MIAT Is an Upstream Regulator of NMYC and the Disruption of the MIAT/NMYC Axis Induces Cell Death in NMYC Amplified Neuroblastoma Cell Lines
Abstract
1. Introduction
2. Results
2.1. MIAT Expression Is Upregulated in Neuroblastoma Cell Lines and Tissues
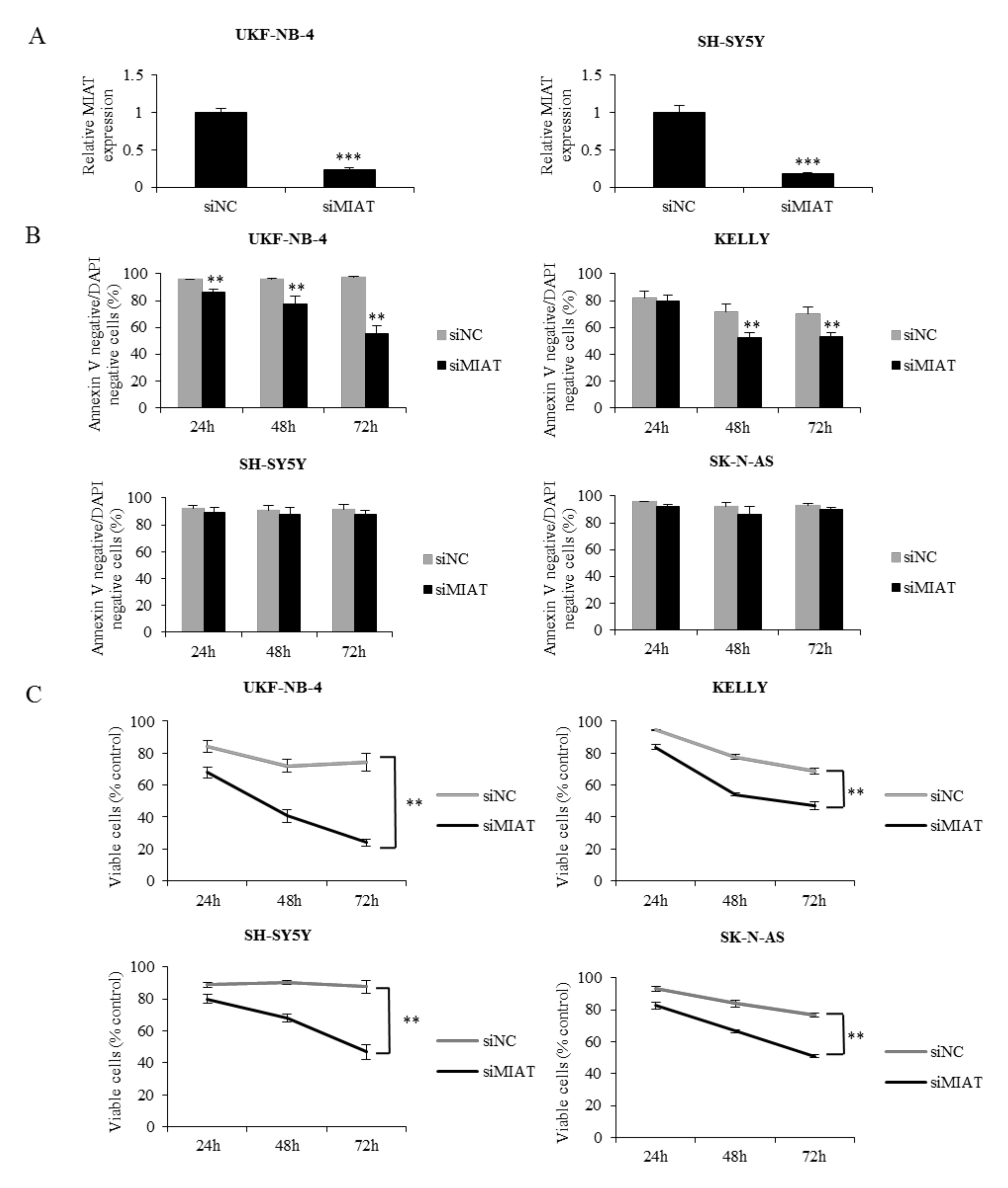
2.2. MIAT Knockdown Inhibits Neuroblastoma Cell Proliferation and Induces Apoptosis in Cell Lines with NMYC Amplification
2.3. MIAT Regulates NMYC and c-Myc Expression at the mRNA Level
2.4. Downregulation of MIAT Causes Changes in Cell Cycle Distribution and Inhibits Neuroblastoma Cells Migration
2.5. MIAT Knockdown Decreases Glycolytic and Respiratory Capacity of Neuroblastoma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Cell Viability Assay
4.3. Annexin V/DAPI Staining Assay
4.4. Cell Cycle Analysis
4.5. RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
4.6. Cell Migration
4.7. Cellular Energetics
4.8. SDS-PAGE and Western Blot Analysis
4.9. Tissue Collection and RNA In Situ Hybridization
4.10. Immunofluorescence Staining
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luksch, R.; Castellani, M.R.; Collini, P.; De Bernardi, B.; Conte, M.; Gambini, C.; Gandola, L.; Garaventa, A.; Biasoni, D.; Podda, M.; et al. Neuroblastoma (Peripheral neuroblastic tumours). Crit. Rev. Oncol. Hematol. 2016, 107, 163–181. [Google Scholar] [CrossRef]
- Huang, M.; Weiss, W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, C. Coding or noncoding, the converging concepts of RNAs. Front. Genet. 2019, 10. [Google Scholar] [CrossRef]
- Peinado, P.; Herrera, A.; Baliñas, C.; Martín-Padrón, J.; Boyero, L.; Cuadros, M.; Coira, I.F.; Rodriguez, M.I.; Reyes-Zurita, F.J.; Rufino-Palomares, E.E.; et al. Long Noncoding RNAs as Cancer Biomarkers. In Cancer and Noncoding RNAs; Elsevier: Amsterdam, The Netherlands, 2018; pp. 95–114. [Google Scholar]
- Sahu, D.; Ho, S.Y.; Juan, H.F.; Huang, H.C. High-risk, expression-based prognostic long noncoding RNA signature in neuroblastoma. JNCI Cancer Spectr. 2018, 2, pky015. [Google Scholar] [CrossRef] [PubMed]
- Swier, L.J.Y.M.; Dzikiewicz-Krawczyk, A.; Winkle, M.; van den Berg, A.; Kluiver, J. Intricate crosstalk between MYC and non-coding RNAs regulates hallmarks of cancer. Mol. Oncol. 2019, 13, 26–45. [Google Scholar] [CrossRef] [PubMed]
