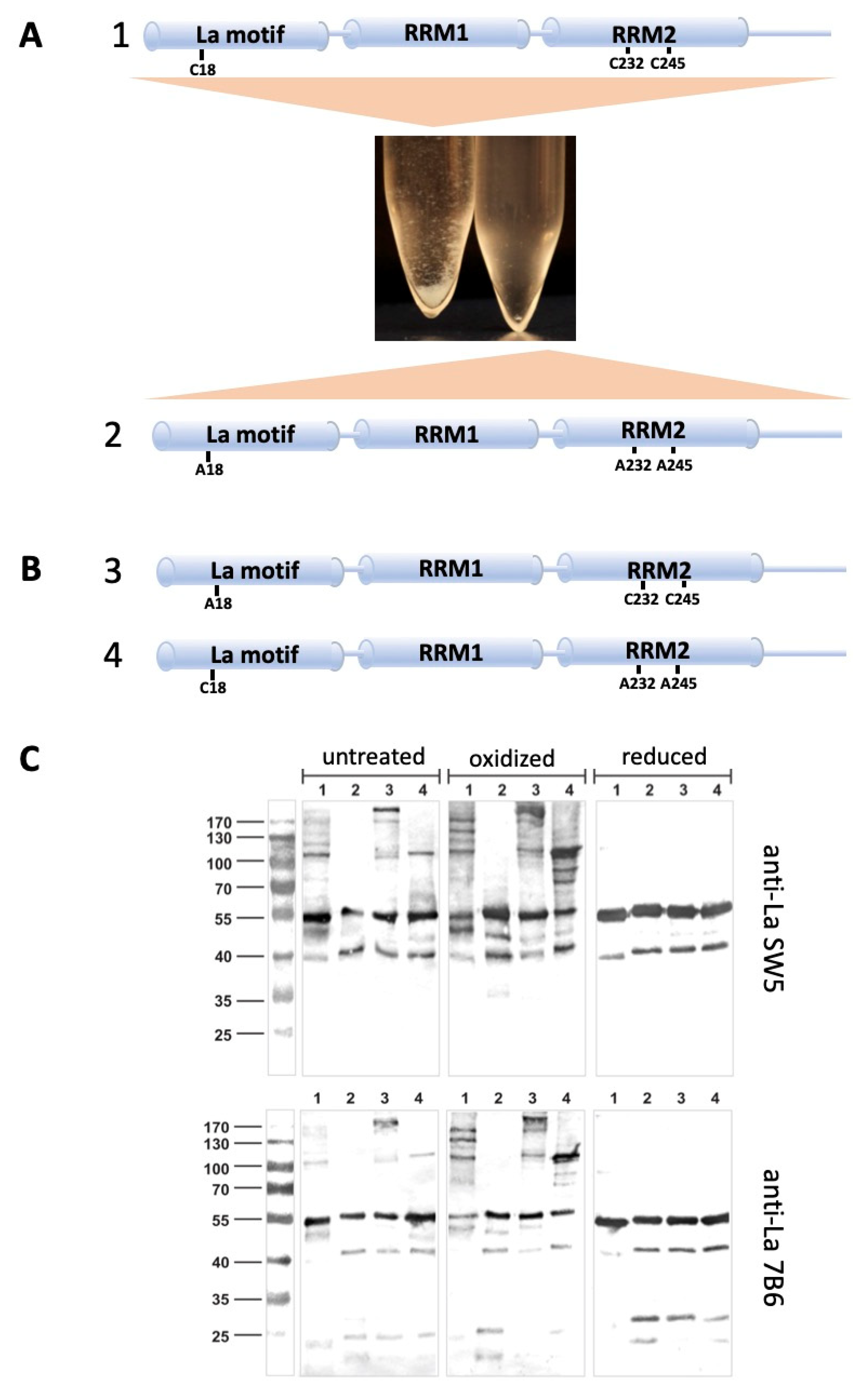
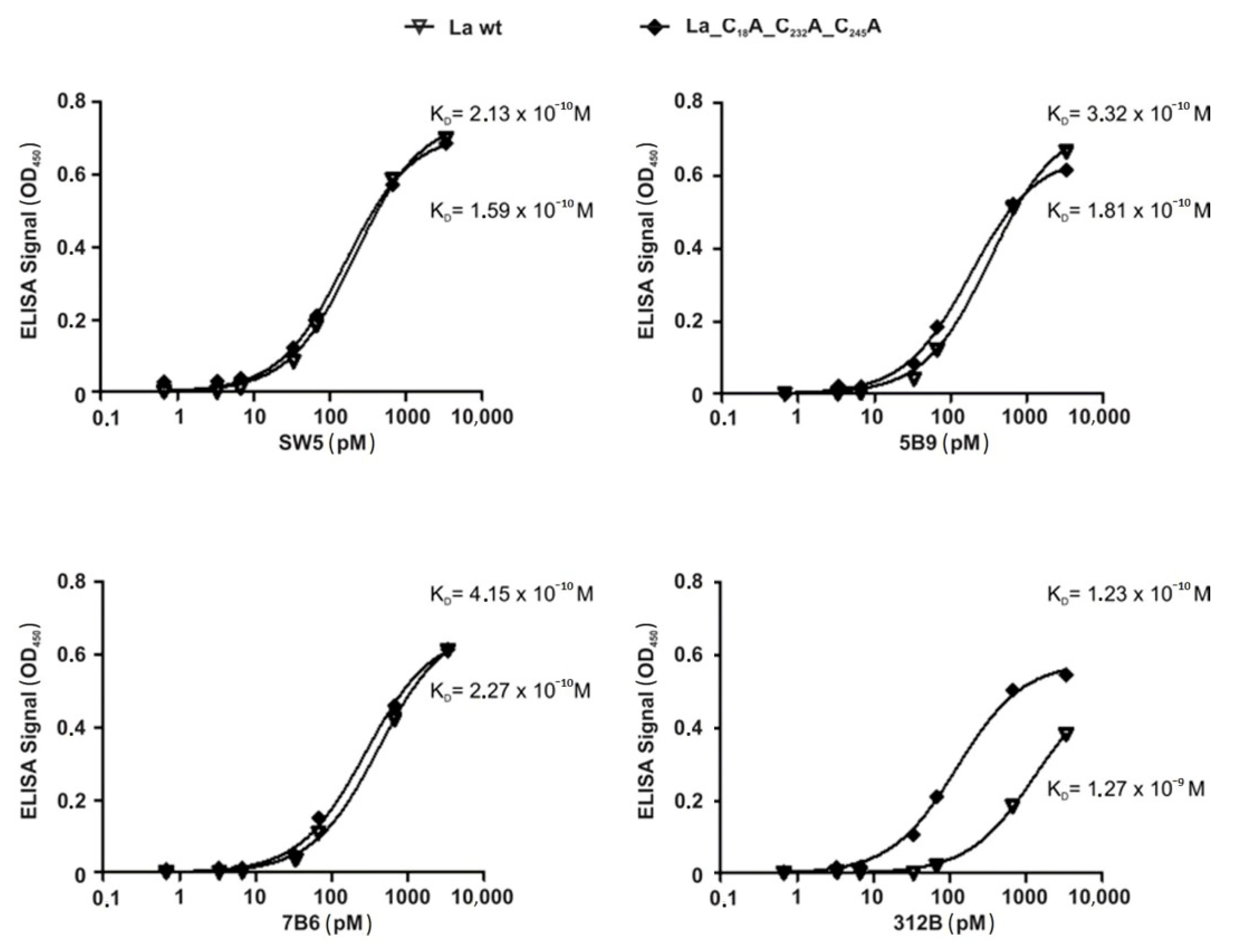
2.6. La Protein Forms Dimers and Oligomers That Can Be Stabilized by Disulfide Bridges
2.7. Antibodies Recognizing the Reduced Form of La Protein
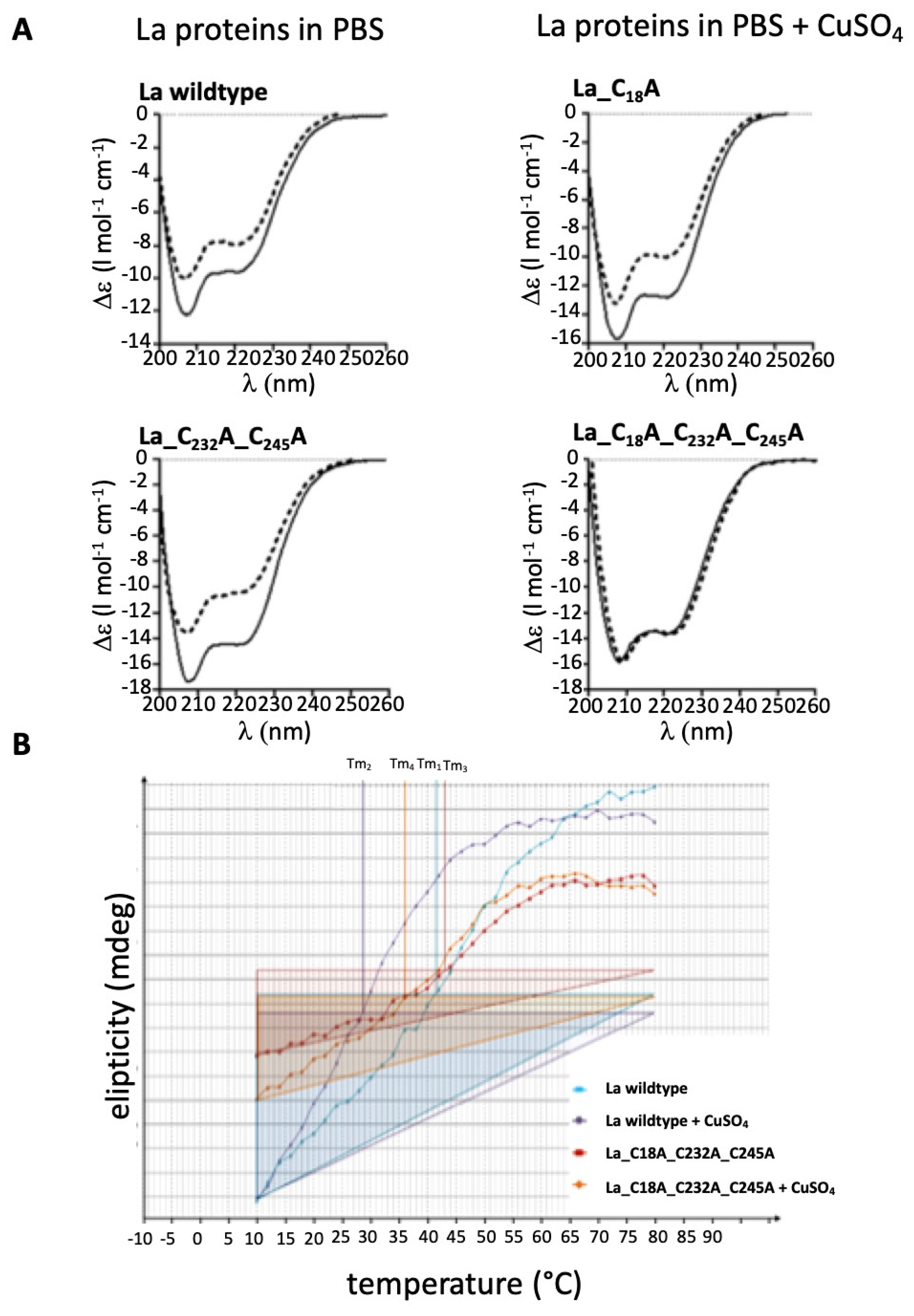
2.8. Evidence for Redox Dependent Conformational Changes of La Protein
2.9. According to the Melting Temperature, the Reduced Form of La Protein Has the Highest Stability
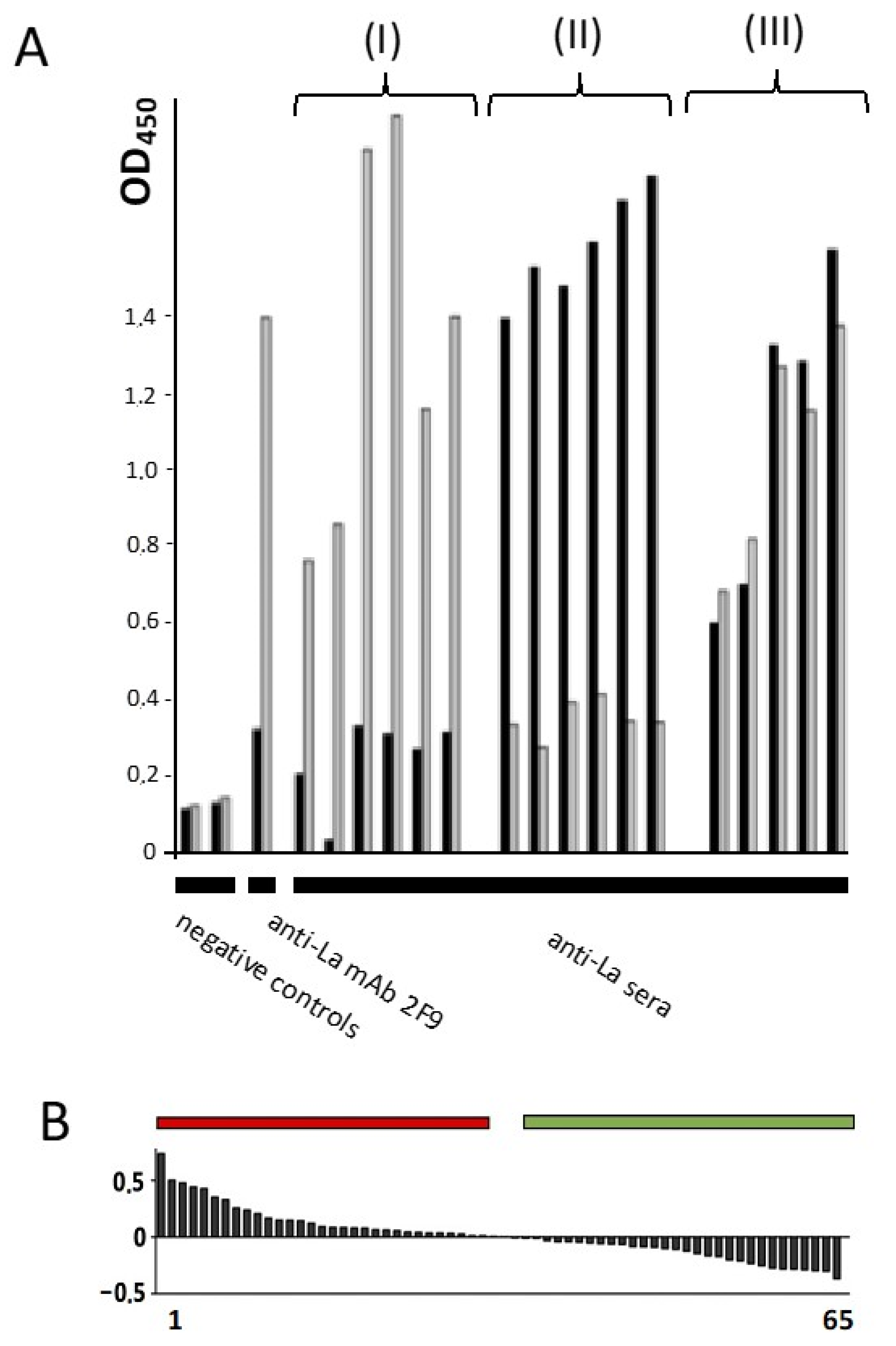
2.10. Redox Dependent Anti-La Antibodies Are Present in Sera of Autoimmune Patients
3. Discussion
4. Materials and Methods
4.1. Recombinant Human La Protein Expression and Characterization
4.2. Anti-La mAbs
4.3. Redox ELISA
4.4. Estimation of CD Spectra
4.5. Surface Plasmon Resonance
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, E.M. Antinuclear Antibodies: Diagnostic Markers for Autoimmune Diseases and Probes for Cell Biology. Adv. Immunol. 1989, 44, 93–151. [Google Scholar] [CrossRef]
- Harley, J.B.; Alexander, E.L.; Bias, W.B.; Fox, O.F.; Provost, T.T.; Reichlin, M.; Yamagata, H.; Arnett, F.C. Anti-Ro (SS-A) and Anti-La (SS-B) in patients with Sjögren’s syndrome. Arthritis Rheum. 1986, 29, 196–206. [Google Scholar] [CrossRef]
- Damoiseaux, J.; Andrade, L.E.C.; Carballo, O.G.; Conrad, K.; Francescantonio, P.L.C.; Fritzler, M.J.; De La Torre, I.G.; Herold, M.; Klotz, W.; Cruvinel, W.D.M.; et al. Clinical relevance of HEp-2 indirect immunofluorescent patterns: The International Consensus on ANA patterns (ICAP) perspective. Ann. Rheum. Dis. 2019, 78, 879–889. [Google Scholar] [CrossRef]
- Stefano, J.E. Purified lupus antigen la recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 1984, 36, 145–154. [Google Scholar] [CrossRef]
- Wolin, S.L.; Cedervall, T. The La Protein. Annu. Rev. Biochem. 2002, 71, 375–403. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.; Pfeifer, K.; Schröder, H.C.; Müller, W.E.; Schrdder, H.-C. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell 1990, 60, 85–93. [Google Scholar] [CrossRef]
- Huhn, P. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 1997, 25, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Sommer, G.; Sendlmeier, C.; Heise, T. Salt-Dependent Modulation of the RNA Chaperone Activity of RNA-Binding Protein La. Methods Mol. Biol. 2019, 2106, 121–136. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Meerovitch, K.; Lee, H.S.; Dholakia, J.N.; Kenan, D.J.; Agol, V.I.; Sonenberg, N. Internal translation initiation on poliovirus RNA: Further characterization of La function in poliovirus translation in vitro. J. Virol. 1994, 68, 1544–1550. [Google Scholar] [CrossRef]
- Ali, N.; Pruijn, G.J.M.; Kenan, D.J.; Keene, J.D.; Siddiqui, A. Human La Antigen Is Required for the Hepatitis C Virus Internal Ribosome Entry Site-mediated Translation. J. Biol. Chem. 2000, 275, 27531–27540. [Google Scholar] [CrossRef]
- Ali, N.; Siddiqui, A. The La antigen binds 5’ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 1997, 94, 2249–2254. [Google Scholar] [CrossRef]
