Abstract
DNA can adopt various structures besides the B-form. Among them, cruciform structures are formed on inverted repeat (IR) sequences. While cruciform formable IRs (CFIRs) are sometimes found in regulatory regions of transcription, their function in transcription remains elusive, especially in eukaryotes. We found a cluster of CFIRs within the mouse Pou5f1 enhancer. Here, we demonstrate that this cluster or some member(s) plays an active role in the transcriptional regulation of not only Pou5f1, but also Sox2, Nanog, Klf4 and Esrrb. To clarify in vivo function of the cluster, we performed genome editing using mouse ES cells, in which each of the CFIRs was altered to the corresponding mirror repeat sequence. The alterations reduced the level of the Pou5f1 transcript in the genome-edited cell lines, and elevated those of Sox2, Nanog, Klf4 and Esrrb. Furthermore, transcription of non-coding RNAs (ncRNAs) within the enhancer was also upregulated in the genome-edited cell lines, in a similar manner to Sox2, Nanog, Klf4 and Esrrb. These ncRNAs are hypothesized to control the expression of these four pluripotency genes. The CFIRs present in the Pou5f1 enhancer seem to be important to maintain the integrity of ES cells.
Keywords:
inverted repeat (IR) sequence; cruciform; super-enhancer; eRNA; mouse ES cells; Pou5f1 (Oct3/4); Sox2; Nanog; Klf4; Esrrb 1. Introduction
Genomes contain many unusual DNA structures, including cruciform structures, left-handed DNA helices, curved DNA structures, triplex structures, and G-quadruplex (G4) structures. Among them, cruciform structures have the longest history of study. In 1955, two years after Watson and Crick revealed the right-handed double helical structure of DNA, Platt proposed the presence of cruciform structures [1]. Cruciforms can be formed where a given DNA has a complete inverted repeat sequence, which reads the same from 5′ to 3′ in either strand, or an incomplete inverted repeat (IR) that contains an appropriate length of spacer DNA between the repeating units. Numerous studies have been performed to understand the functional significance of cruciform and/or IR sequences. Although their implications in DNA replication [2,3,4], transcription [5,6,7,8,9,10,11], and recombination [12,13,14,15] have been suggested, their functional or mechanistic role in each process remains elusive.
To gain insight into the biological role of cruciform structures, we recently performed genome-wide analyses of the Escherichia coli and Saccharomyces cerevisiae genomes, constructed comprehensive maps of IRs, and classified them depending on their structures and positions, as compared with gene positions. In E. coli, cruciform formable IRs (CFIRs) were significantly enriched in five regions, including the adjacent regions downstream of stop codon-coding sites and on and around the positions corresponding to mRNA ends. Furthermore, most of the CFIRs with a repeat unit length of ≥8 bp and a spacer size of ≤8 were found to be parts of the intrinsic terminators [10]. Regarding S. cerevisiae, the close vicinity of the DNA positions corresponding to poly(A) sites was the most statistically significant region of IR enrichment, and these IRs strongly correlated with the poly(A) signal. In addition, the majority of the IRs caused low nucleosome occupancy [11]. These studies suggested that in both organisms, the IRs actively participate in the mechanism of transcription termination. However, these studies could not suggest any plausible mechanism for the functions of IRs or CFIRs in the regulation of transcription initiation or modulation of transcription.
To our knowledge, no report has shown the relevance of IR(s) or CFIR(s) in the functions of enhancers. Using the mouse genome, we recently performed a screening of CFIRs and found that mouse Pou5f1 (Oct3/4) has a CFIR cluster within its enhancer. Based on the FANTOM5 cap analysis of gene expression (CAGE) database [16,17], the Pou5f1 enhancer produces enhancer RNAs (eRNAs), which are a signature of active enhancers [18,19,20]. Two types of eRNAs have been identified: those transcribed bidirectionally and those transcribed unidirectionally. The former is short and not adenylated, whilst the latter is long and adenylated [18,21,22]. Judging from the FANTOM5 CAGE database [16,17], the eRNAs transcribed from this enhancer belong to the former type. Another notable point is that the enhancer of Pou5f1 was originally regarded as a “general” enhancer [23,24,25], but it was later found to be a part of a super-enhancer (SE) [26]. The SEs are defined as the regions where multiple enhancers are clustered together, and they are associated with genes involved in determining cell identity in both the physiological and pathological states [27].
To determine the function of the CFIR cluster present in the Pou5f1 enhancer, we performed genome editing in which each of the CFIRs was replaced with the corresponding mirror repeat (MR). This alteration downregulated the Pou5f1 transcription, but upregulated the eRNA transcription and Sox2, Nanog, Klf4 and Esrrb transcription. The eRNAs are hypothesized to control the expression of these four pluripotency genes. A delicate transcriptional regulation network seems to exist among these genes, in which CFIRs apparently play an active role.
