Bio-Engineered Nisin with Increased Anti-Staphylococcus and Selectively Reduced Anti-Lactococcus Activity for Treatment of Bovine Mastitis
Abstract
:1. Introduction
2. Results
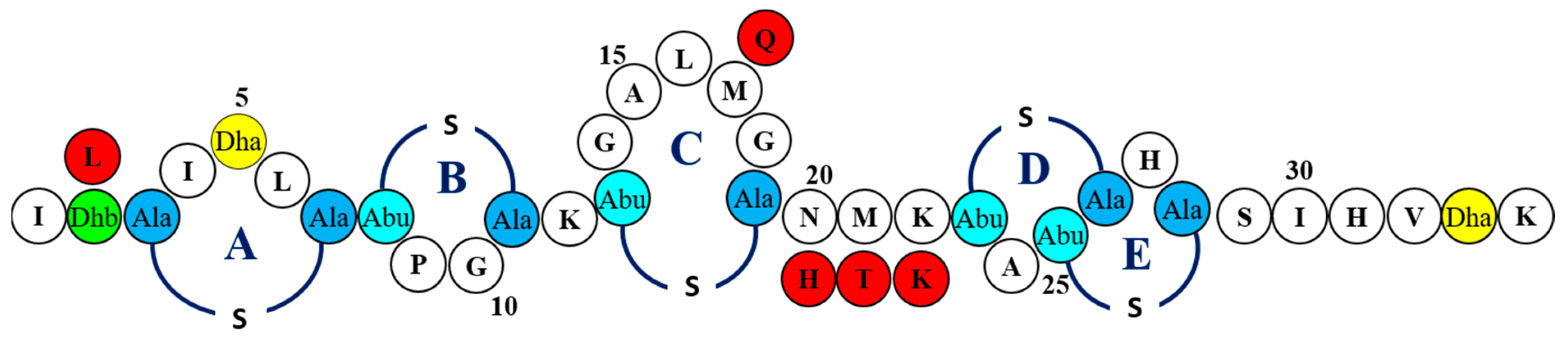
2.1. Screening of nisin Derivatives for Enhanced Antimicrobial Activity Against S. aureus Strains Associated with Bovine Mastitis
2.2. MIC-Based Investigations Demonstrate Enhanced Specific Activity of nisin Derivatives against Bovine Mastitis-Associated S. aureus
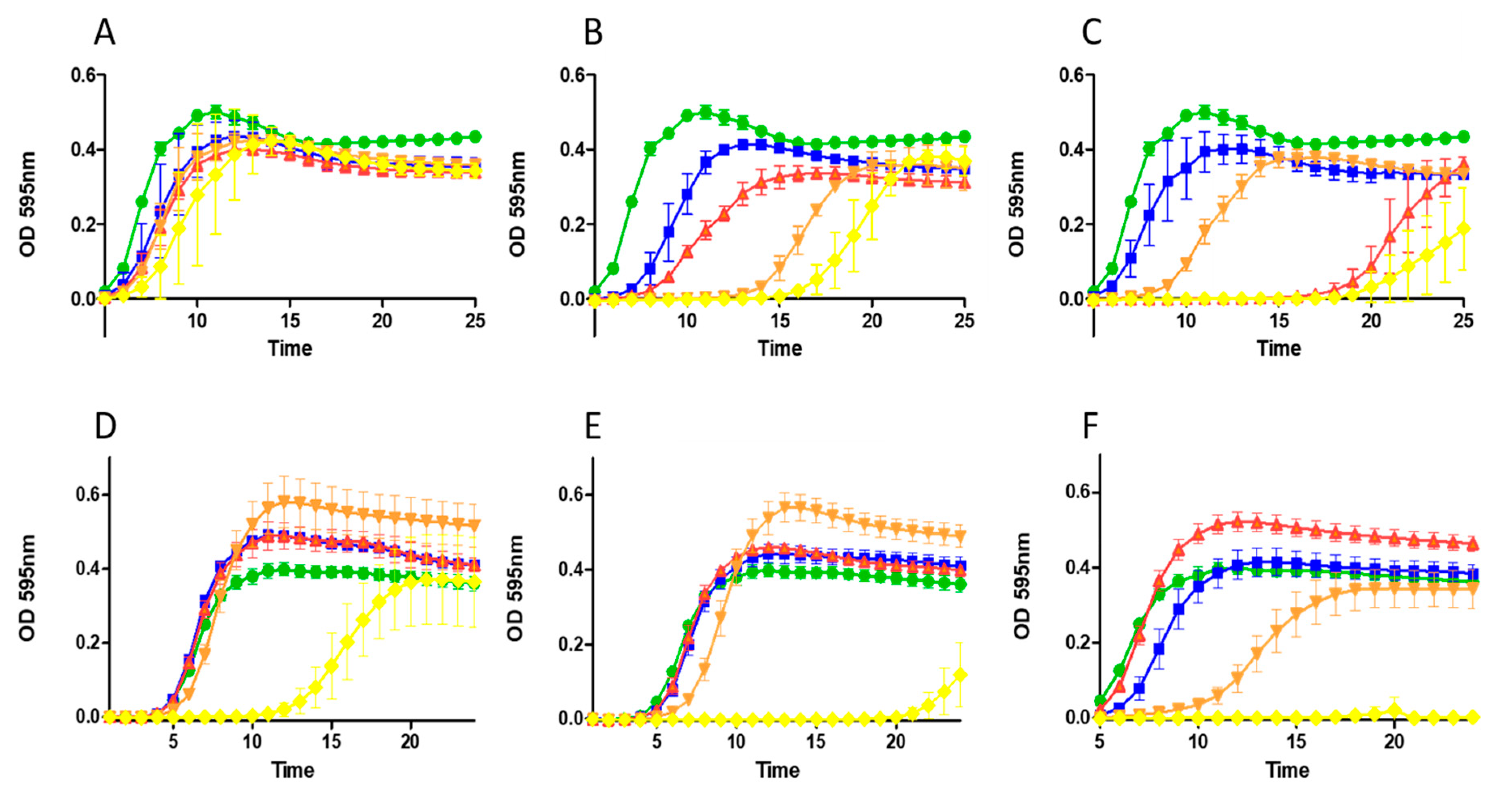
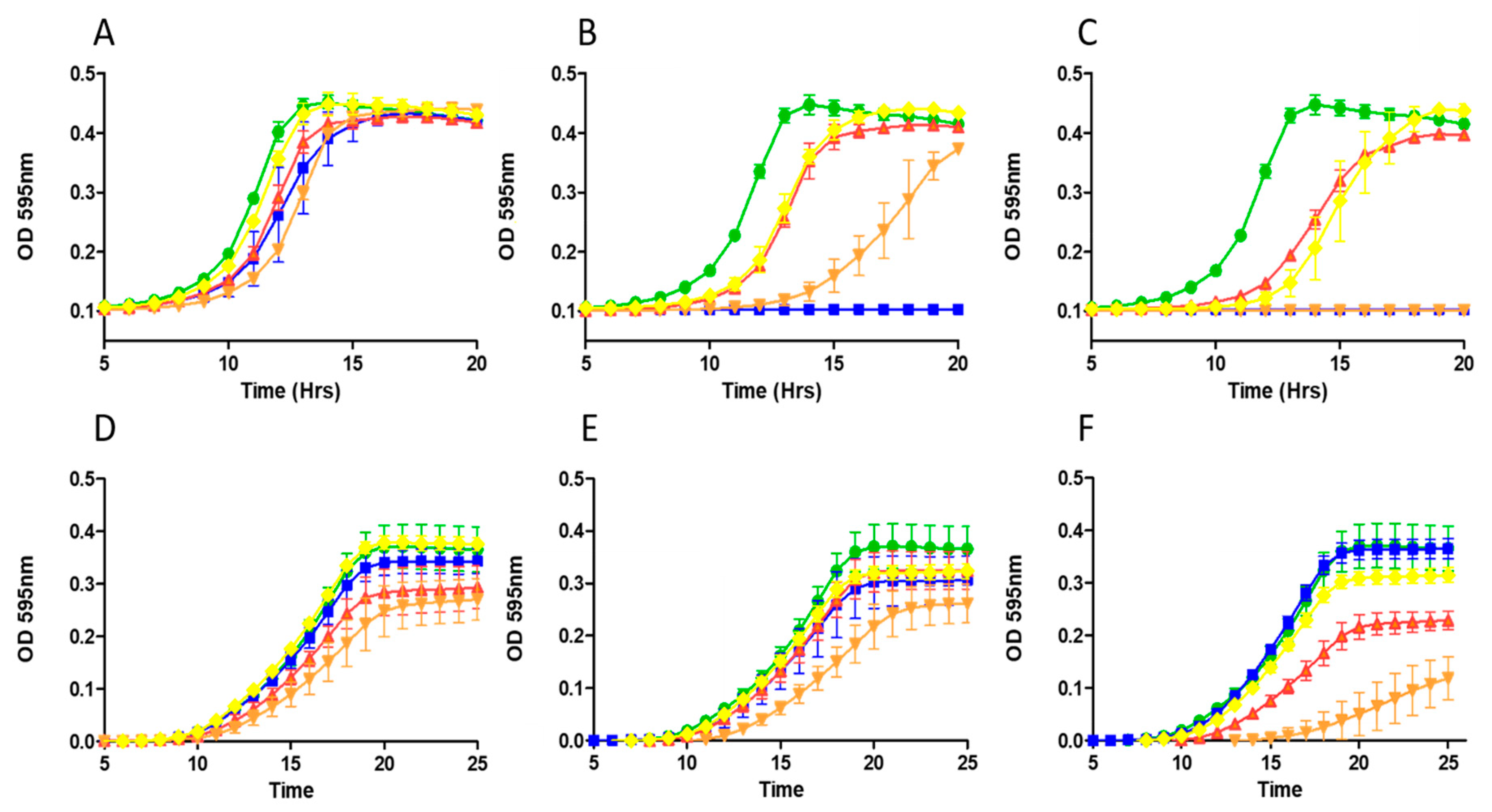
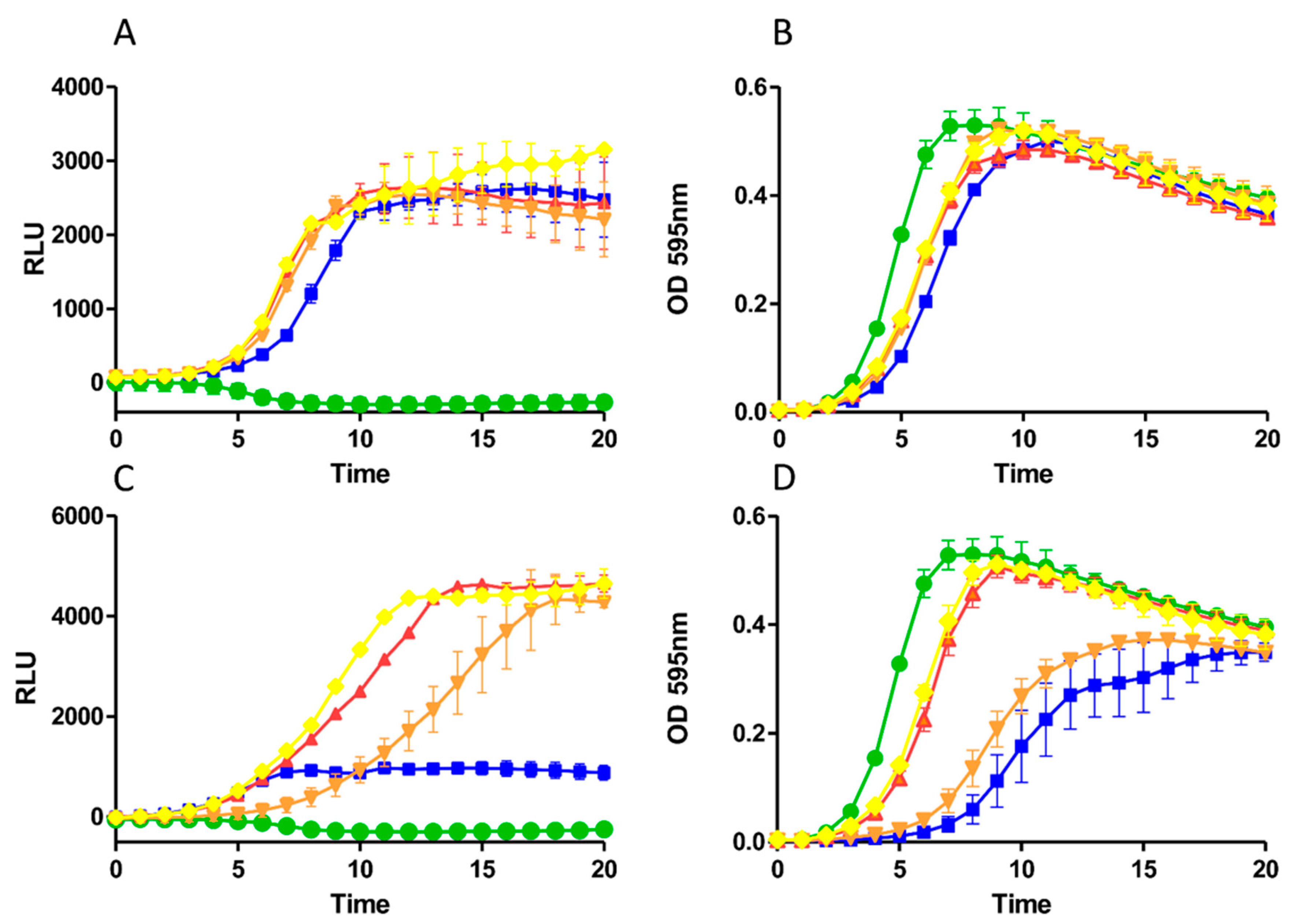
2.3. Growth Curve Based Assessment
2.4. Impact of Derivatives on Nisin Induction and Production
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Growth Conditions
5.2. Generation and Assessment of a Bank of Nisin Derivatives
5.3. Identification of Nisin Derivatives
5.4. Mass Spectrometry
5.5. Nisin Purification
5.6. Minimum Inhibitory Concentration Assays
5.7. Growth Curve Analysis
5.8. GFP Assays
5.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Viguier, C.; Arora, S.; Gilmartin, N.; Welbeck, K.; O’Kennedy, R. Mastitis Detection: Current Trends and Future Perspectives. Trends Biotechnol. 2009, 27, 486–493. [Google Scholar] [CrossRef]
- Zadoks, R.; Fitzpatrick, J. Changing Trends in Mastitis. Ir. Veter J. 2009, 62, S59–S70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghamohammadi, M.; Haine, D.; Kelton, D.F.; Barkema, H.W.; Hogeveen, H.; Keefe, G.P.; Dufour, S. Herd-Level Mastitis-Associated Costs on Canadian Dairy Farms. Front. Veter Sci. 2018, 5, 100. [Google Scholar] [CrossRef]
- Kelton, D.F.; Lissemore, K.D.; Martin, R.E. Recommendations for Recording and Calculating the Incidence of Selected Clinical Diseases of Dairy Cattle. J. Dairy Sci. 1998, 81, 2502–2509. [Google Scholar] [CrossRef]
