Structural and Mechanismic Studies of Lactophoricin Analog, Novel Antibacterial Peptide
Abstract
:1. Introduction
2. Results
2.1. Design and Antibacterial Activity Test of Various LPcin Analogs
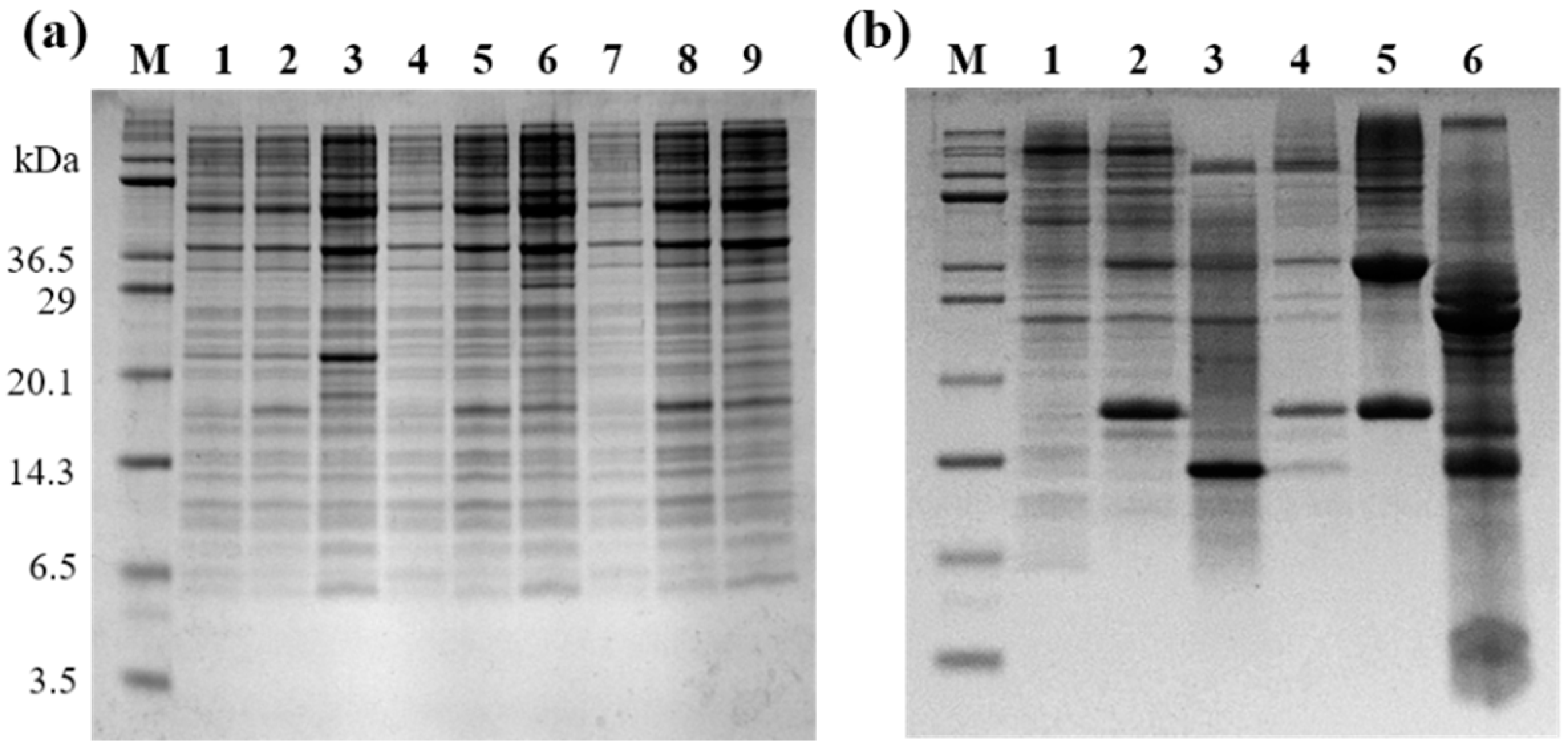
2.2. Expression of Selected LPcin Analog
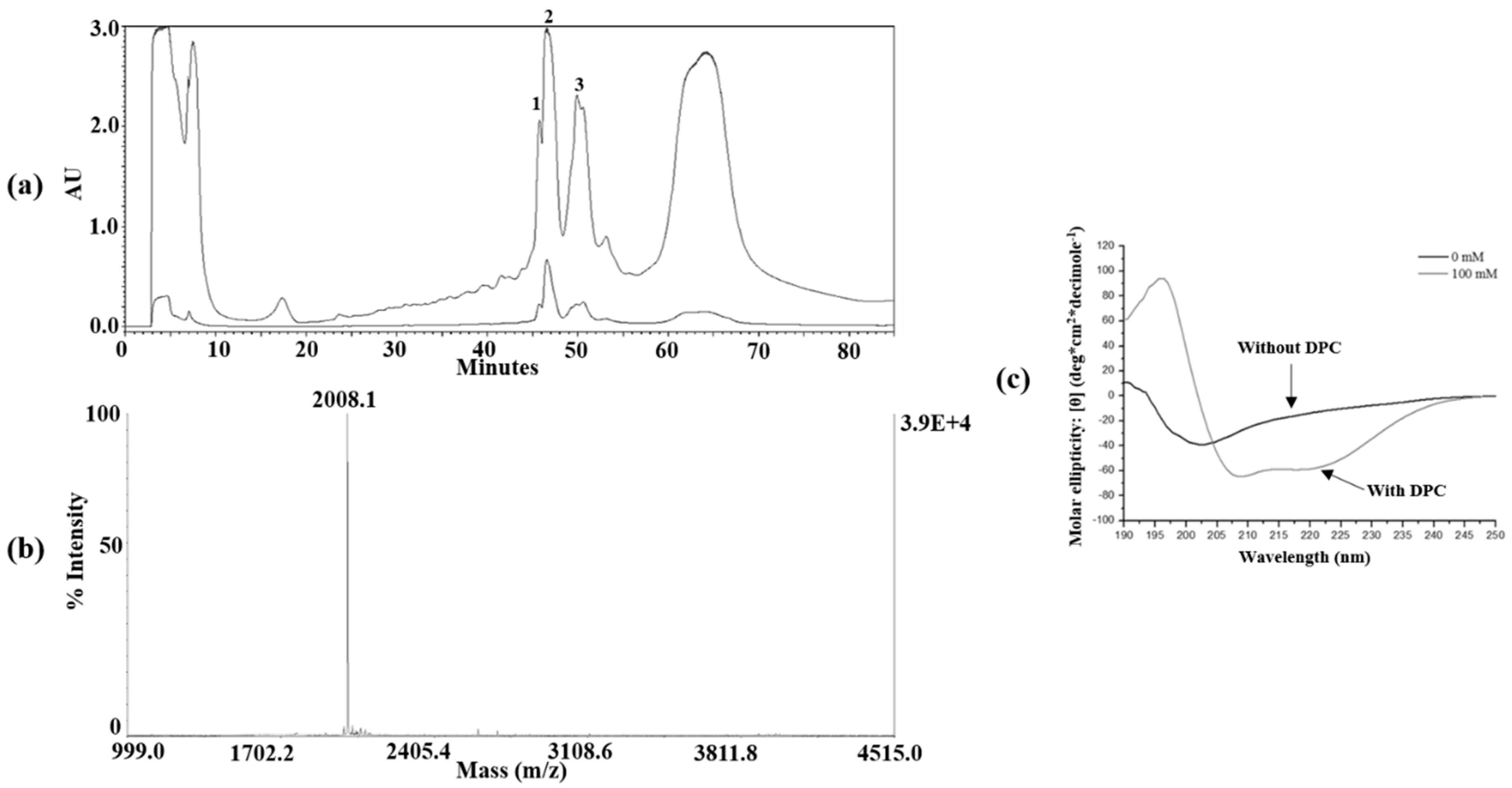