2. Results
Using our IR identifier CIRI created in-house [10], we screened the mouse genome and found that the upstream region of Pou5f1 contains 12 IRs with repeat unit (R) lengths greater than or equal to 6 bp, spacer (S) lengths between 0 and 6 bp, and an entire IR length equal to or longer than 15 bp (i.e., IRs with R ≥ 6, S ≤ 6 and 2R + S ≥ 15) (Figure 1). Each of these IRs is thought to have the potential to form a cruciform [10,11], and thus they were named CFIR1 to CFIR12, respectively. Notably, six CFIRs (CFIR2 to CFIR7) are located in the region spanning from ~−2000 to ~−1600 relative to the transcription start site (TSS; +1) of Pou5f1. This region is within the distal enhancer (DE) of the gene (Figure 1a and Table 1). Among the 12 CFIRs, only CFIR2 to CFIR7 are located in this important region for the Pou5f1 transcription. Thus, to determine whether this cluster has any function in Pou5f1 transcription, using mouse ES cells, we substituted each of the six CFIRs with the corresponding MR sequence by 2H2OP method [28]-based genome editing, and examined the resulting effect on the transcription of the gene. This substitution did not change the nucleotide composition of each CFIR, but the resulting MR lost the cruciform-forming potential (Figure 1b). Finally, we established two cell lines, MR/WT (heterozygous editing) and MR/MR (homozygous editing) (Figure 1c). A sequence analysis of the region containing −2266 to −1546 (data not shown) and quantitative real-time PCR (qPCR) analyses confirmed that the substitution cassette was only present in the targeted region (Figure 1d).
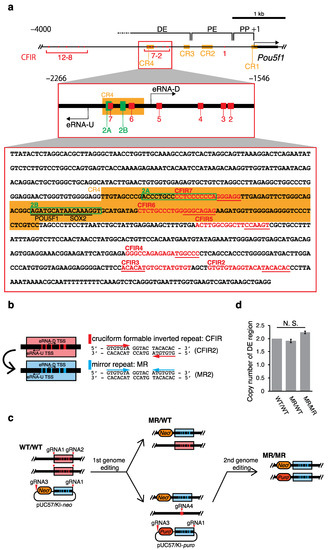
Figure 1.
Regulatory region of Pou5f1 and cruciform formable inverted repeats (CFIRs) as genome editing targets. (a) Map of the CFIRs and partial sequence of the distal enhancer (DE). The DNA sequence is from positions −2266 to −1546 relative to the TSS (+1) of Pou5f1. In the sequence, CFIRs are indicated with red characters, and the 2A and 2B sequences are enclosed within green rectangles. POU5F1 and SOX2 binding sites are underlined. The CR1 to CR4 regions (CR: conserved region) [29] are shaded in orange. The sequences underlined in red in the CFIR motifs were altered in the editing (Table 1). (b) Alteration of CFIRs into mirror repeats (MRs). CFIRs 2–7 were each altered to MRs 2–7, which maintained the GC content of each CFIR but removed its potential to form a cruciform. As an example, the alteration of CFIR2 to MR2 is shown on the right. (c) Pathway to establish genome-edited cell lines. The red triangles and star indicate target sites of guide RNAs (gRNAs) and repair junction, respectively. (d) Validation of the editing as examined by qPCR. Validation was performed by determining the copy number of DE, in which a part of Pou5f1 was used for the copy number reference. Values are mean ± SD. The statistical analysis was performed using one-way ANOVA and Tukey’s post hoc analysis (n = 3).

Table 1.
Sequences and structures of CFIRs 2–7.
2.1. Effects of the Genome Editing on Pou5f1 and eRNA Transcription
Using WT/WT (wild-type), MR/WT and MR/MR cells, effects of the genome editing on Pou5f1 transcription were first examined by qPCR. As shown in Figure 2a, the Pou5f1 mRNA level was reduced in both the MR/WT and MR/MR cells, and the effect was larger in MR/MR than MR/WT. The transcription levels of the gene in MR/WT and MR/MR were ~85% and ~75% of that in the wild-type cells, respectively. Since the leukemia inhibitory factor (LIF) is required to maintain mouse ES cells in an undifferentiated state [30,31], we also examined the effects of LIF depletion. Although the transcription levels of Pou5f1 were gradually reduced in all of the cell lines as time elapsed, the extents were higher in MR/WT and MR/MR cells, especially in the latter (Figure 2b). Regarding the data at day 4, the Pou5f1 transcription levels in the former and the latter were ~80% and ~60% of that in the wild-type cells, respectively. Thus, the data shown in Figure 2a,b suggested that CFIRs 2–7 play an active role in Pou5f1 transcription.
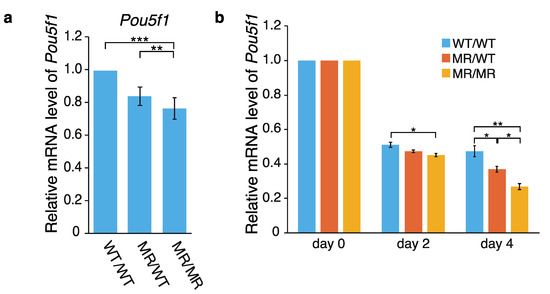
Figure 2.
Effect of CFIR-to-MR editing on Pou5f1 expression. Quantification was performed by qPCR. The level of Pou5f1 mRNA in each cell line was normalized against that of Gapdh. (a) Pou5f1 expression in the wild-type and genome-edited cells. The mean mRNA level in the wild-type WT/WT cells was set to 1.0, and the relative mRNA levels in the genome-edited MR/WT and MR/MR cells are shown. The values are represented as mean ± SD (n = 7). The statistical analysis was performed using one-way ANOVA and Tukey’s post hoc analysis. * p < 0.05; ** p < 0.01; *** p < 0.001. (b) Effects of leukemia inhibitory factor (LIF) depletion on Pou5f1 expression. For quantification by qPCR, total RNAs were prepared immediately after LIF depletion (“day 0”), or two or four days after the depletion (“day 2” and “day 4”, respectively). In each cell line, the mean mRNA level at day 0 was set to 1.0 and relative transcript levels at day 2 and day 4 are shown. The values are represented as mean ± SD (n = 7). The statistical analysis was performed using one-way ANOVA and Tukey’s post hoc analysis. * p < 0.05; ** p < 0.01; *** p < 0.001.