- Szweda, P.; Schielmann, M.; Frankowska, A.; Kot, B.; Zalewska, M. Antibiotic Resistance in Staphylococcus aureus Strains Isolated from Cows with Mastitis in Eastern Poland and Analysis of Susceptibility of Resistant Strains to Alternative Nonantibiotic Agents: Lysostaphin, Nisin and Polymyxin B. J. Veter Med. Sci. 2014, 76, 355–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawada-Matsuo, M.; Watanabe, A.; Arii, K.; Oogai, Y.; Noguchi, K.; Miyawaki, S.; Hayashi, T.; Komatsuzawa, H. Staphylococcus aureus Virulence Affected by an Alternative Nisin A Resistance Mechanism. Appl. Environ. Microbiol. 2020, 86, e00182-20. [Google Scholar] [CrossRef]
- Park, S.; Ronholm, J. Staphylococcus aureus in Agriculture: Lessons in Evolution from a Multispecies Pathogen. Clin. Microbiol. Rev. 2021, 34. [Google Scholar] [CrossRef]
- Doyle, M.P.; Loneragan, G.H.; Scott, H.M.; Singer, R.S. Antimicrobial Resistance: Challenges and Perspectives. Compr. Rev. Food Sci. Food Saf. 2013, 12, 234–248. [Google Scholar] [CrossRef]
- Pieterse, R.; Todorov, S.D. Bacteriocins-Exploring Alternatives to Antibiotics in Mastitis Treatment. Braz. J. Microbiol. 2010, 41, 542–562. [Google Scholar] [CrossRef] [Green Version]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. Molecular Approaches to Analysing the Microbial Composition of Raw Milk and Raw Milk Cheese. Int. J. Food Microbiol. 2011, 150, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.; Hill, C.; Ross, R. Bacterial Lantibiotics: Strategies to Improve Therapeutic Potential. Curr. Protein Pept. Sci. 2005, 6, 61–75. [Google Scholar] [CrossRef]
- Ongey, E.L.; Yassi, H.; Pflugmacher, S.; Neubauer, P. Pharmacological and Pharmacokinetic Properties of Lanthipeptides Undergoing Clinical Studies. Biotechnol. Lett. 2017, 39, 473–482. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A Viable Alternative to Antibiotics? Nat. Rev. Genet. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Breukink, E.; De Kruijff, B. Lipid II as a Target for Antibiotics. Nat. Rev. Drug Discov. 2006, 5, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Sandiford, S.K. Perspectives on Lantibiotic Discovery-Where Have We Failed and What Improvements Are Required? Expert Opin. Drug Discov. 2015, 10, 315–320. [Google Scholar] [CrossRef]