- Rombaut, D.; Chiu, H.S.; Decaesteker, B.; Everaert, C.; Yigit, N.; Peltier, A.; Janoueix, I.; Bartenhagen, C.; Fischer, M.; Roberts, S.; et al. Integrative analysis identifies lincRNAs up- and downstream of neuroblastoma driver genes. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Song, F.; Yang, Y.; Liu, J. Long non-coding RNA MIAT promotes the proliferation and invasion of laryngeal squamous cell carcinoma cells by sponging microRNA-613. Exp. Ther. Med. 2021, 21, 1. [Google Scholar] [CrossRef]
- Xu, H.; Zhou, J.; Tang, J.; Min, X.; Yi, T.; Zhao, J.; Ren, Y. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J. Clin. Lab. Anal. 2020, 34, e23323. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Xiao, H.; Xiao, W.; Xiong, Z.; Hu, W.; Gao, Y.; Ru, Z.; Wang, C.; Bao, L.; Wang, K.; et al. Upregulation of MIAT Regulates LOXL2 Expression by Competitively Binding miR-29c in Clear Cell Renal Cell Carcinoma. Cell. Physiol. Biochem. 2018, 48, 1075–1087. [Google Scholar] [CrossRef]
- Jin, H.; Jin, X.; Chai, W.; Yin, Z.; Li, Y.; Dong, F.; Wang, W. Long non-coding RNA MIAT competitively binds miR-150-5p to regulate ZEB1 expression in osteosarcoma. Oncol. Lett. 2019, 17, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.L.; Xu, S.Q.; Li, Q.; Zhao, Y.B.; Li, X.; Yang, M.P. Long noncoding RNA MIAT promotes the growth and metastasis of non-small cell lung cancer by upregulating TDP43. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3383–3389. [Google Scholar] [PubMed]
- Li, T.F.; Liu, J.; Fu, S.J. The interaction of long non-coding RNA MIAT and miR-133 play a role in the proliferation and metastasis of pancreatic carcinoma. Biomed. Pharmacother. 2018, 104, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, C.; Luo, Y.; Li, L.; Liu, J.; Gui, R. Silencing of long non-coding RNA MIAT sensitizes lung cancer cells to gefitinib by epigenetically regulating miR-34a. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gao, Y.; Qin, J.; Lu, S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 314, G559–G565. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, Y.; Sun, H.; Pan, Y.; Wu, M.; Zhang, J. Deregulation of miR-520d-3p promotes hepatocellular carcinoma development via lncRNA MIAT regulation and EPHA2 signaling activation. Biomed. Pharmacother. 2019, 109, 1630–1639. [Google Scholar] [CrossRef]