- Holcik, M.; Korneluk, R.G. Functional Characterization of the X-Linked Inhibitor of Apoptosis (XIAP) Internal Ribosome Entry Site Element: Role of La Autoantigen in XIAP Translation. Mol. Cell. Biol. 2000, 20, 4648–4657. [Google Scholar] [CrossRef]
- Petz, M.; Them, N.; Huber, H.; Beug, H.; Mikulits, W. La enhances IRES-mediated translation of laminin B1 during ma-lignant epithelial to mesenchymal transition. Nucleic Acids Res. 2012, 40, 290–302. [Google Scholar] [CrossRef][Green Version]
- Craig, A.W.; Svitkin, Y.V.; Lee, H.S.; Belsham, G.J.; Sonenberg, N. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol. Cell. Biol. 1997, 17, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Jacks, A.; Babon, J.; Kelly, G.; Manolaridis, I.; Cary, P.D.; Curry, S.; Conte, M.R. Structure of the C-Terminal Domain of Human La Protein Reveals a Novel RNA Recognition Motif Coupled to a Helical Nuclear Retention Element. Structres 2003, 11, 833–843. [Google Scholar] [CrossRef]
- Horke, S.; Reumann, K.; Schweizer, M.; Will, H.; Heise, T. Nuclear Trafficking of La Protein Depends on a Newly Identified Nucleolar Localization Signal and the Ability to Bind RNA. J. Biol. Chem. 2004, 279, 26563–26570. [Google Scholar] [CrossRef]
- Maraia, R.J.; Mattijssen, S.; Cruz-Gallardo, I.; Conte, M.R. The La and related RNA-binding proteins (LARPs): Structures, functions, and evolving perspectives. Wiley Interdiscip. Rev. RNA 2017, 8, e1430. [Google Scholar] [CrossRef]
- Alfano, C.; Sanfelice, D.; Babon, J.; Kelly, G.; Jacks, A.; Curry, S.; Conte, M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004, 11, 323–329. [Google Scholar] [CrossRef]
- Kotik-Kogan, O.; Valentine, E.R.; Sanfelice, D.; Conte, M.R.; Curry, S. Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3′ Ends by the Human La Protein. Structures 2008, 16, 852–862. [Google Scholar] [CrossRef]
- Sanfelice, D.; Kelly, G.; Curry, S.; Conte, M.R. NMR assignment of the N-terminal region of human La free and in complex with RNA. Biomol. NMR Assign. 2008, 2, 107–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huth, J.R.; Mendoza, R.; Olejniczak, E.T.; Johnson, R.W.; Cothron, D.A.; Liu, Y.; Lerner, C.G.; Chen, A.J.; Hajduk, P.J. ALARM NMR: A Rapid and Robust Experimental Method to Detect Reactive False Positives in Biochemical Screens. J. Am. Chem. Soc. 2005, 127, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Huth, J.R.; Song, D.; Mendoza, R.R.; Black-Schaefer, C.L.; Mack, J.C.; Dorwin, S.A.; Ladror, U.S.; Severin, J.M.; Walter, K.A.; Bartley, D.M.; et al. Toxicological Evaluation of Thiol-Reactive Compounds Identified Using a La Assay to Detect Reactive Molecules by Nuclear Magnetic Resonance. Chem. Res. Toxicol. 2007, 20, 1752–1759. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, J.L.; Cuellar, M.; Singh, G.; Nelson, K.M.; Strasser, J.M.; Rappe, T.; Xia, Y.; Veglia, G.; Walters, M.A. ALARM NMR for HTS Triage and Chemical Probe Validation. Curr. Protoc. Chem. Biol. 2018, 10, 91–117. [Google Scholar] [CrossRef]