- Delves-Broughton, J. Nisin as a Food Preservative. Food Aust. 2005, 57, 525–527. [Google Scholar]
- Sears, P.; Smith, B.; Stewart, W.; Gonzalez, R.; Rubino, S.; Gusik, S.; Kulisek, E.; Projan, S.; Blackburn, P. Evaluation of a Nisin-Based Germicidal Formulation on Teat Skin of Live Cows. J. Dairy Sci. 1992, 75, 3185–3190. [Google Scholar] [CrossRef]
- Wu, J.; Hu, S.; Cao, L. Therapeutic Effect of Nisin Z on Subclinical Mastitis in Lactating Cows. Antimicrob. Agents Chemother. 2007, 51, 3131–3135. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Wu, J.; Xie, F.; Hu, S.; Mo, Y. Efficacy of Nisin in Treatment of Clinical Mastitis in Lactating Dairy Cows. J. Dairy Sci. 2007, 90, 3980–3985. [Google Scholar] [CrossRef]
- Healy, B.; Field, D.; O’Connor, P.M.; Hill, C.; Cotter, P.D.; Ross, R.P. Intensive Mutagenesis of the Nisin Hinge Leads to the Rational Design of Enhanced Derivatives. PLoS ONE 2013, 8, e79563. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; Teng, K.; Wang, J.; Zhao, F.; Wang, F.; Zhang, J.; Zhong, J. Ligand Determinants of Nisin for Its Induction Activity. J. Dairy Sci. 2016, 99, 5022–5031. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Montalban-Lopez, M.; Kuipers, O.P. Increasing the Antimicrobial Activity of Nisin-Based Lantibiotics against Gram-Negative Pathogens. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Van Heel, A.J.; Montalban-Lopez, M.; Kuipers, O.P. Potentiating the Activity of Nisin against Escherichia coli. Front. Cell Dev. Biol. 2016, 4, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, D.; Connor, P.M.O.; Cotter, P.D.; Hill, C.; Ross, R.P. The Generation of Nisin Variants with Enhanced Activity against Specific Gram-Positive Pathogens. Mol. Microbiol. 2008, 69, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Begley, M.; O’Connor, P.M.; Daly, K.M.; Hugenholtz, F.; Cotter, P.D.; Hill, C.; Ross, R.P. Bioengineered Nisin a Derivatives with Enhanced Activity against Both Gram Positive and Gram Negative Pathogens. PLoS ONE 2012, 7, e46884. [Google Scholar] [CrossRef] [Green Version]