Active enhancers are transcribed to generate ncRNAs, which are referred to as eRNAs [18,19,20,32]. According to the FANTOM5 CAGE data [16,17], two eRNA species are transcribed bi-directionally within DE (Figure 1a). Therefore, to determine whether the alteration of CFIRs 2–7 to MRs 2–7 affected the transcription of these eRNA species, their relative transcription levels were analyzed by qPCR (Figure 3). Production of eRNA-U (eRNA transcribed toward upstream) was increased by ~10-fold and ~7-fold in the MR/WT and MR/MR cells, respectively, as compared to that in the wild-type cells. In the eRNA-U transcription, CFIRs 2–7 are all located upstream of the TSS for the RNA. Thus, it seems that some or all of the CFIRs 2–7 presumably play some repressive role in the eRNA-U transcription in the wild-type cells. On the other hand, the alteration of CFIRs 2–7 to MRs 2–7 had a smaller effect on eRNA-D (eRNA transcribed toward downstream) transcription: the activation was ~4-fold in MR/WT and only slight in MR/MR, as compared to the eRNA-D amount in the wild-type cells. Among CFIRs 2–7, only CFIRs 6 and 7 are located upstream of the eRNA-D TSS. Thus, the slightly different phenomena observed between eRNA-U and eRNA-D transcription may originate from the number of valid CFIRs in transcription: six in the former and two in the latter (Figure 1a). The result that the levels of eRNA-U and eRNA-D were both lower in MR/MR cells, as compared to those in MR/WT cells, may suggest the presence of a negative feedback mechanism in their production. Regardless of the hypothesis, the results suggest that at least some of CFIRs 2–7 have a negative influence on the transcription of the two eRNA species. Another important point is that the effects on transcription were opposite between Pou5f1 and the two eRNA species.
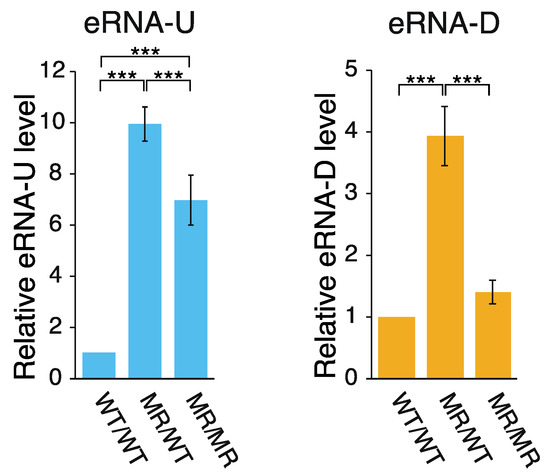
Figure 3.
Effect of CFIR-to-MR editing on eRNA expression. The amount of eRNAs in each cell line was quantified by qPCR and normalized against that of Gapdh mRNA. The mean eRNA-U level (left) or mean eRNA-D level (right) in the WT/WT cells was set to 1.0 and the relative eRNA levels in MR/WT and MR/MR cells are shown. The values are represented as mean ± SD (n = 7). The statistical analysis was performed using one-way ANOVA and Tukey’s post hoc analysis. *** p < 0.001.
2.2. Correlation between the Transcription of eRNA and That of Genes Controlled by a Super-Enhancer
The ~3 kb region upstream of Pou5f1 reportedly acts as a super-enhancer [26], in which two notable sites were identified: one coincides with the 2B sequence and the other is within the proximal enhancer (PE) (Figure 1a). These are binding sites for transcription factors (TFs). The CFIR to MR alterations may affect the gene expression controlled by this super-enhancer. The relevant genes are Pou5f1, Sox2, Nanog, Klf4, and Esrrb [26]. We examined the effects of the genome editing on the transcription of Sox2, Nanog, Klf4, and Esrrb. Interestingly, the qPCR analysis showed that the transcription levels of these genes all exhibited the same changes in the following sense: the mRNA levels increased in both genome-edited cell lines, and those in MR/WT were higher than those in MR/MR (Figure 4). Extents of the transcription activation in the genome-edited cells were generally smaller for the four genes, as compared to the data for the eRNAs (Figure 3 and Figure 4), but the profiles of the changes were very similar to each other, especially between the four genes and eRNA-U. Although this study could not clarify whether eRNA-U or eRNA-D or both influenced the transcription of the four genes, the data shown in Figure 3 and Figure 4 strongly indicated that the eRNA(s) is (are) involved in the transcription of each of the four genes.
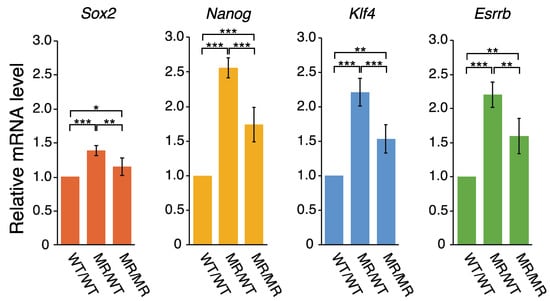
Figure 4.