- Kwak, M.S.; Kim, H.S.; Lee, B.; Kim, Y.H.; Son, M.; Shin, J.-S. Immunological Significance of HMGB1 Post-Translational Modification and Redox Biology. Front. Immunol. 2020, 11, 1189. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, N. Intermolecular disulfide bond to modulate protein function as a redox-sensing switch. Amino Acids 2010, 41, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Clancey, C.J.; Gilbert, H.F. Thiol/disulfide exchange in the thioredoxin-catalyzed reductive activation of spinach chlo-roplast fructose-1,6-bisphosphatase. Kinetics and thermodynamics. J. Biol. Chem. 1987, 262, 13545–13549. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, H.C.; Blackburn, E.C.; Freedman, R.B. Comparison of the activities of protein disulphide-isomerase and thioredoxin in catalysing disulphide isomerization in a protein substrate. Biochem. J. 1991, 275, 349–353. [Google Scholar] [CrossRef]
- Zheng, M.; Åslund, F.; Storz, G. Activation of the OxyR Transcription Factor by Reversible Disulfide Bond Formation. Science 1998, 279, 1718–1722. [Google Scholar] [CrossRef]
- Sohn, J.; Rudolph, J. Catalytic and Chemical Competence of Regulation of Cdc25 Phosphatase by Oxidation/Reduction†. Biochemestry 2003, 42, 10060–10070. [Google Scholar] [CrossRef]
- Mikkelsen, R.; Mutenda, K.E.; Mant, A.; Schürmann, P.; Blennow, A. -Glucan, water dikinase (GWD): A plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc. Natl. Acad. Sci. USA 2005, 102, 1785–1790. [Google Scholar] [CrossRef]
- Sevier, C.S.; Kaiser, C.A. Disulfide Transfer between Two Conserved Cysteine Pairs Imparts Selectivity to Protein Oxidation by Ero1. Mol. Biol. Cell 2006, 17, 2256–2266. [Google Scholar] [CrossRef]
- Yang, D.; Han, Z.; Oppenheim, J.J. Alarmins and immunity. Immunol. Rev. 2017, 280, 41–56. [Google Scholar] [CrossRef]
- Zandarashvili, L.; Sahu, D.; Lee, K.; Lee, Y.S.; Singh, P.; Rajarathnam, K.; Iwahara, J. Real-time Kinetics of High-mobility Group Box 1 (HMGB1) Oxidation in Extracellular Fluids Studied by in Situ Protein NMR Spectroscopy. J. Biol. Chem. 2013, 288, 11621–11627. [Google Scholar] [CrossRef]
- Bachmann, M.; Pfeifer, K.; Schröder, H.C.; Müller, W.E. The La antigen shuttles between the nucleus and the cytoplasm in CV-1 cells. Mol. Cell. Biochem. 1989, 85, 103–114. [Google Scholar] [CrossRef]
- Bachmann, M.; Chang, S.; Slor, H.; Kukulies, J.; Müller, W.E. Shuttling of the autoantigen La between nucleus and cell surface after uv irradiation of human keratinocytes. Exp. Cell Res. 1990, 191, 171–180. [Google Scholar] [CrossRef]
- Bachmann, M.; Falke, D.; Schröder, H.-C.; Müller, W.E.G. Intracellular Distribution of the La Antigen in CV-1 Cells after Herpes Simplex Virus Type 1 Infection Compared with the Localization of U Small Nuclear Ribonucleoprotein Particles. J. Gen. Virol. 1989, 70, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Bbachmann, M.; Althoff, H.; Tröster, H.; Selenka, C.; Falke, D.; Müller, W.E.G. Translocation of the Nuclear Autoantigen La to the Cell Surface of Herpes Simplex Virus Type 1 Infected Cells. Autoimmunology 1992, 12, 37–45. [Google Scholar] [CrossRef]
- Bachmann, M.; Schroder, H.; Falke, D.; Muller, W. Alteration of the intracellular localization of the La protein compared with the localization of U snRNPs. Cell Biol. Int. Rep. 1988, 12, 765–789. [Google Scholar] [CrossRef]