- Field, D.; Quigley, L.; O’Connor, P.M.; Rea, M.C.; Daly, K.; Cotter, P.D.; Hill, C.; Ross, R.P. Studies with Bioengineered Nisin Peptides Highlight the Broad-Spectrum Potency of Nisin V. Microb. Biotechnol. 2010, 3, 473–486. [Google Scholar] [CrossRef] [Green Version]
- Field, D.; Gaudin, N.; Lyons, F.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Ross, R.P. A Bioengineered Nisin Derivative to Control Biofilms of Staphylococcus pseudintermedius. PLoS ONE 2015, 10, e0119684. [Google Scholar] [CrossRef] [Green Version]
- Field, D.; Blake, T.; Mathur, H.; O’Connor, P.M.; Cotter, P.D.; Ross, R.P.; Hill, C. Bioengineering Nisin to Overcome the Nisin Resistance Protein. Mol. Microbiol. 2018, 111, 717–731. [Google Scholar] [CrossRef]
- Sun, Z.; Zhong, J.; Liang, X.; Liu, J.; Chen, X.; Huan, L. Novel Mechanism for Nisin Resistance via Proteolytic Degradation of Nisin by the Nisin Resistance Protein NSR. Antimicrob. Agents Chemother. 2009, 53, 1964–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouse, S.; Field, D.; Daly, K.M.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Ross, R.P. Bioengineered Nisin Derivatives with Enhanced Activity in Complex Matrices. Microb. Biotechnol. 2012, 5, 501–508. [Google Scholar] [CrossRef]
- Weidenmaier, C.; Peschel, A.; Xiong, Y.; Kristian, S.A.; Dietz, K.; Yeaman, M.R.; Bayer, A.S. Lack of Wall Teichoic Acids in Staphylococcus aureus Leads to Reduced Interactions with Endothelial Cells and to Attenuated Virulence in a Rabbit Model of Endocarditis. J. Infect. Dis. 2005, 191, 1771–1777. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, M.; Field, D.; Grainger, A.; O’Connor, P.M.; Draper, L.; Ross, R.P.; Hill, C. Nisin M: A Bioengineered Nisin A Variant That Retains Full Induction Capacity but Has Significantly Reduced Antimicrobial Activity. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Islam, M.R.; Shioya, K.; Nagao, J.; Nishie, M.; Jikuya, H.; Zendo, T.; Nakayama, J.; Sonomoto, K. Evaluation of Essential and Variable Residues of Nukacin ISK-1 by NNK Scanning. Mol. Microbiol. 2009, 72, 1438–1447. [Google Scholar] [CrossRef]