2.3. Peptide Isolation and Purification
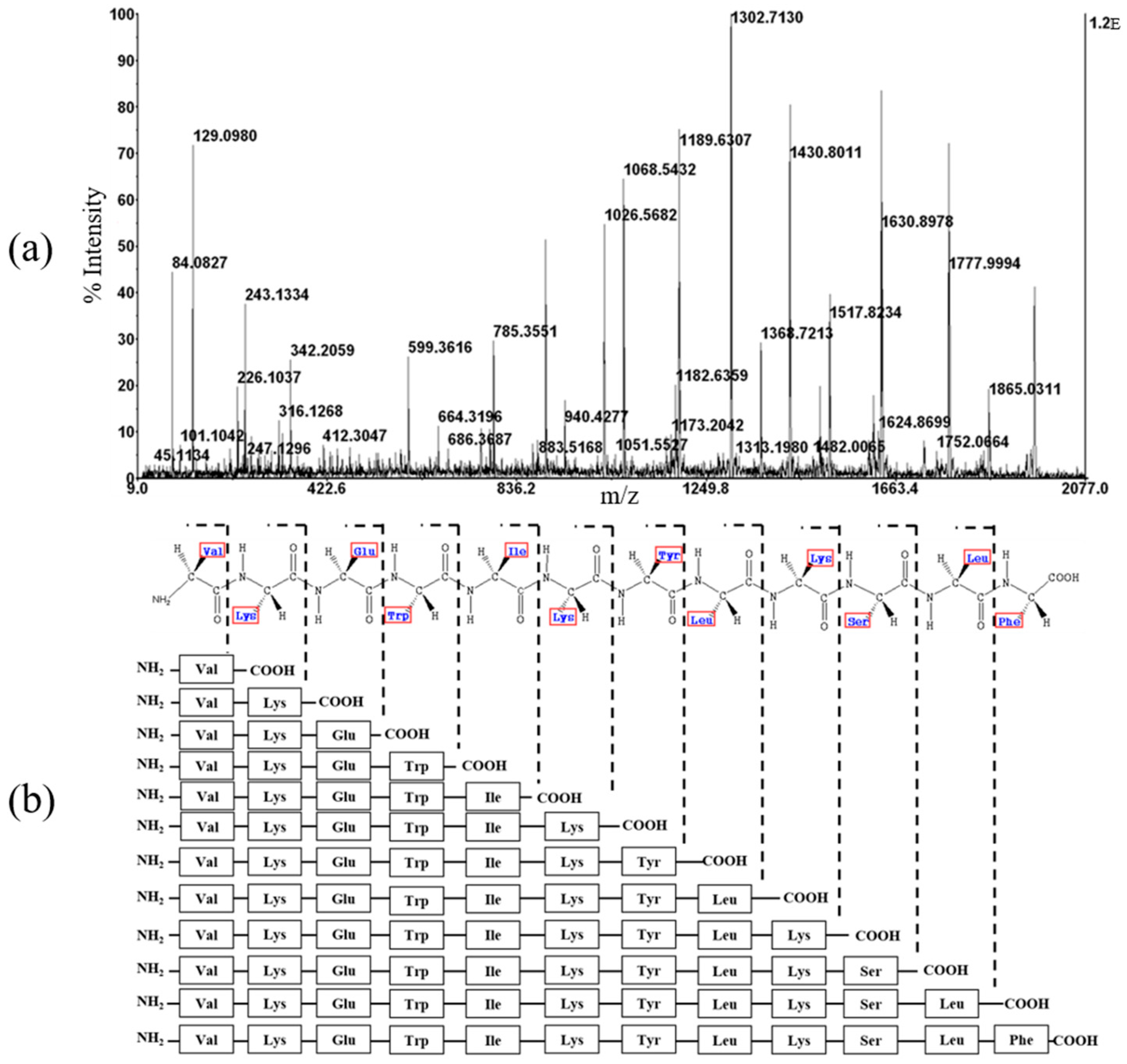
2.4. Mass Spectrometry and CD Spectroscopy
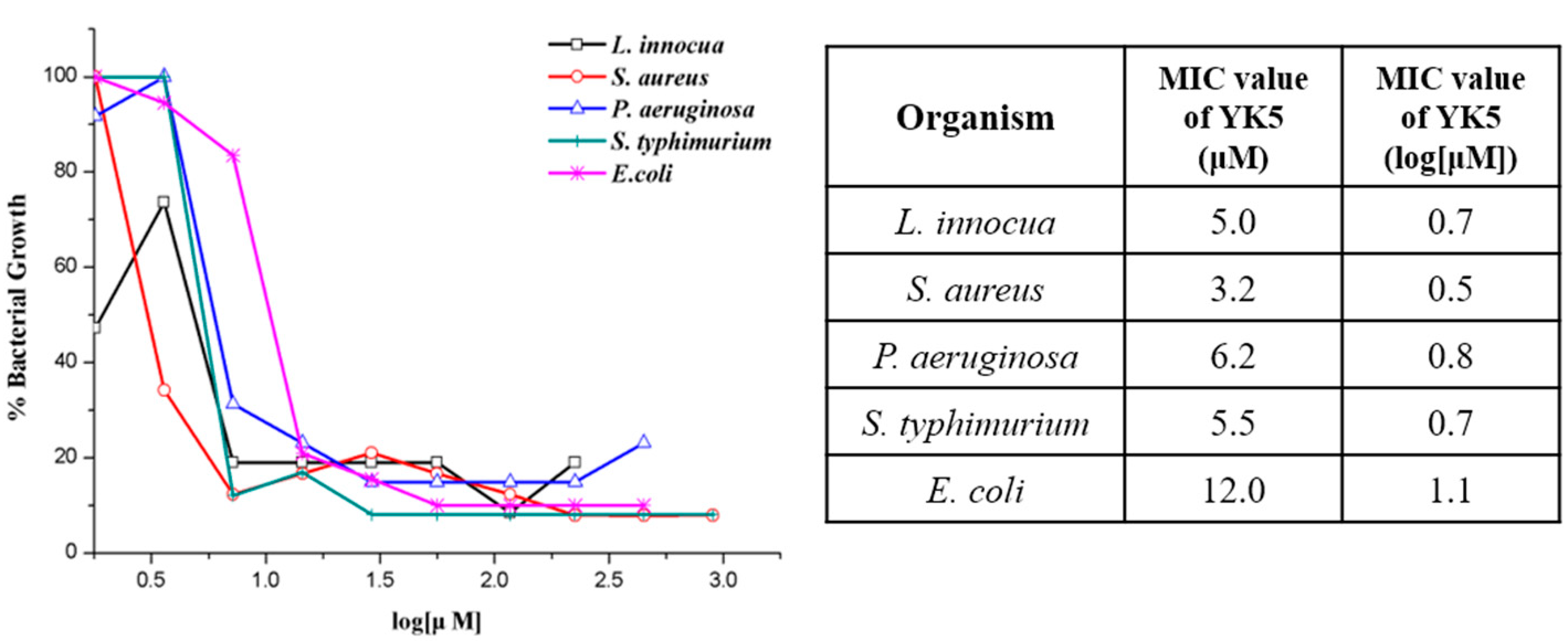
2.5. Antibacterial Assays
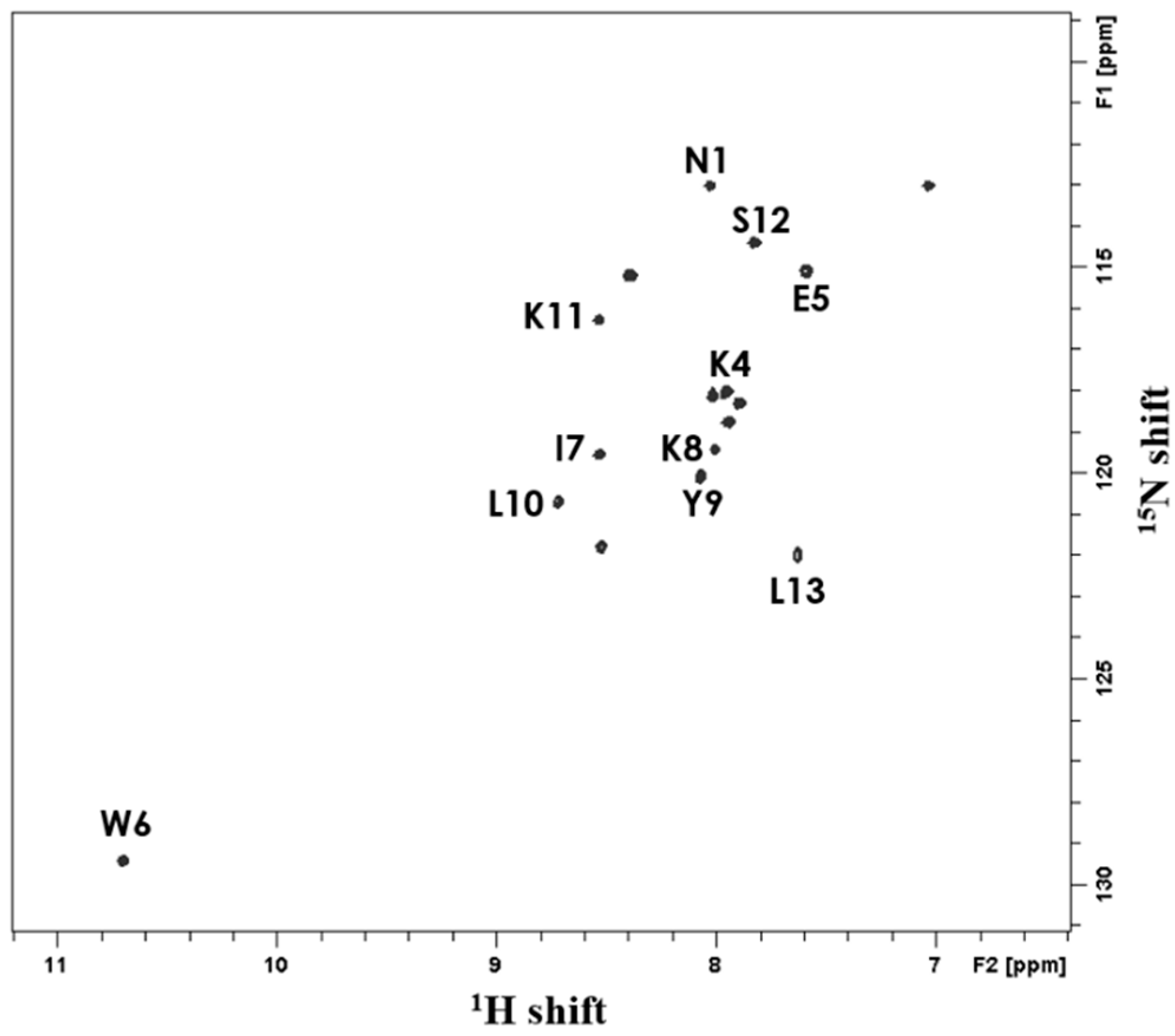
2.6. Solution-State NMR Spectroscopy
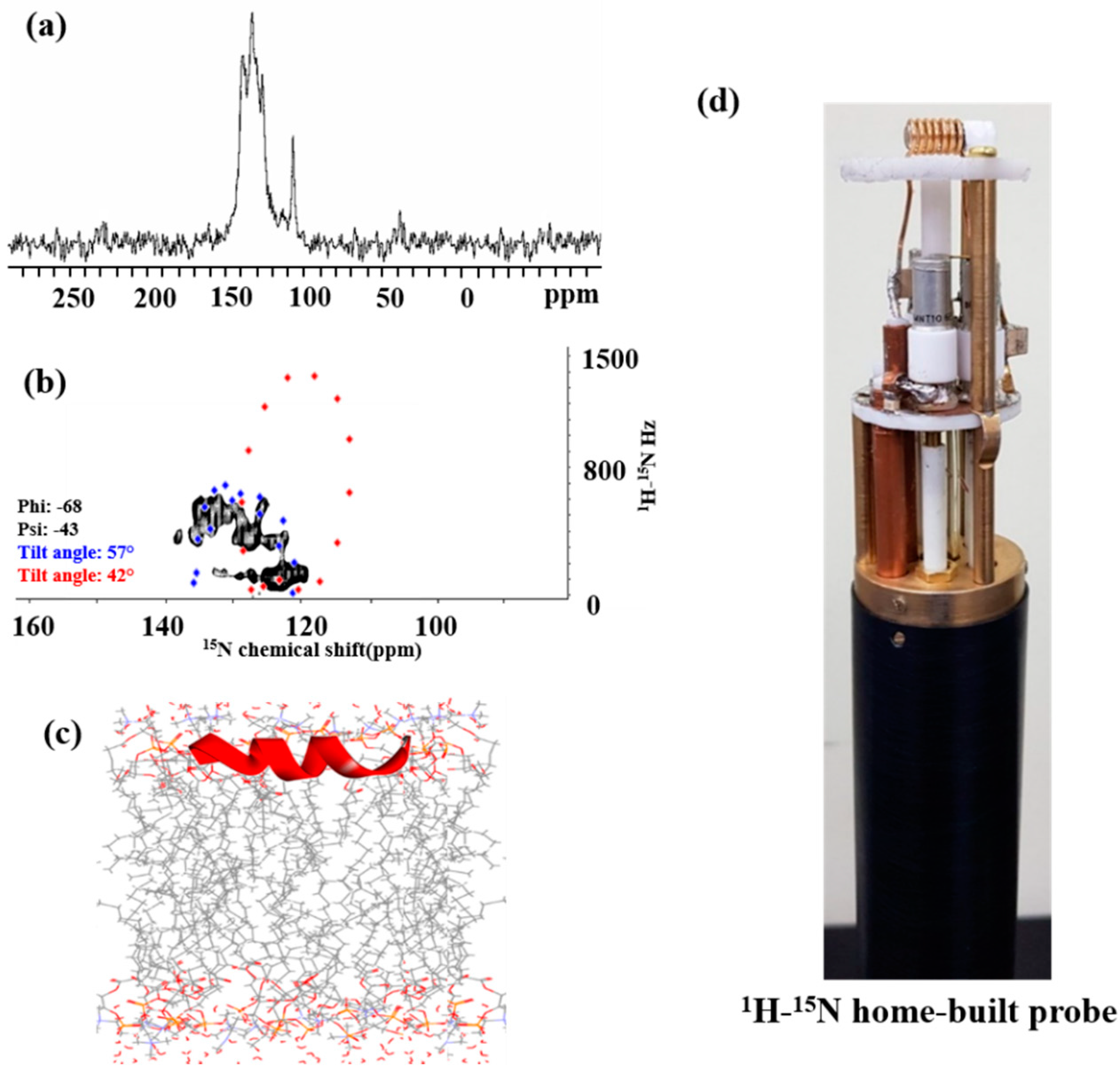
