Reconstitution of Cytokinin Signaling in Rice Protoplasts
Abstract
:1. Introduction
2. Results
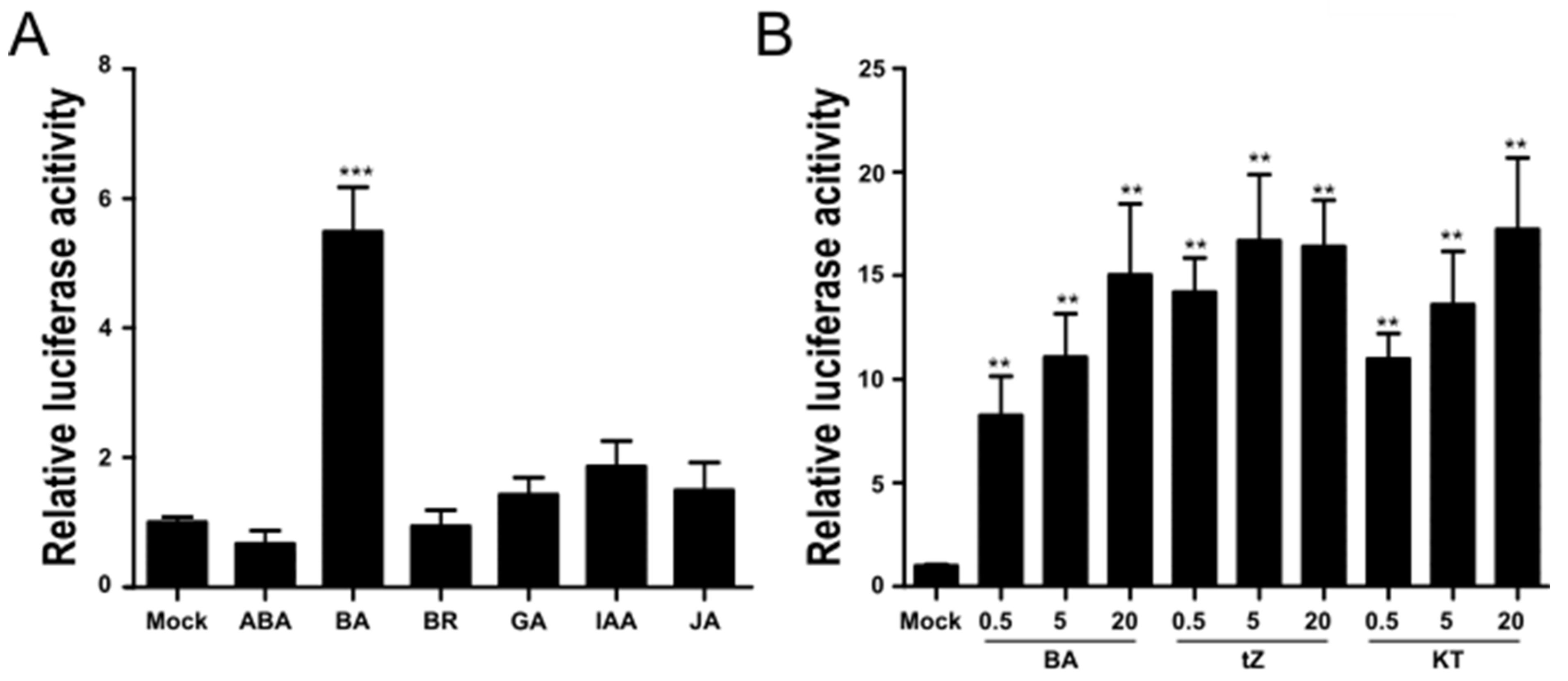
2.1. TCSn::fLUC Is a CK-Responsive Reporter in Rice Protoplasts
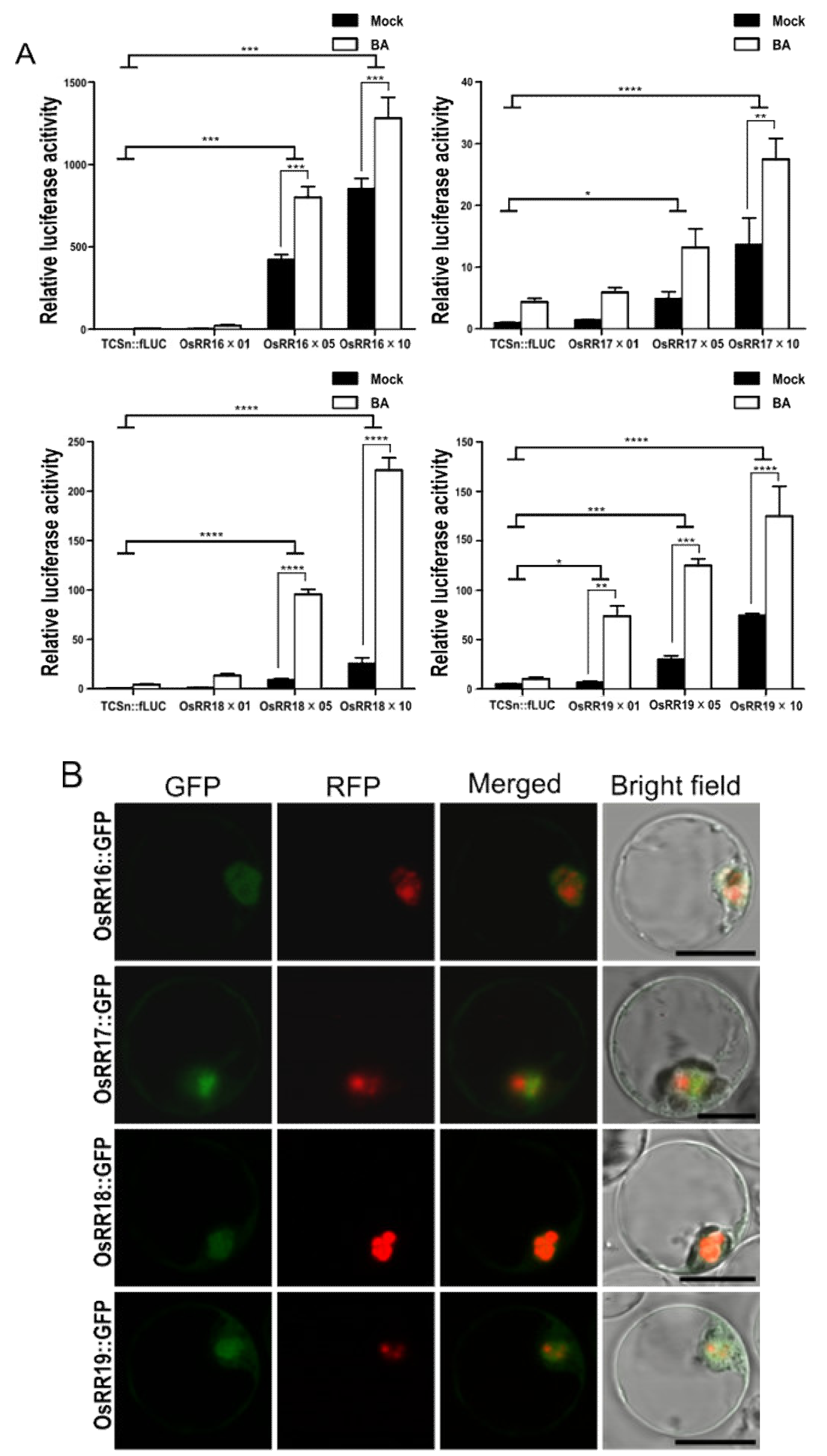
2.2. Type-B OsRRs Have Different Trans Activities and CK Responsiveness for TCSn Promoter
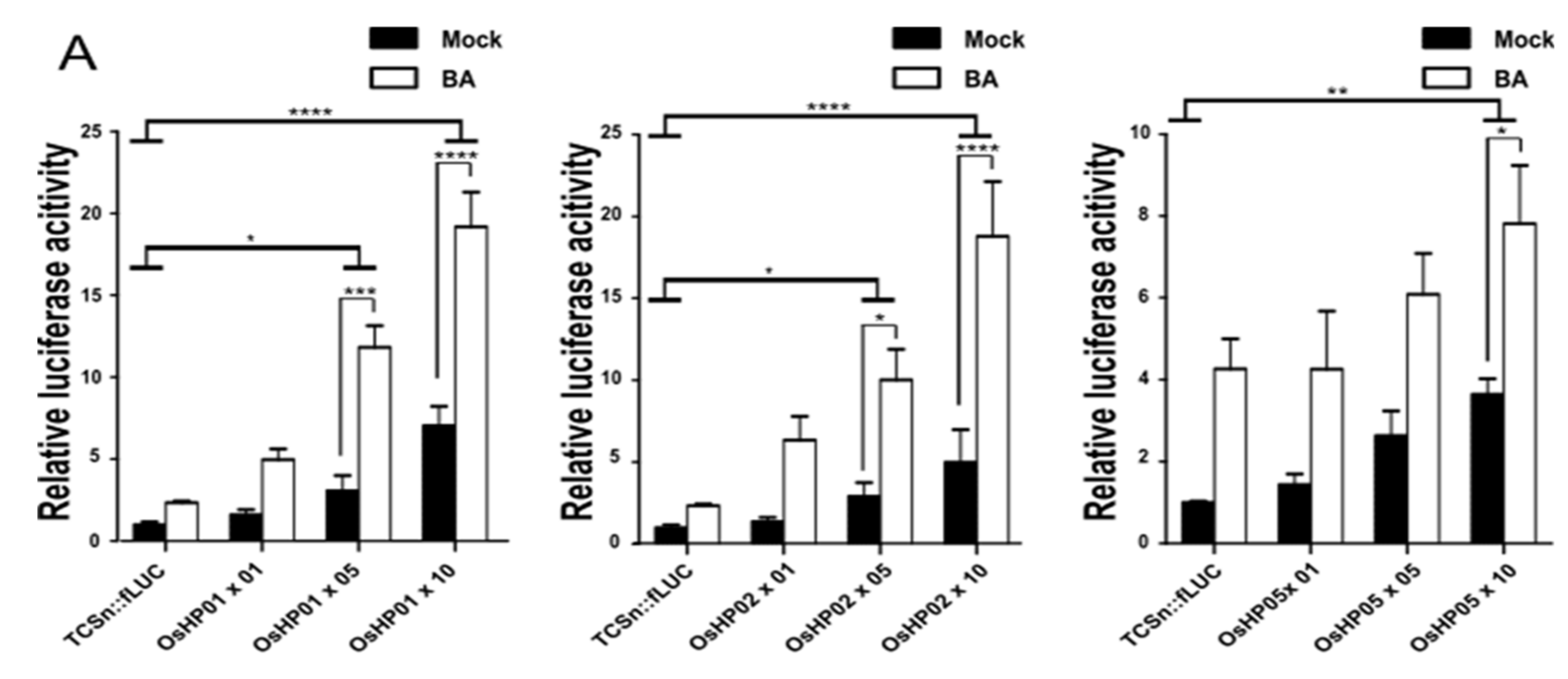
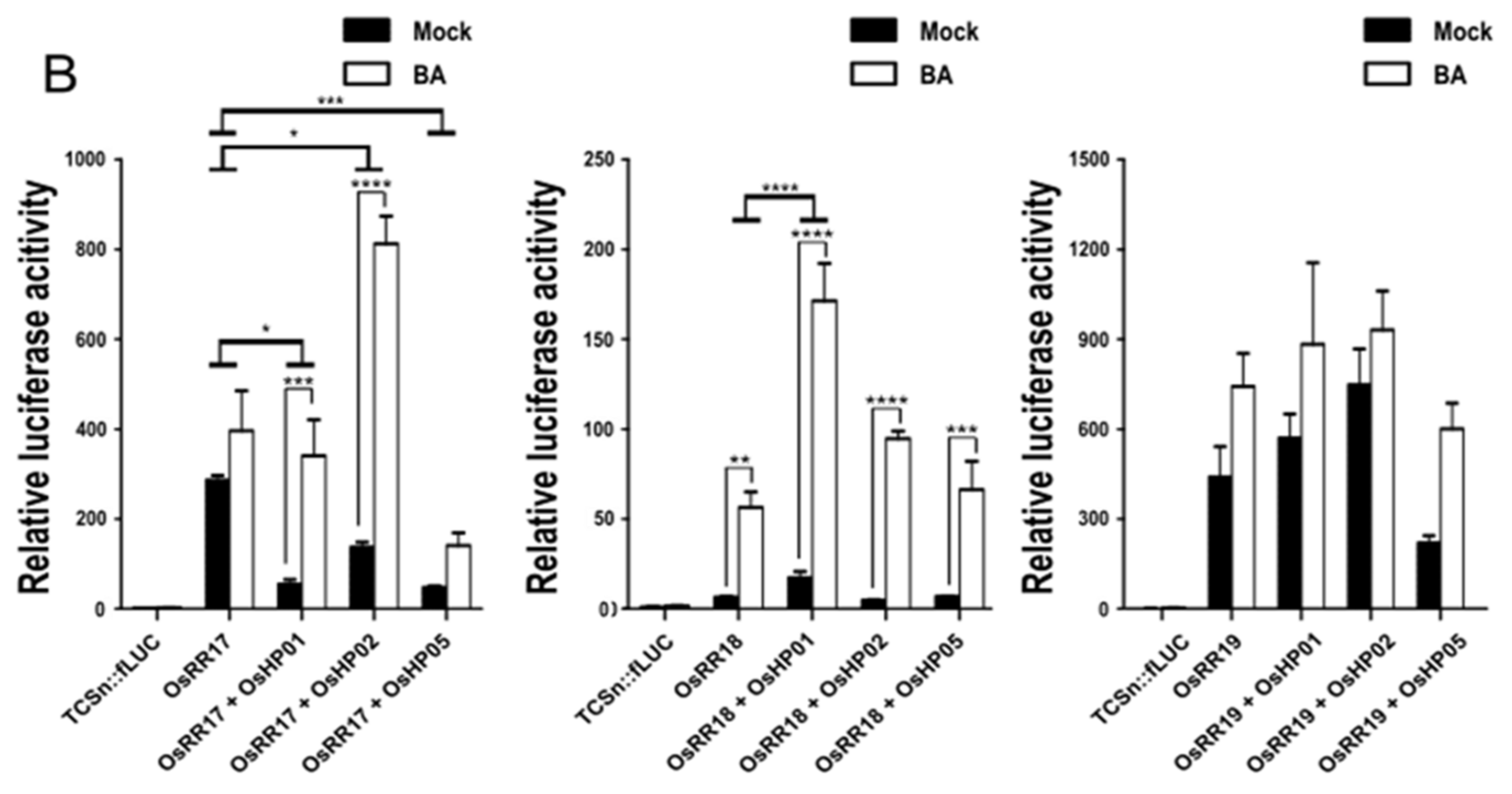
2.3. Each HP Protein Increases the Transcriptional Activation of the TCSn Promoter by Type-B OsRRs
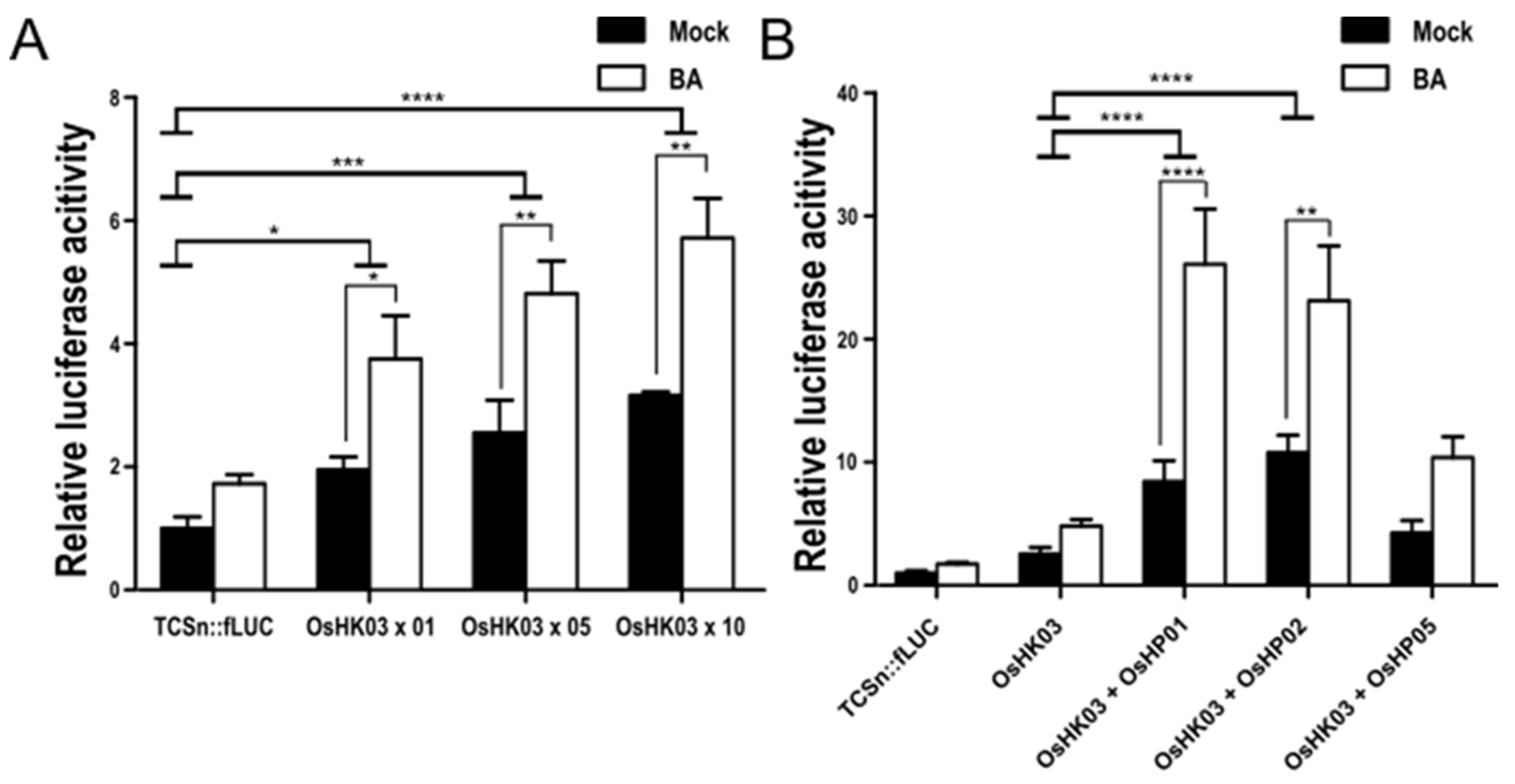
2.4. The CK Receptor OsHK03 Requires OsHPs to Induce the TCSn:fLUC Reporter
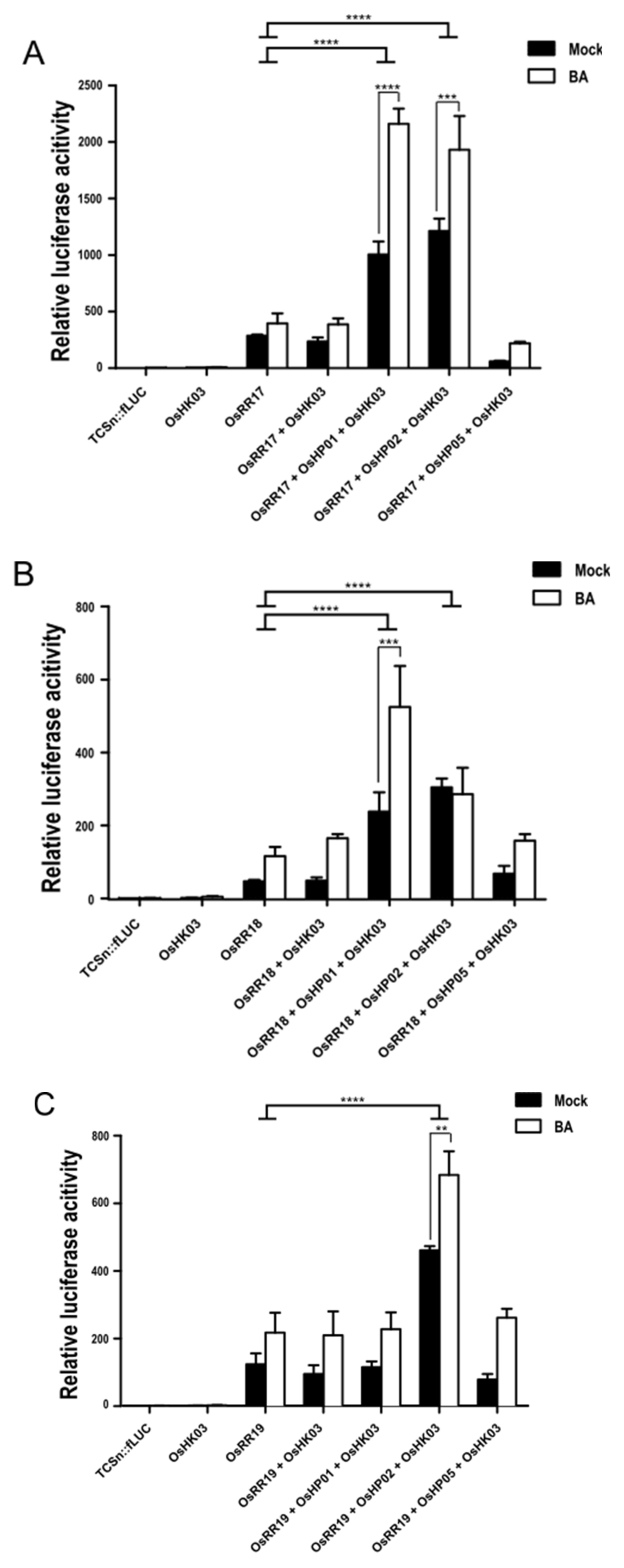
2.5. Reconstitution of the CK Signaling Pathway with Type-B OsRR, OsHP, and OsHK in Rice Protoplasts
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Isolation of Rice Protoplasts
4.3. Transient Gene Expression in Rice Protoplast
4.4. Construction of Vectors
4.5. Dual Luciferase Assays