- Appleyard, A.N.; Choi, S.; Read, D.M.; Lightfoot, A.; Boakes, S.; Hoffmann, A.; Chopra, I.; Bierbaum, G.; Rudd, B.A.; Dawson, M.J.; et al. Dissecting Structural and Functional Diversity of the Lantibiotic Mersacidin. Chem. Biol. 2009, 16, 490–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, J.; Caetano, T.; Mösker, E.; Süssmuth, R.; Mendo, S. Lichenicidin Rational Site-Directed Mutagenesis library: A Tool to Generate Bioengineered Lantibiotics. Biotechnol. Bioeng. 2019, 116, 3053–3062. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, O.P.; Beerthuyzen, M.M.; de Ruyter, P.G.G.; Luesink, E.J.; de Vos, W.M. Autoregulation of Nisin Biosynthesis in Lactococcus lactis by Signal Transduction. J. Biol. Chem. 1995, 270, 27299–27304. [Google Scholar] [CrossRef] [Green Version]
- Twomey, E.; Hill, C.; Field, D.; Begley, M. Bioengineered Nisin Derivative M17Q Has Enhanced Activity against Staphylococcus epidermidis. Antibiotics 2020, 9, 305. [Google Scholar] [CrossRef]
- Hiron, A.; Falord, M.; Valle, J.; Débarbouillé, M.; Msadek, T. Bacitracin and Nisin Resistance in Staphylococcus aureus: A Novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 2011, 81, 602–622. [Google Scholar] [CrossRef] [PubMed]
- Draper, L.A.; Cotter, P.D.; Hill, C.; Ross, R.P. Lantibiotic Resistance. Microbiol. Mol. Biol. Rev. 2015, 79, 171–191. [Google Scholar] [CrossRef] [Green Version]
- Randall, C.P.; Gupta, A.; Utley-Drew, B.; Lee, S.Y.; Morrison-Williams, G.; O’Neill, A.J.; Andersson, D.; Gebhard, S.; Miller, K. Acquired Nisin Resistance in Staphylococcus aureus Involves Constitutive Activation of an Intrinsic Peptide Antibiotic Detoxification Module. mSphere 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Medeiros-Silva, J.; Jekhmane, S.; Paioni, A.L.; Gawarecka, K.; Baldus, M.; Swiezewska, E.; Breukink, E.; Weingarth, M. High-Resolution NMR Studies of Antibiotics in Cellular Membranes. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Molloy, E.M.; Field, D.; O’Connor, P.M.; Cotter, P.D.; Hill, C.; Ross, R.P. Saturation Mutagenesis of Lysine 12 Leads to the Identification of Derivatives of Nisin A with Enhanced Antimicrobial Activity. PLoS ONE 2013, 8, e58530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.-G. Specific Binding of Nisin to the Peptidoglycan Precursor Lipid II Combines Pore Formation and Inhibition of Cell Wall Biosynthesis for Potent Antibiotic Activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Yin, Z.; Breukink, E.; Moll, G.N.; Kuipers, O.P. An Engineered Double Lipid II Binding Motifs-Containing Lantibiotic Displays Potent and Selective Antimicrobial Activity against Enterococcus faecium. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rink, R.; Wierenga, J.; Kuipers, A.; Kluskens, L.D.; Driessen, A.J.M.; Kuipers, O.P.; Moll, G.N. Dissection and Modulation of the Four Distinct Activities of Nisin by Mutagenesis of Rings A and B and by C-Terminal Truncation. Appl. Environ. Microbiol. 2007, 73, 5809–5816. [Google Scholar] [CrossRef] [Green Version]
- Jensen, C.; Li, H.; Vestergaard, M.; Dalsgaard, A.; Frees, D.; Leisner, J.J. Nisin Damages the Septal Membrane and Triggers DNA Condensation in Methicillin-Resistant Staphylococcus aureus. Front. Microbiol. 2020, 11, 1007. [Google Scholar] [CrossRef]
- Wouters, J.T.; Ayad, E.H.; Hugenholtz, J.; Smit, G. Microbes from Raw Milk for Fermented Dairy Products. Int. Dairy J. 2002, 12, 91–109. [Google Scholar] [CrossRef]
- Fernández, L.; Delgado, S.; Herrero, H.; Maldonado, A.; Rodríguez, J.M. The Bacteriocin Nisin, an Effective Agent for the Treatment of Staphylococcal Mastitis During Lactation. J. Hum. Lact. 2008, 24, 311–316. [Google Scholar] [CrossRef]
- Roy, S.M.; Riley, M.A.; Crabb, J.H. Treating Bovine Mastitis with Nisin: A Model for the Use of Protein Antimicrobials in Veterinary Medicine. In The Bacteriocins: Current Knowledge and Future Prospects; Caister Academic Press: Poole, UK, 2016; pp. 127–140. [Google Scholar] [CrossRef]