- Bachmann, M.; Chang, S.; Bernd, A.; Mayet, W.; Büschenfelde, K.H.M.Z.; Muller, W.E.G. Translocation of the Nuclear Auto Antigen La to cell Surface: Assembly and Disassembly with the Extracellular Matrix. Autoimmunology 1991, 9, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Koristka, S.; Cartellieri, M.; Arndt, C.; Bippes, C.C.; Feldmann, A.; Michalk, I.; Wiefel, K.; Stamova, S.; Schmitz, M.; Ehninger, G.; et al. Retargeting of regulatory T cells to surface-inducible autoantigen La/SS-B. J. Autoimmun. 2013, 42, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Grölz, D.; Bartsch, H.; Tröster, H.; Bachmann, M. The nuclear autoantigen La/SS-B: Mapping and sequencing of the gene and the three retropseudogenes. Gene 1997, 191, 23–29. [Google Scholar] [CrossRef]
- Semsei, I.; Tröster, H.; Bartsch, H.; Schwemmle, M.; Igloi, G.L.; Bachmann, M. Isolation of rat cDNA clones coding for the autoantigen SS-B/La: Detection of species-specific variations. Gene 1993, 126, 265–268. [Google Scholar] [CrossRef]
- Grölz, D.; Laubinger, J.; Wilmer, F.; Tröster, H.; Bachmann, M. Transfection Analysis of Expression of mRNA Isoforms Encoding the Nuclear Autoantigen La/SS-B. J. Biol. Chem. 1997, 272, 12076–12082. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.; Grölz, D.; Bartsch, H.; Klein, R.R.; Tröster, H. Analysis of expression of an alternative La (SS-B) cDNA and localization of the encoded N- and C-terminal peptides. Biochim. Biophys. Acta (BBA) Bioenerg. 1997, 1356, 53–63. [Google Scholar] [CrossRef]
- Bachmann, M.; Bartsch, T.; Bippes, C.; Bachmann, D.; Puentes-Cala, E.; Bachmann, J.; Bartsch, H.; Arndt, C.; Koristka, S.; Loureiro, L.; et al. T Cell Mediated Conversion of a Non-Anti-La Reactive B Cell to an Autoreactive Anti-La B Cell by Somatic Hypermutation. Int. J. Mol. Sci. 2021, 22, 1198. [Google Scholar] [CrossRef] [PubMed]
- Tröster, H.; Bartsch, H.; Klein, R.; Metzger, T.E.; Pollak, G.; Semsei, I.; Schwemmle, M.; Pruijn, G.J.M.; Van Venrooij, W.J.; Bachmann, M. Activation of a murine autoreactive B cell by immunization with human recombinant autoantigen La/SS-B: Characterization of the autoepitope. J. Autoimmun. 1995, 8, 825–842. [Google Scholar] [CrossRef][Green Version]
- Von Arnim, A.G.; Deng, X.-W.; Stacey, M.G. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 1998, 221, 35–43. [Google Scholar] [CrossRef]
- Broekhuis, C.H.D.; Neubauer, G.; Van Der Heijden, A.; Mann, M.; Proud, C.G.; Van Venrooij, W.J.; Pruijn, G.J.M. Detailed Analysis of the Phosphorylation of the Human La (SS-B) Autoantigen. (De)phosphorylation Does Not Affect Its Subcellular Distribution†. Biochemestry 2000, 39, 3023–3033. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.R.; Williams, D.G.; Venables, P.J.; Maini, R.N.; Maimi, R.N. Monoclonal antibodies to the Sjögren’s syndrome associated antigen SS-B (La). J. Immunol. Methods 1985, 77, 63–76. [Google Scholar] [CrossRef]
- Pruijn, G.J.M.; Thijssen, J.P.H.; Smith, P.R.; Williams, D.G.; Venrooij, W.J. Anti-La Monoclonal Antibodies Recognizing Epitopes Within the RNA-Binding Domain of the La Protein Show Differential Capacities to Immunoprecipitate RNA-Associated La Protein. JBIC J. Biol. Inorg. Chem. 1995, 232, 611–619. [Google Scholar] [CrossRef]