Effect on the expression of other genes regulated by the eRNAs. Quantification was performed by qPCR. The mRNA levels of Sox2, Nanog, Klf4, and Esrrb in each cell line were normalized against that of Gapdh. The mean mRNA level of Sox2, Nanog, Klf4, or Esrrb in the WT/WT cells was set to 1.0 and the mRNA levels relative to it in the MR/WT and MR/MR cells are shown. The values are represented as mean ± SD (n = 7). The statistical analysis was performed using one-way ANOVA and Tukey’s post hoc analysis. * p < 0.05; ** p < 0.01; *** p < 0.001.
3. Discussion
3.1. Hypothetical Mechanism Underlying the Editing-Elicited Downregulation of Pou5f1 Expression
POU5F1 (OCT3/4) is a key TF for maintaining pluripotency in ES cells and iPS cells [33,34,35,36,37], and Pou5f1 transcription is regulated by proximal promoter (PP), PE, and DE (Figure 1a) [23,24,25,34,38,39]. These three regions were determined by conventional approaches and some TF binding sites were identified, including the POU5F1 and SOX2 binding sites [40,41]. Analyses of the epigenetic features of these regions suggested that DNA methylation and histone modifications are strong regulators of Pou5f1 expression [42,43,44,45,46]. However, no study has been performed to determine whether the CFIR cluster (CFIRs 2–7) is used in some regulation mechanism of Pou5f1 expression.
A sequence homology analysis revealed that there are four conserved regions, named CR1 to CR4, in the upstream of the human, bovine and mouse Pou5f1 TSSs [29]. In the mouse genome, only CR3 and CR4 are within DE, which contains the 2A and 2B sequences [41,47] and CFIRs 6 and 7 (Figure 1a). The 2B sequence harbors the POU5F1 and SOX2 binding sites [40,41]. The current study showed that the alteration of the cluster CFIRs 2–7 into MRs 2–7 within DE reduced the transcription of Pou5f1. Although the negative effects of MRs 2–7 were not large, the presence of the effect was obvious and ~20% transcriptional reduction occurred in MR/MR cells (Figure 2). Among the sequence alterations, those from CFIRs 6 and 7 to MRs 6 and 7 may be most relevant to the reduction, because they are in the vicinity of the 2A and 2B sequences.
The 2B sequence is located between CFIRs 6 and 7 and does not overlap with them (Figure 1a). Distances between 2B and CFIR7 and 2B and CFIR6 are 22 bp and 9 bp, respectively. In the alteration of CFIR6 to MR6, however, the upstream repeat unit (8 bp) and spacer sequence (3 bp) remained unchanged. Thus, the 17 bp 2B sequence has 20 or 22 bp intact flanking sequences on either side, suggesting that the transcriptional reduction of Pou5f1 was not due to the sequence alteration itself, but rather to some other effect. One possible explanation is that the inhibition of the cruciform formation caused by the CFIR-to-MR alteration might have negatively influenced POU5F1 (and SOX2) binding to the 2B sequence, which may have impaired some mechanism in Pou5f1 expression (POU5F1 binding-implicated positive feedback mechanism may exist). This scenario seems plausible if the protein binding induces DNA bending that entails unwinding. In this hypothetical case, a torsional stress imposed on the 2B region by the binding of POU5F1 (and SOX2) can be alleviated by the cruciforms formed on CFIR6 and/or CFIR7, which can rewind structurally unfavorable local DNA unwinding (melting), but MRs 6 and 7 cannot do this. Thus, this editing could have destabilized the binding of these TFs or even inhibited their binding. Many reports have described protein-induced DNA bending and unwinding [48,49,50,51,52]. After the E. coli promoter is wrapped around RNA polymerase (DNA is “intensely bent” on the enzyme), it also induces DNA unwinding [53]. These DNA unwinding phenomena are considered to be prerequisites for the subsequent step(s) to proceed. The same may be true for POU5F1 (and SOX2) binding.
The 18 bp 2A sequence is 100% conserved in the human and bovine alignment, and it shares 55.6% homology with the mouse 2A sequence [29]. Deletion of the sequence greatly reduced the Pou5f1 expression in mouse cells [34,41], indicating that this sequence also plays a positive regulatory role in Pou5f1 expression, at least in mice. Although CFIR7 partially shares a sequence with 2A, the alteration of CFIR7 to MR7 did not impair the 2A sequence itself (Figure 1a). Thus, transcriptional reduction of Pou5f1 may be irrelevant to the function of the 2A sequence or it can be also explained in terms of protein-induced DNA bending and unwinding described above. However, to our knowledge, there has been no report that identified the 2A-binding protein [54]. Clearly, this issue needs further study.
3.2. eRNAs Transcribed from DE Are Presumably Implicated in the Transcriptional Regulation of Sox2, Nanog, Klf4, and Esrrb
The sequence alteration of CFIRs 2–7 to MRs 2–7 generated different effects on Pou5f1 and eRNA transcription. Contrary to the Pou5f1 transcription, this alteration upregulated eRNA transcription (Figure 2 and Figure 3), indicating that CFIRs 2–7 have some repressive effect on the eRNA transcription in mouse ES cells. Furthermore, the eRNA transcription profiles (Figure 3) correlated well with those of Sox2, Nanog, Klf4, and Esrrb (Figure 4). The upstream ~3kb region of Pou5f1 act as an SE and it regulates the expression of these pluripotency genes [26].
Two notable sites, which are binding sites for TFs, were identified in this SE: one coincides with the 2B sequence and the other is in the PE [26]. However, only the 2B sequence is relevant to the current study, and we hypothesized that CFIRs 6 and 7 may facilitate or stabilize POU5F1 (and SOX2) binding. On the other hand, stable binding of these TFs may be an obstacle in the transcription of eRNA-U and eRNA-D, which presumably act in trans to activate Sox2, Nanog, Klf4, and Esrrb expression (Figure 5). The different editing-caused effects on transcription observed between Pou5f1 and the other genes may be explained in terms of the stoichiometry of their products in cells.