| Indicator | Nisin A (mm) | HTK (mm) | M17Q (mm) | T2L (mm) |
|---|---|---|---|---|
| S. aureus RF122 | 10.44 ± 0.69 | 14.89 ± 0.47 * | 15.39 ± 1.7 * | 10.15 ± 0.11 |
| S. aureus NCDO1499 | 11.73 ± 0.54 | 14.75 ± 0.48 * | 16.51 ± 0.16 * | 10.20 ± 0.13 * |
| S. aureus DPC 5243 | 12.71 ± 0.33 | 16.06 ± 0.37 * | 16.76 ± 0.55 * | 10.04 ± 0.43 * |
| S. aureus ST528 (MRSA) | 12.25 ± 0.30 | 14.91 ± 0.26 * | 16.32 ± 1.2 * | 10.09 ± 0.64 * |
| S. aureus ST534 (MRSA) | 7.51 ± 0.19 | 10.41 ± 0.88 * | 12.17 ± 0.36 * | 7.26 ± 0.66 |
| S. aureus SA113 | 6.90 ± 0.16 | 8.34 ± 0.57 * | 9.14 ± 0.51 * | 6.28 ± 0.08 |
| S. aureus SA113 mprf | 10.16 ± 0.52 | 13.34 ± 0.25 * | 12.98 ± 0.53 * | 7.83 ± 0.1 * |
| S. aureus SA113 dltA | 13.09 ± 0.84 | 15.66 ± 0.54 * | 17.51 ± 1.40 * | 12.75 ± 0.75 |
| S. uberis DPC5344 | 17.59 ± 0.37 | 18.66 ± 0.69 | 18.27 ± 0.47 | 16.93 ± 1.50 |
| S. dysgal ATCC43078 | 10.55 ± 0.48 | 15.51 ± 0.27 * | 11.99 ± 0.47 * | 11.28 ± 0.32 * |
| S. agalactiae ATCC13813 | 9.34 ± 0.15 | 9.36 ± 0.08 | 12.25 ± 0.11 * | 9.08 ± 0.12 |
| L. lactis MG1363 | 9.39 ± 0.04 | 9.12 ± 0.13 | 11.25 ± 0.24 * | 7.54 ± 0.39 * |
| L. lactis HP | 23.65 ± 0.13 | 23.17 ± 0.42 | 27.69 ± 0.70 * | 20.33 ± 0.66 * |
| L. lactis KH | 17.43 ± 1.89 | 17.71 ± 2.03 | 20.36 ± 2.2 | 14.72 ± 2.06 |
| L. lactis IP5 | 17.14 ± 2.2 | 18.13 ± 3.1 | 20.71 ± 2.2 | 14.13 ± 3.2 |
| Lb. acidophilus ATCC 4356 | 20.17 ± 0.25 | 17.75 ± 3.5 | 27.54 ± 1.2 * | 17.43 ± 2.0 * |
| Indicator | Nisin A µg/mL (µM) | HTK µg/mL (µM) | M17Q µg/mL (µM) | T2L µg/mL (µM) |
|---|---|---|---|---|
| S. aureus NCDO1499 | 2 (0.625) | 1 (0.312) | 1 (0.312) | 0.5 (0.156) |
| S. aureus RF122 | 1 (0.312) | 0.12 (0.039) | 0.25 (0.078) | 0.25 (0.078) |
| S. aureus DPC 5243 | 1 (0.312) | 1 (0.312) | 2 (0.625) * | 0.5 (0.156) |
| S. aureus DPC 5245 | 1 (0.312) | 1 (0.312) | 1 (0.312) | 0.5(0.156) |
| S. aureus DPC 5247 | 1 (0.312) | 1 (0.312) | 1 (0.312) | 0.5(0.156) |
| S. aureus SA113 | 8 (2.5) | 8 (2.5) | 8 (2.5) | 1 (0.312) |