- Bachmann, M. The UniCAR system: A modular CAR T cell approach to improve the safety of CAR T cells. Immunol. Lett. 2019, 211, 13–22. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 2008, 8, 370–380. [Google Scholar] [CrossRef]
- Cartellieri, M.; Koristka, S.; Arndt, C.; Feldmann, A.; Stamova, S.; Von Bonin, M.; Töpfer, K.; Krüger, T.; Geib, M.; Michalk, I.; et al. A Novel Ex Vivo Isolation and Expansion Procedure for Chimeric Antigen Receptor Engrafted Human T Cells. PLoS ONE 2014, 9, e93745. [Google Scholar] [CrossRef] [PubMed]
- Arndt, C.; Bachmann, M.; Bergmann, R.; Berndt, N.; Feldmann, A.; Koristka, S. Theranostic CAR T cell targeting: A brief review. J. Label. Compd. Radiopharm. 2019, 62, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Jureczek, J.; Bergmann, R.; Berndt, N.; Koristka, S.; Kegler, A.; Puentes-Cala, E.; Soto, J.A.; Arndt, C.; Bachmann, M.; Feldmann, A. An oligo-His-tag of a targeting module does not influence its biodistribution and the retargeting capabilities of UniCAR T cells. Sci. Rep. 2019, 9, 10547. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.; Arndt, C.; Feldmann, A.; Bergmann, R.; Bachmann, D.; Koristka, S.; Ludwig, F.; Ziller-Walter, P.; Kegler, A.; Gärtner, S.; et al. A novel nanobody-based target module for retargeting of T lymphocytes to EGFR-expressing cancer cells via the modular UniCAR platform. OncoImmunology 2017, 6, e1287246. [Google Scholar] [CrossRef]
- Feldmann, A.; Hoffmann, A.; Bergmann, R.; Koristka, S.; Berndt, N.; Arndt, C.; Loureiro, L.R.; Kittel-Boselli, E.; Mitwasi, N.; Kegler, A.; et al. Versatile chimeric antigen receptor platform for controllable and combinatorial T cell therapy. OncoImmunology 2020, 9, 1785608. [Google Scholar] [CrossRef] [PubMed]
- Arndt, C.; Fasslrinner, F.; Loureiro, L.R.; Koristka, S.; Feldmann, A.; Bachmann, M. Adaptor CAR Platforms—Next Generation of T Cell-Based Cancer Immunotherapy. Cancers 2020, 12, 1302. [Google Scholar] [CrossRef]
- Koristka, S.; Cartellieri, M.; Feldmann, A.; Arndt, C.; Loff, S.; Michalk, I. Flexible antigen-specific redirection of human regulatory T cells via a novel universal chimeric antigen receptor system. Blood 2014, 124, 3494. [Google Scholar] [CrossRef]
- Cartellieri, M.; Loff, S.; von Bonin, M.; Bejestani, E.P.; Ehninger, A.; Feldmann, A. Unicar: A novel modular retargeting platform technology for CAR T cells. Blood 2015, 126, 5549. [Google Scholar] [CrossRef]
- Koristka, S.; Kegler, A.; Bergmann, R.; Arndt, C.; Feldmann, A.; Albert, S.; Cartellieri, M.; Ehninger, A.; Ehninger, G.; Middeke, J.M.; et al. Engrafting human regulatory T cells with a flexible modular chimeric antigen receptor technology. J. Autoimmun. 2018, 90, 116–131. [Google Scholar] [CrossRef]
- Feldmann, A.; Arndt, C.; Bergmann, R.; Loff, S.; Cartellieri, M.; Bachmann, D.; Aliperta, R.; Hetzenecker, M.; Ludwig, F.; Albert, S.; et al. Retargeting of T lymphocytes to PSCA- or PSMA positive prostate cancer cells using the novel modular chimeric antigen receptor platform technology “UniCAR”. Oncotarget 2017, 8, 31368–31385. [Google Scholar] [CrossRef] [PubMed]