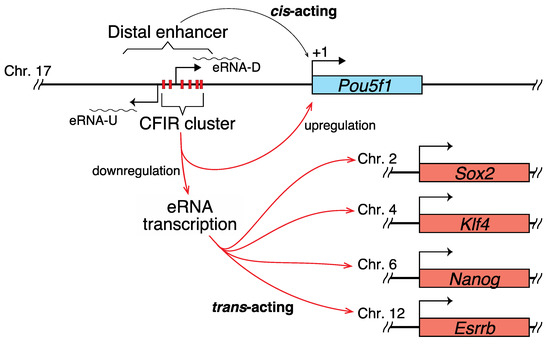
Figure 5.
Hypothetical role of the CFIR cluster. The CFIR cluster focused on in the current study or some member of it is hypothesized to function as a modulator of the expression balance of the pluripotency genes Pou5f1, Sox2, Nanog, Klf4, and Esrrb, by upregulating Pou5f1 expression while downregulating the other genes. In the downregulation mechanism for the latter, eRNA-U and eRNA-D, with expression that is also downregulated by the CFIR member(s), are strongly suggested to act as trans-acting molecules.
The genes that are involved in determining cell identity in both physiological and pathological states are generally regulated by SEs [27]. The SE focused on in the current study is known to influence the fate of pluripotent cells [26,55]. Considering this, the eRNA-U and eRNA-D expression levels may be strictly regulated by CFIRs to control the expression levels of Sox2, Nanog, Klf4 and Esrrb and/or the balance between these gene products and that of Pou5f1, which may be important for maintaining the integrity of ES cells (Figure 5). Although some reports have suggested the mechanisms by which eRNAs activate transcription [56,57,58,59,60,61], the molecular mechanisms underlying the results obtained in the current study remain to be determined.
3.3. The Cluster of CFIRs Can Also Act as an Absorber of Negative Supercoils
We have mainly focused on CFIRs 6 and 7 to explain the phenomena caused by the editing. Finally, we must also discuss the possible functions of all six CFIRs. An important hint may be that cruciform formation can absorb the increased negative superhelicity that leads to DNA strand separation [62,63,64]. The six CFIRs exist in a small, 466 bp region spanning from the upstream end of CR4 to the downstream end of CFIR2. The negative superhelicity of this region may increase at some step preceding the eRNA and Pou5f1 transcription or in the process of transcription, and should be generated upon nucleosome decomposition (detachment of histones). If this is the case, then most or all of these CFIRs may be used as “absorbers” of the increased negative superhelical density in the region, which seems beneficial in Pou5f1 transcription, at least. Multiple or consecutive cruciforms may be further stabilized by inter-loop base-pairing, such as that found between the D-loop and TΨC-loop in tRNA [65,66].
4. Materials and Methods
4.1. Cell Culture and Transfection
The mouse ES cell line E14Tg2a [67] was maintained as described previously [68]. Briefly, the cells were maintained in G-MEM (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% fetal bovine serum (Biosera, Nuaille, France), 0.1 mM 2-mercaptoethanol, 1x MEM non-essential amino acid solution (Thermo Fisher Scientific, Waltham, MA, USA), 1 mM sodium pyruvate (Thermo Fisher Scientific, Waltham, MA, USA), and 1000 units/mL of LIF (Cell Guidance Systems, Cambridge, UK), on gelatin-coated dishes without feeder cells at 37 °C in 5% CO2. For the 2H2OP method [28]-based genome editing, vectors and oligodeoxyribonucleotides (ODNs) were co-transfected into ES cells. Transfection was performed using Lipofectamine 3000 according to the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA, USA), in which 250 ng of px330 vector, 250 ng of donor vector, 50 ng of ODN1 and 50 ng of ODN2 (ODN: oligodeoxyribonucleotide, Table S1) were used. Forty-eight h after the transfection, 200 µg/mL G418 (Enzo Life Sciences Inc., Farmingdale, NY, USA) or 0.1 µg/mL puromycin (Nacalai Tesque, Kyoto, Japan) was added to the culture for the selection. The cells were then cultured for two weeks. In the LIF depletion experiment, each cell line was cultured for two or four days after the depletion, and then total RNA was isolated.
4.2. Construction of Donor and Px330 Vectors
The pUC57/KI-neo plasmid, which carries the substitution donor DNA and the neomycin-resistance gene within the EcoRV site of pUC57, was prepared by Genewiz, Inc. (South Plainfield, NJ, USA). To generate pUC57/KI-puro, a DNA fragment encoding the puromycin-resistance gene in pSF-MinCMV-Puro (Oxford Genetics, Oxford, UK) was obtained by PCR, using the primers puro-fw-NcoI and puro-rv-XhoI (Table S1). The resulting fragment was inserted between the NcoI and XhoI sites of pUC57/KI-neo. The CRISPRdirect web tool (https://crispr.dbcls.jp/) [69] was used to design the gRNA sequences. After annealing the corresponding ODNs (Table S1), the resulting fragments were inserted within the BbsI site of px330-U6-Chimeric-BB-CBh-hSpCas9 (Addgene, Watertown, MA, USA), to generate px330-gRNA1 to px330-gRNA4. The region containing gRNA expression cassettes in each construct was amplified by PCR, using the primers U6-gRNA-Fw-AflIII and U6-gRNA-Rv-HindIII or U6-gRNA-Fw-HindIII and U6-gRNA-Rv-AflIII (Table S1), and inserted into the AflIII site of px330-gRNA1. The resulting constructs px330-gRNA1-2-3 and px330-gRNA1-3-4 generated two sets of gRNAs: gRNA1, gRNA2 and gRNA3, and gRNA1, gRNA3 and gRNA4, respectively.