| S. aureus SA113Δmprf | 4 (1.25) | 4 (1.25) | 4 (1.25) | 0.25 (0.078) |
| S. aureus SA113ΔdltA | 2 (0.625) | 2 (0.625) | 2 (0.625) | 0.06 (0.019) |
| S. aureus 513 | 6 (1.875) | 6 (1.875) | 6 (1.875) | 3 (0.937) |
| S. aureus 272 | 3 (0.937) | 6 (1.875) * | 1.5 (0.468) | 0.75 (0.234) |
| S. aureus ST528(MRSA) | 0.5 (0.156) | 0.25 (0.078) | 0.25 (0.078) | 0.125 (0.039) |
| S. aureus ST534(MRSA) | 1 (0.312) | 1 (0.312) | 1 (0.312) | 0.25 (0.078) |
| S. dysgalactiae ATCC43078 | 4 (1.25) | 4 (1.25) | 8 (2.5) * | 8 (2.5) * |
| L. lactis MG1363 | 0.2 (0.06) | 0.1 (0.03) * | 0.1 (0.03) * | 0.2 (0.06) |
| L. lactis HP | 0.2 (0.06) | 0.4 (0.125) | 0.1 (0.03) * | 0.4 (0.125) |
| L. lactis KH | 0.05 (0.015) | 0.05 (0.015) | 0.05 (0.015) | (0.031) |
| L. lactis IP5 | 0.25 (0.078) | 0.25 (0.078) | 0.25 (0.078) | 1 (0.312) |
| B. longum UCC 44b | 0.06 (0.019) | 0.24 (0.072) | 0.06 (0.019) | 0.12 (0.039) |
| Lb. acidophilus ATCC 4356 | 0.03 (0.010) | 0.12 (0.039) | 0.06 (0.022) | 0.12 (0.039) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Field, D.; Considine, K.; O’Connor, P.M.; Ross, R.P.; Hill, C.; Cotter, P.D. Bio-Engineered Nisin with Increased Anti-Staphylococcus and Selectively Reduced Anti-Lactococcus Activity for Treatment of Bovine Mastitis. Int. J. Mol. Sci. 2021, 22, 3480. https://doi.org/10.3390/ijms22073480
Field D, Considine K, O’Connor PM, Ross RP, Hill C, Cotter PD. Bio-Engineered Nisin with Increased Anti-Staphylococcus and Selectively Reduced Anti-Lactococcus Activity for Treatment of Bovine Mastitis. International Journal of Molecular Sciences. 2021; 22(7):3480. https://doi.org/10.3390/ijms22073480
Chicago/Turabian StyleField, Des, Kiera Considine, Paula M. O’Connor, R. Paul Ross, Colin Hill, and Paul D. Cotter. 2021. "Bio-Engineered Nisin with Increased Anti-Staphylococcus and Selectively Reduced Anti-Lactococcus Activity for Treatment of Bovine Mastitis" International Journal of Molecular Sciences 22, no. 7: 3480. https://doi.org/10.3390/ijms22073480