- Arndt, C.; Feldmann, A.; Koristka, S.; Schäfer, M.; Bergmann, R.; Mitwasi, N.; Berndt, N.; Bachmann, D.; Kegler, A.; Schmitz, M.; et al. A theranostic PSMA ligand for PET imaging and retargeting of T cells expressing the universal chimeric antigen receptor UniCAR. OncoImmunology 2019, 8, 1659095. [Google Scholar] [CrossRef] [PubMed]
- Jureczek, J.; Feldmann, A.; Bergmann, R.; Arndt, C.; Berndt, N.; Koristka, S.; Loureiro, L.R.; Mitwasi, N.; Hoffmann, A.; Kegler, A.; et al. Highly Efficient Targeting of EGFR-Expressing Tumor Cells with UniCAR T Cells via Target Modules Based on Cetuximab®. OncoTargets Ther. 2020, 13, 5515–5527. [Google Scholar] [CrossRef]
- Bachmann, D.; Aliperta, R.; Bergmann, R.; Feldmann, A.; Koristka, S.; Arndt, C.; Loff, S.; Welzel, P.; Albert, S.; Kegler, A.; et al. Retargeting of UniCAR T cells with an in vivo synthesized target module directed against CD19 positive tumor cells. Oncotarget 2017, 9, 7487–7500. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, L.R.; Feldmann, A.; Bergmann, R.; Koristka, S.; Berndt, N.; Arndt, C.; Pietzsch, J.; Novo, C.; Videira, P.; Bachmann, M. Development of a novel target module redirecting UniCAR T cells to Sialyl Tn-expressing tumor cells. Blood Cancer J. 2018, 8, 1–6. [Google Scholar] [CrossRef]
- Loureiro, L.R.; Feldmann, A.; Bergmann, R.; Koristka, S.; Berndt, N.; Máthé, D.; Hegedüs, N.; Szigeti, K.; Videira, P.A.; Bachmann, M.; et al. Extended half-life target module for sustainable UniCAR T-cell treatment of STn-expressing cancers. J. Exp. Clin. Cancer Res. 2020, 39, 77. [Google Scholar] [CrossRef] [PubMed]
- Mitwasi, N.; Feldmann, A.; Arndt, C.; Koristka, S.; Berndt, N.; Jureczek, J.; Loureiro, L.R.; Bergmann, R.; Máthé, D.; Hegedüs, N.; et al. “UniCAR”-modified off-the-shelf NK-92 cells for targeting of GD2-expressing tumour cells. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cartellieri, M.; Feldmann, A.; Koristka, S.; Arndt, C.; Loff, S.; Ehninger, A.; Von Bonin, M.; Bejestani, E.P.; Bachmann, M.P. Switching CAR T cells on and off: A novel modular platform for retargeting of T cells to AML blasts. Blood Cancer J. 2016, 6, e458. [Google Scholar] [CrossRef]
- Feldmann, A.; Arndt, C.; Koristka, S.; Berndt, N.; Bergmann, R.; Bachmann, M.P. Conventional CARs versus modular CARs. Cancer Immunol. Immunother. 2019, 68, 1713–1719. [Google Scholar] [CrossRef]
- Koristka, S.; Ziller-Walter, P.; Bergmann, R.; Arndt, C.; Feldmann, A.; Kegler, A.; Cartellieri, M.; Ehninger, A.; Ehninger, G.; Bornhäuser, M.; et al. Anti-CAR-engineered T cells for epitope-based elimination of autologous CAR T cells. Cancer Immunol. Immunother. 2019, 68, 1401–1415. [Google Scholar] [CrossRef] [PubMed]
- Arndt, C.; Loureiro, L.R.; Feldmann, A.; Jureczek, J.; Bergmann, R.; Máthé, D.; Hegedüs, N.; Berndt, N.; Koristka, S.; Mitwasi, N.; et al. UniCAR T cell immunotherapy enables efficient elimination of radioresistant cancer cells. OncoImmunology 2020, 9, 1743036. [Google Scholar] [CrossRef] [PubMed]