4.6. Subcellular Localization and Bimolecular Fluorescence Complementation (BiFC) Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Aoyama, T.; Oka, A. Cytokinin signal transduction in plant cells. J. Plant Res. 2003, 116, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, M.A.; Nelson, D.C.; Weijers, D. Evolution of Plant Hormone Response Pathways. Annu. Rev. Plant Biol. 2020, 71, 327–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McSteen, P.; Zhao, Y. Plant hormones and signaling: Common themes and new developments. Dev. Cell 2008, 14, 467–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Liu, Y.; Li, S.S.; Han, G.Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e3345. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.; Moon, S.J.; Min, M.K.; Choi, E.H.; Kim, J.A.; Koh, E.Y.; Yoon, I.; Byun, M.O.; Yoo, S.D.; Kim, B.G. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front. Plant Sci. 2015, 6, 614. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruschhaupt, M.; Mergner, J.; Mucha, S.; Papacek, M.; Doch, I.; Tischer, S.V.; Hemmler, D.; Chiasson, D.; Edel, K.H.; Kudla, J.; et al. Rebuilding core abscisic acid signaling pathways of Arabidopsis in yeast. EMBO J. 2019, 38, e101859. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, B. Generic signal-specific responses: Cytokinin and context-dependent cellular responses. J. Exp. Bot. 2011, 62, 3273–3288. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. Phosphorelay and transcription control in cytokinin signal transduction. Science 2002, 296, 1650–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef] [PubMed]
- To, J.P.; Kieber, J.J. Cytokinin signaling: Two-components and more. Trends Plant Sci. 2008, 13, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Mira-Rodado, V. New Insights into Multistep-Phosphorelay (MSP)/ Two-Component System (TCS) Regulation: Are Plants and Bacteria that Different? Plants 2019, 8, 590. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Jiao, F.; Chu, J.; Jin, G.; Chen, M.; Wu, P. The two-component signal system in rice (Oryza sativa L.): A genome-wide study of cytokinin signal perception and transduction. Genomics 2007, 89, 697–707. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.H.; Schaller, G.E.; Kieber, J.J. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef] [Green Version]
- Romanov, G.A.; Lomin, S.N.; Schmulling, T. Cytokinin signaling: From the ER or from the PM? That is the question! New Phytol. 2018, 218, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [Green Version]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arab. Book 2008, 6, e0112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramireddy, E.; Brenner, W.G.; Pfeifer, A.; Heyl, A.; Schmulling, T. In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence. Plant Cell Physiol. 2013, 54, 1079–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, B.; Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 2008, 453, 1094–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zürcher, E.; Tavor-Deslex, D.; Lituiev, D.; Enkerli, K.; Tarr, P.T.; Müller, B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013, 161, 1066–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, J.; Sun, H.; Gu, P.; Liang, Z.; Chen, X.; Lou, J.; Xu, G.; Zhang, Y. A sensitive synthetic reporter for visualizing cytokinin signaling output in rice. Plant Methods 2017, 13, 89. [Google Scholar] [CrossRef] [Green Version]
- Steiner, E.; Israeli, A.; Gupta, R.; Schwartz, I.; Nir, I.; Leibman-Markus, M.; Tal, L.; Farber, M.; Amsalem, Z.; Ori Naomi Müller, B.; et al. Characterization of the cytokinin sensor TCSv2 in arabidopsis and tomato. Plant Methods 2020, 16, 1–12. [Google Scholar] [CrossRef]