- Li, D.; Hu, X.; Yu, S.; Deng, S.; Yan, M.; Sun, F.; Song, J.; Tang, L. Silence of lncRNA MIAT-mediated inhibition of DLG3 promoter methylation suppresses breast cancer progression via the Hippo signaling pathway. Cell. Signal. 2020, 73, 109697. [Google Scholar] [CrossRef]
- Li, Y.; Wang, K.; Wei, Y.; Yao, Q.; Zhang, Q.; Qu, H.; Zhu, G. lncRNA-MIAT regulates cell biological behaviors in gastric cancer through a mechanism involving the miR-29a-3p/HDAC4 axis. Oncol. Rep. 2017, 38, 3465–3472. [Google Scholar] [CrossRef] [PubMed]
- Aprea, J.; Prenninger, S.; Dori, M.; Ghosh, T.; Monasor, L.S.; Wessendorf, E.; Zocher, S.; Massalini, S.; Alexopoulou, D.; Lesche, M.; et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013, 32, 3145–3160. [Google Scholar] [CrossRef]
- Bountali, A.; Tonge, D.P.; Mourtada-Maarabouni, M. RNA sequencing reveals a key role for the long non-coding RNA MIAT in regulating neuroblastoma and glioblastoma cell fate. Int. J. Biol. Macromol. 2019, 130, 878–891. [Google Scholar] [CrossRef] [PubMed]
- Prochazka, P.; Hrabeta, J.; Vicha, A.; Cipro, S.; Stejskalova, E.; Musil, Z.; Vodicka, P.; Eckschlager, T. Changes in MYCN expression in human neuroblastoma cell lines following cisplatin treatment may not be related to MYCN copy numbers. Oncol. Rep. 2013, 29, 2415–2421. [Google Scholar] [CrossRef] [PubMed]
- Aygun, N.; Altungoz, O. MYCN is amplified during S phase, and c-myb is involved in controlling MYCN expression and amplification in MYCN-amplified neuroblastoma cell lines. Mol. Med. Rep. 2019, 19, 345–361. [Google Scholar] [CrossRef]
- Wang, H.; Mannava, S.; Grachtchouk, V.; Zhuang, D.; Soengas, M.S.; Gudkov, A.V.; Prochownik, E.V.; Nikiforov, M.A. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene 2008, 27, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, H.; Liu, J.; Tong, J. Long Non-coding RNA MIAT Knockdown Prevents the Formation of Intracranial Aneurysm by Downregulating ENC1 via MYC. Front. Physiol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Raul, F. Revival of 2-(difluoromethyl)ornithine (DFMO), an inhibitor of polyamine biosynthesis, as a cancer chemopreventive agent. Biochem. Soc. Trans. 2007, 35, 353–355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hogarty, M.D.; Norris, M.D.; Davis, K.; Liu, X.; Evageliou, N.F.; Hayes, C.S.; Pawel, B.; Guo, R.; Zhao, H.; Sekyere, E.; et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008, 68, 9735–9745. [Google Scholar] [CrossRef]