- Fok, V.; Friend, K.; Steitz, J.A. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell Biol. 2006, 173, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Simons, F.H.; Broers, F.J.; Van Venrooij, W.J.; Pruijn, G.J. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996, 224, 224–236. [Google Scholar] [CrossRef]
- Rosenblum, J.S.; Pemberton, L.F.; Bonifaci, N.; Blobel, G. Nuclear Import and the Evolution of a Multifunctional RNA-binding Protein. J. Cell Biol. 1998, 143, 887–899. [Google Scholar] [CrossRef]
- Shiroki, K.; Isoyama, T.; Kuge, S.; Ishii, T.; Ohmi, S.; Hata, S.; Suzuki, K.; Takasaki, Y.; Nomoto, A. Intracellular Redistribution of Truncated La Protein Produced by Poliovirus 3Cpro-Mediated Cleavage. J. Virol. 1999, 73, 2193–2200. [Google Scholar] [CrossRef]
- Wójcik, P.; Gęgotek, A.; Žarković, N.; Skrzydlewska, E. Oxidative Stress and Lipid Mediators Modulate Immune Cell Functions in Autoimmune Diseases. Int. J. Mol. Sci. 2021, 22, 723. [Google Scholar] [CrossRef]
- Bona, N.; Pezzarini, E.; Balbi, B.; Daniele, S.M.; Rossi, M.F.; Monje, A.L.; Basiglio, C.L.; Pelusa, H.F.; Arriaga, S.M.M. Oxidative stress, inflammation and disease activity biomarkers in lupus nephropathy. Lupus 2020, 29, 311–323. [Google Scholar] [CrossRef]
- Sam, N.B.; Li, B.-Z.; Leng, R.-X.; Pan, H.-F.; Ye, D.-Q. Circulating antioxidant levels in systemic lupus erythematosus patients: A systematic review and meta-analysis. Biomark. Med. 2019, 13, 1137–1152. [Google Scholar] [CrossRef]
- Otsuki, N.; Konno, T.; Kurahashi, T.; Suzuki, S.; Lee, J.; Okada, F.; Iuchi, Y.; Homma, T.; Fujii, J. The SOD1 transgene expressed in erythroid cells alleviates fatal phenotype in congenic NZB/NZW-F1 mice. Free Radic. Res. 2016, 50, 793–800. [Google Scholar] [CrossRef]
- Smallwood, M.J.; Nissim, A.; Knight, A.R.; Whiteman, M.; Haigh, R.; Winyard, P.G. Oxidative stress in autoimmune rheumatic diseases. Free Radic. Biol. Med. 2018, 125, 3–14. [Google Scholar] [CrossRef]
- Scofield, R.H.; Kurien, B.T.; Ganick, S.; McClain, M.T.; Pye, Q.; James, J.A.; Schneider, R.I.; Broyles, R.H.; Bachmann, M.; Hensley, K. Modification of lupus-associated 60-kDa Ro protein with the lipid oxidation product 4-hydroxy-2-nonenal increases antigenicity and facilitates epitope spreading. Free Radic. Biol. Med. 2005, 38, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Kurien, B.T.; Scofield, R.H. Free radical mediated peroxidative damage in systemic lupus erythematosus. Life Sci. 2003, 73, 1655–1666. [Google Scholar] [CrossRef]
- Kurien, B.T.; Hensley, K.; Bachmann, M.; Scofield, R.H. Oxidatively modified autoantigens in autoimmune diseases. Free Radic. Biol. Med. 2006, 41, 549–556. [Google Scholar] [CrossRef]
- Feldmann, A.; Stamova, S.; Bippes, C.C.; Bartsch, H.; Wehner, R.; Schmitz, M.; Temme, A.; Cartellieri, M.; Bachmann, M. Retargeting of T cells to prostate stem cell antigen expressing tumor cells: Comparison of different antibody formats. Prostate 2010, 71, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Grölz, D.; Bachmann, M. An Altered Intracellular Distribution of the Autoantigen La/SS-B When Translated from a La mRNA Isoform. Exp. Cell Res. 1997, 234, 329–335. [Google Scholar] [CrossRef]