- Dortray, H.; Mehnert, N.; Burkle, L.; Schmulling, T.; Hey, A. Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 2006, 273, 4631–4644. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Leyva-Gonzalez, M.A.; Ha, C.V.; Fujita, Y.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Herrera-Estrella, L.; et al. Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. USA 2013, 110, 4840–4845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaller, G.E.; Shiu, S.; Armitage, P.A. Two-Component Systems and Their Co-Option for Eukaryotic Signal Transduction. Curr. Biol. 2011, 21, R320–R330. [Google Scholar] [CrossRef] [Green Version]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Zhang, Q.; Wu, J.; Zhang, L.; Jiao, X.; Zhang, S.; Zhang, Z.; Sun, D.; Lu, T.; Sun, Y. Two rice authentic histidine phosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice. Plant Physiol. 2014, 165, 335–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.; Kim, J. Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 2013, 161, 408–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mähönen, A.P.; Bishopp, A.; Higuchi, M.; Nieminen, K.M.; Kinoshita, K.; Tormakangas, K.; Ikeda, Y.; Oka, A.; Kakimoto, T.; Helariutta, Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 2006, 311, 94–98. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ga, E.; Song, J.; Min, M.K.; Ha, J.; Park, S.; Lee, S.B.; Lee, J.-Y.; Kim, B.-G. Reconstitution of Cytokinin Signaling in Rice Protoplasts. Int. J. Mol. Sci. 2021, 22, 3647. https://doi.org/10.3390/ijms22073647
Ga E, Song J, Min MK, Ha J, Park S, Lee SB, Lee J-Y, Kim B-G. Reconstitution of Cytokinin Signaling in Rice Protoplasts. International Journal of Molecular Sciences. 2021; 22(7):3647. https://doi.org/10.3390/ijms22073647
Chicago/Turabian StyleGa, Eunji, Jaeeun Song, Myung Ki Min, Jihee Ha, Sangkyu Park, Saet Buyl Lee, Jong-Yeol Lee, and Beom-Gi Kim. 2021. "Reconstitution of Cytokinin Signaling in Rice Protoplasts" International Journal of Molecular Sciences 22, no. 7: 3647. https://doi.org/10.3390/ijms22073647