- Bell, E.; Chen, L.; Liu, T.; Marshall, G.M.; Lunec, J.; Tweddle, D.A. MYCN oncoprotein targets and their therapeutic potential. Cancer Lett. 2010, 293, 144–157. [Google Scholar] [CrossRef]
- Yildirim, O.; Izgu, E.C.; Damle, M.; Chalei, V.; Ji, F.; Sadreyev, R.I.; Szostak, J.W.; Kingston, R.E. S-phase Enriched Non-coding RNAs Regulate Gene Expression and Cell Cycle Progression. Cell Rep. 2020, 31, 107629. [Google Scholar] [CrossRef]
- Ren, P.; Yue, M.; Xiao, D.; Xiu, R.; Gan, L.; Liu, H.; Qing, G. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J. Pathol. 2015, 235, 90–100. [Google Scholar] [CrossRef]
- Zirath, H.; Frenzel, A.; Oliynyk, G.; Segerström, L.; Westermark, U.K.; Larsson, K.; Persson, M.M.; Hultenby, K.; Lehtiö, J.; Einvik, C.; et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. USA 2013, 110, 10258–10263. [Google Scholar] [CrossRef]
- Qing, G.; Skuli, N.; Mayes, P.A.; Pawel, B.; Martinez, D.; Maris, J.M.; Simon, M.C. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1α. Cancer Res. 2010, 70, 10351–10361. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feriancikova, B.; Feglarova, T.; Krskova, L.; Eckschlager, T.; Vicha, A.; Hrabeta, J. MIAT Is an Upstream Regulator of NMYC and the Disruption of the MIAT/NMYC Axis Induces Cell Death in NMYC Amplified Neuroblastoma Cell Lines. Int. J. Mol. Sci. 2021, 22, 3393. https://doi.org/10.3390/ijms22073393
Feriancikova B, Feglarova T, Krskova L, Eckschlager T, Vicha A, Hrabeta J. MIAT Is an Upstream Regulator of NMYC and the Disruption of the MIAT/NMYC Axis Induces Cell Death in NMYC Amplified Neuroblastoma Cell Lines. International Journal of Molecular Sciences. 2021; 22(7):3393. https://doi.org/10.3390/ijms22073393
Chicago/Turabian StyleFeriancikova, Barbara, Tereza Feglarova, Lenka Krskova, Tomas Eckschlager, Ales Vicha, and Jan Hrabeta. 2021. "MIAT Is an Upstream Regulator of NMYC and the Disruption of the MIAT/NMYC Axis Induces Cell Death in NMYC Amplified Neuroblastoma Cell Lines" International Journal of Molecular Sciences 22, no. 7: 3393. https://doi.org/10.3390/ijms22073393
APA StyleFeriancikova, B., Feglarova, T., Krskova, L., Eckschlager, T., Vicha, A., & Hrabeta, J. (2021). MIAT Is an Upstream Regulator of NMYC and the Disruption of the MIAT/NMYC Axis Induces Cell Death in NMYC Amplified Neuroblastoma Cell Lines. International Journal of Molecular Sciences, 22(7), 3393. https://doi.org/10.3390/ijms22073393