- Arndt, C.; Koristka, S.; Feldmann, A.; Bachmann, M. Native Polyacrylamide Gels. Adv. Struct. Saf. Stud. 2018, 1855, 87–91. [Google Scholar] [CrossRef]
- Bachmann, M.P.; Bartsch, H.; Gross, J.K.; Maier, S.M.; Gross, T.F.; Workman, J.L.; James, J.A.; Farris, A.D.; Jung, B.; Franke, C.; et al. Autoimmunity as a Result of Escape from RNA Surveillance. J. Immunol. 2006, 177, 1698–1707. [Google Scholar] [CrossRef]
- Johnson, W.C. Extending Circular Dichroism Spectra into the Vacuum UV and its Application to Proteins. Photochem. Photobiol. 1986, 44, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.C. Protein secondary structure and circular dichroism: A practical guide. Proteins Struct. Funct. Bioinform. 1990, 7, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins Struct. Funct. Genet. 1999, 35, 307–312. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Scofield, R.; Kurien, B.; Zhang, F.; Mehta, P.; Kaufman, K.; Gross, T.; Bachmann, M.; Gordon, T.; Harley, J. Protein–protein interaction of the Ro-ribonucleoprotein particle using multiple antigenic peptides. Mol. Immunol. 1999, 36, 1093–1106. [Google Scholar] [CrossRef]

| Wildtype La | La_C18A | La_C232A_C245A | La_C18A_C232A_C245A | |||||
|---|---|---|---|---|---|---|---|---|
| helix 1 | 55 | 29 | 50 | 58 | 63 | 38 | 44 | 42 |
| strand | 16 | 26 | 21 | 17 | 13 | 23 | 15 | 17 |
| turns | 7 | 2 | 8 | 5 | 6 | 9 | 11 | 1 |
| random | 21 | 25 | 25 | 19 | 17 | 2 | 21 | 22 |
| total | 99 | 100 | 99 | 99 | 99 | 99 | 100 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berndt, N.; Bippes, C.C.; Michalk, I.; Bachmann, D.; Bachmann, J.; Puentes-Cala, E.; Bartsch, T.; Loureiro, L.R.; Kegler, A.; Bergmann, R.; et al. Two Be or Not Two Be: The Nuclear Autoantigen La/SS-B Is Able to Form Dimers and Oligomers in a Redox Dependent Manner. Int. J. Mol. Sci. 2021, 22, 3377. https://doi.org/10.3390/ijms22073377
Berndt N, Bippes CC, Michalk I, Bachmann D, Bachmann J, Puentes-Cala E, Bartsch T, Loureiro LR, Kegler A, Bergmann R, et al. Two Be or Not Two Be: The Nuclear Autoantigen La/SS-B Is Able to Form Dimers and Oligomers in a Redox Dependent Manner. International Journal of Molecular Sciences. 2021; 22(7):3377. https://doi.org/10.3390/ijms22073377
Chicago/Turabian StyleBerndt, Nicole, Claudia C. Bippes, Irene Michalk, Dominik Bachmann, Jennifer Bachmann, Edinson Puentes-Cala, Tabea Bartsch, Liliana R. Loureiro, Alexandra Kegler, Ralf Bergmann, and et al. 2021. "Two Be or Not Two Be: The Nuclear Autoantigen La/SS-B Is Able to Form Dimers and Oligomers in a Redox Dependent Manner" International Journal of Molecular Sciences 22, no. 7: 3377. https://doi.org/10.3390/ijms22073377
APA StyleBerndt, N., Bippes, C. C., Michalk, I., Bachmann, D., Bachmann, J., Puentes-Cala, E., Bartsch, T., Loureiro, L. R., Kegler, A., Bergmann, R., Gross, J. K., Gross, T., Kurien, B. T., Scofield, R. H., Farris, A. D., James, J. A., Schmitz, M., Fahmy, K., Feldmann, A., ... Bachmann, M. P. (2021). Two Be or Not Two Be: The Nuclear Autoantigen La/SS-B Is Able to Form Dimers and Oligomers in a Redox Dependent Manner. International Journal of Molecular Sciences, 22(7), 3377. https://doi.org/10.3390/ijms22073377