4.3. Sequence Analysis
The genomic DNA was purified using a conventional method. The sequence of the edited region was amplified by PCR with the primers KIcheck-fw and KIcheck-rv (Table S1). The resulting products were confirmed by Sanger sequencing, using a Big Dye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol.
4.4. RNA Isolation and Reverse Transcription
Total RNA was isolated from the cells with a RNeasy Mini Kit (Qiagen, Venlo, The Netherlands) and QIAshredder (Qiagen, Venlo, Netherlands), and treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA), according to the manufacturers’ protocols. The resulting sample was subjected to reverse transcription with ReverTra Ace (Toyobo, Osaka, Japan). Briefly, 1 µg of purified total RNA was used in the reaction in the presence of 5 pmol of oligo(dT)20 (Toyobo, Osaka, Japan), and 25 pmol of random primer (Toyobo, Osaka, Japan).
4.5. qPCR Analysis
The qPCR was performed using the primer sets shown in Table S1, with a StepOnePlus system (Thermo Fisher Scientific, Waltham, MA, USA) and THUNDERBIRD SYBR qPCR Mix (Toyobo, Osaka, Japan), according to Toyobo’s protocol. After the reverse transcription described above, reverse transcripts from 1.5 ng-equivalents (to quantify mRNAs) or those of 25 ng-equivalents (to quantify ncRNAs) of total RNA were subjected to PCR. For the gene copy analysis, 0.8 ng of genomic DNA was used. For qPCR, we used the following conditions: 95 °C, 2 min; 40 cycles of 98 °C, 10 sec and 68 °C, 1 min. To assess the specificity of the amplification, a melting analysis (from 60 °C to 95 °C with a 0.3 °C increment every 15 s) was performed at the end of the PCR cycles.
Supplementary Materials
The following is available online at https://www.mdpi.com/article/10.3390/ijms22073399/s1, Table S1: Primers used in the study.
Author Contributions
Y.Y. and T.O. conceived and designed the study; Y.Y. performed the experiments; O.M. scrutinized the results from the bioinformatic analysis; Y.Y. and T.O. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science & Technology-JAPAN (MEXT; to T.O., 19H04391).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Platt, J.R. Possible separation of intertwined nucleic acid chains by transfer-twist. Proc. Natl. Acad. Sci. USA 1955, 41, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Pearson, C.E.; Zorbas, H.; Price, G.B.; Zannis-Hadjopoulos, M. Inverted repeats, stem-loops, and cruciforms: Significance for initiation of DNA replication. J. Cell. Biochem. 1996, 63, 1–22. [Google Scholar] [CrossRef]
- Zannis-Hadjopoulos, M.; Yahyaoui, W.; Callejo, M. 14-3-3 Cruciform-binding proteins as regulators of eukaryotic DNA replication. Trends Biochem. Sci. 2008, 33, 44–50. [Google Scholar] [CrossRef]
- Brázda, V.; Laister, R.C.; Jagelská, E.B.; Arrowsmith, C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 2011, 12, 33. [Google Scholar] [CrossRef]
- Dai, X.; Greizerstein, M.B.; Nadas-Chinni, K.; Rothman-Denes, L.B. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc. Natl. Acad. Sci. USA 1997, 94, 2174–2179. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Rothman-Denes, L.B. Sequence and DNA structural determinants of N4 virion RNA polymerase-promoter recognition. Genes Dev. 1998, 12, 2782–2790. [Google Scholar] [CrossRef]
- Jagelská, E.B.; Pivoňková, H.; Fojta, M.; Brázda, V. The potential of the cruciform structure formation as an important factor influencing p53 sequence-specific binding to natural DNA targets. Biochem. Biophys. Res. Commun. 2010, 391, 1409–1414. [Google Scholar] [CrossRef]
- Brázda, V.; Coufal, J.; Liao, J.C.C.; Arrowsmith, C.H. Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochem. Biophys. Res. Commun. 2012, 422, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Coufal, J.; Jagelská, E.B.; Liao, J.C.C.; Brázda, V. Preferential binding of p53 tumor suppressor to p21 promoter sites that contain inverted repeats capable of forming cruciform structure. Biochem. Biophys. Res. Commun. 2013, 441, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Miura, O.; Ogake, T.; Ohyama, T. Requirement or exclusion of inverted repeat sequences with cruciform-forming potential in Escherichia coli revealed by genome-wide analyses. Curr. Genet. 2018, 64, 945–958. [Google Scholar] [CrossRef]
- Miura, O.; Ogake, T.; Yoneyama, H.; Kikuchi, Y.; Ohyama, T. A strong structural correlation between short inverted repeat sequences and the polyadenylation signal in yeast and nucleosome exclusion by these inverted repeats. Curr. Genet. 2019, 65, 575–590. [Google Scholar] [CrossRef]
- Lin, C.T.; Lyu, Y.L.; Liu, L.F. A cruciform-dumbbell model for inverted dimer formation mediated by inverted repeats. Nucleic Acids Res. 1997, 25, 3009–3016. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtenko, L.S.; Hsieh, P.; Grigoriev, M.; Potaman, V.N.; Sinden, R.R.; Lyubchenko, Y.L. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J. Mol. Biol. 2000, 296, 1169–1173. [Google Scholar] [CrossRef] [PubMed]
- Lobachev, K.S.; Gordenin, D.A.; Resnick, M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 2002, 108, 183–193. [Google Scholar] [CrossRef]
- Wang, Y.; Leung, F.C.C. Long inverted repeats in eukaryotic genomes: Recombinogenic motifs determine genomic plasticity. FEBS Lett. 2006, 580, 1277–1284. [Google Scholar] [CrossRef]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef]
- Forrest, A.R.R.; Kawaji, H.; Rehli, M.; Baillie, J.K.; De Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; et al. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar] [CrossRef] [PubMed]
- De Santa, F.; Barozzi, I.; Mietton, F.; Ghisletti, S.; Polletti, S.; Tusi, B.K.; Muller, H.; Ragoussis, J.; Wei, C.L.; Natoli, G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 2010, 8, e1000384. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Production of spliced long noncoding RNAs specifies regions with increased enhancer activity. Cell Syst. 2018, 7, 537–547.e3. [Google Scholar] [CrossRef] [PubMed]
- Vučićević, D.; Corradin, O.; Ntini, E.; Scacheri, P.C.; Ørom, U.A. Long ncRNA expression associates with tissue-specific enhancers. Cell Cycle 2015, 14, 253–260. [Google Scholar] [CrossRef]
- Pan, G.J.; Chang, Z.Y.I.; Schöler, H.R.; Pei, D. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002, 12, 321–329. [Google Scholar] [CrossRef]
- Niwa, H. How is pluripotency determined and maintained? Development 2007, 134, 635–646. [Google Scholar] [CrossRef]
- Kellner, S.; Kikyo, N. Transcriptional regulation of the Oct4 gene, a master gene for pluripotency. Histol. Histopathol. 2010, 25, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef]
- Wu, M.; Shen, J. From super-enhancer non-coding RNA to immune checkpoint: Frameworks to functions. Front. Oncol. 2019, 9, 1–16. [Google Scholar] [CrossRef]
- Yoshimi, K.; Kunihiro, Y.; Kaneko, T.; Nagahora, H.; Voigt, B.; Mashimo, T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun. 2016, 7, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Nordhoff, V.; Hübner, K.; Bauer, A.; Orlova, I.; Malapetsa, A.; Schöler, H.R. Comparative analysis of human, bovine, and murine Oct-4 upstream promoter sequences. Mamm. Genome 2001, 12, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.G.; Heath, J.K.; Donaldson, D.D.; Wong, G.G.; Moreau, J.; Stahl, M.; Rogers, D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 1988, 336, 688–690. [Google Scholar] [CrossRef]
- Williams, R.L.; Hilton, D.J.; Pease, S.; Willson, T.A.; Stewart, C.L.; Gearing, D.P.; Wagner, E.F.; Metcalf, D.; Nicola, N.A.; Gough, N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 1988, 336, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Tippens, N.D.; Vihervaara, A.; Lis, J.T. Enhancer transcription: What, where, when, and why? Genes Dev. 2018, 32, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Schöler, H.; Smith, A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998, 95, 379–391. [Google Scholar] [CrossRef]
- Yeom, Y.I.; Fuhrmann, G.; Ovitt, C.E.; Brehm, A.; Ohbo, K.; Gross, M.; Hübner, K.; Schöler, H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 1996, 122, 881–894. [Google Scholar]
- Pesce, M.; Schöler, H.R. Oct-4: Control of totipotency and germline determination. Mol. Reprod. Dev. 2000, 55, 452–457. [Google Scholar] [CrossRef]
- Pesce, M.; Schöler, H.R. Oct-4: Gatekeeper in the beginnings of mammalian development. Stem Cells 2001, 19, 271–278. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016, 17, 183–193. [Google Scholar] [CrossRef]
- Liao, J.; He, Y.; Szabó, P.E. The Pou5f1 distal enhancer is sufficient to drive Pou5f1 promoter-EGFP expression in embryonic stem cells. Int. J. Dev. Biol. 2013, 57, 725–729. [Google Scholar] [CrossRef]
- Canon, E.; Jouneau, L.; Blachère, T.; Peynot, N.; Daniel, N.; Boulanger, L.; Maulny, L.; Archilla, C.; Voisin, S.; Jouneau, A.; et al. Progressive methylation of POU5F1 regulatory regions during blastocyst development. Reproduction 2018, 156, 145–161. [Google Scholar] [CrossRef]
- Chew, J.L.; Loh, Y.H.; Zhang, W.; Chen, X.; Tam, W.L.; Yeap, L.S.; Li, P.; Ang, Y.S.; Lim, B.; Robson, P.; et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005, 25, 6031–6046. [Google Scholar] [CrossRef]
- Okumura-Nakanishi, S.; Saito, M.; Niwa, H.; Ishikawa, F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 2005, 280, 5307–5317. [Google Scholar] [CrossRef]
- Feldman, N.; Gerson, A.; Fang, J.; Li, E.; Zhang, Y.; Shinkai, Y.; Cedar, H.; Bergman, Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006, 8, 188–194. [Google Scholar] [CrossRef]
- Hattori, N.; Nishino, K.; Ko, Y.G.; Hattori, N.; Ohgane, J.; Tanaka, S.; Shiota, K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004, 279, 17063–17069. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Pu, M.T.; Hirasawa, R.; Li, B.Z.; Huang, Y.N.; Zeng, R.; Jing, N.H.; Chen, T.; Li, E.; Sasaki, H.; et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell. Biol. 2007, 27, 8748–8759. [Google Scholar] [CrossRef]
- Tachibana, M.; Sugimoto, K.; Nozaki, M.; Ueda, J.; Ohta, T.; Ohki, M.; Fukuda, M.; Takeda, N.; Niida, H.; Kato, H.; et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002, 16, 1779–1791. [Google Scholar] [CrossRef]
- Ben-Shushan, E.; Pikarsky, E.; Klar, A.; Bergman, Y. Extinction of Oct-3/4 gene expression in embryonal carcinoma x fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol. Cell. Biol. 1993, 13, 891–901. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Minucci, S.; Botquin, V.; Yeom, Y.I.; Dey, A.; Sylvester, I.; Zand, D.J.; Ohbo, K.; Ozato, K.; Schöler, H.R. Retinoic acid-mediated down-regulation of Oct3/4 coincides with the loss of promoter occupancy in vivo. EMBO J. 1996, 15, 888–899. [Google Scholar] [CrossRef]
- Aiken, C.R.; Fisher, E.W.; Gumport, R.I. The specific binding, bending, and unwinding of DNA by RsrI endonuclease, an isoschizomer of EcoRI endonuclease. J. Biol. Chem. 1991, 266, 19063–19069. [Google Scholar] [CrossRef]
- Giese, K.; Pagel, J.; Groosshedl, R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc. Natl. Acad. Sci. USA 1997, 94, 12845–12850. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.L.; Nikolov, D.B.; Burley, S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 1993, 365, 520–527. [Google Scholar] [CrossRef]
- Kim, Y.; Geiger, J.H.; Hahn, S.; Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993, 365, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Missura, M.; Buterin, T.; Hindges, R.; Hübscher, U.; Kaspárková, J.; Brabec, V.; Naegeli, H. Double-check probing of DNA bending and unwinding by XPA-RPA: An architectural function in DNA repair. EMBO J. 2001, 20, 3554–3564. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, B.; Burton, Z.F. DNA bending and wrapping around RNA polymerase: A “revolutionary” model describing transcriptional Mmechanisms. Microbiol. Mol. Biol. Rev. 1999, 63, 457–478. [Google Scholar] [CrossRef] [PubMed]
- Nazarov, I.B.; Krasnoborova, V.A.; Mitenberg, A.G.; Chikhirzhina, E.V.; Davidov-Sinitzin, A.P.; Liskovykh, M.A.; Tomilin, A.N. Transcription regulation of Oct4 (Pou5F1) gene by its distal enhancer. Cell Tissue Biol. 2014, 8, 27–32. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934. [Google Scholar] [CrossRef]
- Bose, D.A.; Donahue, G.; Reinberg, D.; Shiekhattar, R.; Bonasio, R.; Berger, S.L. RNA binding to CBP stimulates histone acetylation and transcription. Cell 2017, 168, 135–149.e22. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.L.; Fei, T.; Chen, Y.; Li, T.; Gao, Y.; Wang, X.; Sun, T.; Sweeney, C.J.; Lee, G.S.M.; Chen, S.; et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. USA 2014, 111, 7319–7324. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Orom, U.A.; Cesaroni, M.; Beringer, M.; Taatjes, D.J.; Blobel, G.A.; Shiekhattar, R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013, 494, 497–501. [Google Scholar] [CrossRef]
- Mousavi, K.; Zare, H.; Dell’Orso, S.; Grontved, L.; Gutierrez-Cruz, G.; Derfoul, A.; Hager, G.L.; Sartorelli, V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 2013, 51, 606–617. [Google Scholar] [CrossRef]
- Schaukowitch, K.; Joo, J.Y.; Liu, X.; Watts, J.K.; Martinez, C.; Kim, T.K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 2014, 56, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Spurlock, C.F.; Shaginurova, G.; Tossberg, J.T.; Hester, J.D.; Chapman, N.; Guo, Y.; Crooke, P.S.; Aune, T.M. Profiles of long noncoding RNAs in human naive and memory T cells. J. Immunol. 2017, 199, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Sinden, R.R. DNA Structure and Function; Elsevier: Amsterdam, The Netherlands, 1994; ISBN 9780080571737. [Google Scholar]
- Panayotatos, N.; Wells, R.D. Cruciform structures in supercoiled DNA. Nature 1981, 289, 466–470. [Google Scholar] [CrossRef]
- Hsieh, T.-S.; Wang, J.C. Thermodynamic properties of superhelical DNAs. Biochemistry 1975, 14, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Nishikawa, K.; Takemura, S. Studies on T. Utilis tRNATyr variants with enzymatically altered D-loop sequences. I. deletion of the conserved sequence Gm-G and its effects on aminoacylation and conformation. J. Biochem. 1985, 97, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T. New aspects of magnesium function: A key regulator in nucleosome self-assembly, chromatin folding and phase separation. Int. J. Mol. Sci. 2019, 20, 4232. [Google Scholar] [CrossRef] [PubMed]
- Hooper, M.; Hardy, K.; Handyside, A.; Hunter, S.; Monk, M. HPRT-deficient (Lesch–Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 1987, 326, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Tanase, J.; Mitani, T.; Udagawa, K.; Nishikawa, J.; Ohyama, T. Competence of an artificial bent DNA as a transcriptional activator in mouse ES cells. Mol. Biol. Rep. 2011, 38, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).