2.7. Solid-State NMR Spectroscopy
2.7.1. 15N NMR Spectroscopy
2.7.2. 31P NMR Spectroscopy
3. Materials and Methods
3.1. Design of Advanced LPcin Analogs
3.2. Antibacterial Activity of Peptides
3.3. Expression of Selected Peptide
3.4. Isolation and Purification of Peptide
3.5. Mass Spectrometry and CD Spectroscopy
3.6. Cytotoxicity Test
3.7. Antibacterial Test by SEM Preparation of DPC Micelle
3.8. Preparation of DPC Micelle
3.9. Solution-State NMR Spectroscopy
3.10. Preparation of Phospholipid Bicelles
3.11. Solid-State NMR Spectroscopy
3.11.1. 15N-NMR Spectroscopy
3.11.2. PISA Wheel Pattern Analysis
3.11.3. Molecular Simulations
3.11.4. 31P NMR Spectroscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| PP3 | Proteose peptone component-3; lactophorin |
| NMR | Nuclear magnetic resonance |
| LPcin | Lactophoricin |
| CD | Circular dichroism |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| KSI | Ketosteroid isomerase |
| Ni-NTA | Nickel-nitrilotriacetic acid |
| CNBr | Cyanogen bromide |
| HPLC | High-performance liquid chromatography |
| MALDI-TOF | Matrix-assisted laser desorption/ionization time-of-flight |
| DPC | Dodecylphosphocholine |
| MIC | Minimal inhibitory concentration |
| SEM | Scanning electron microscope |
| HSQC | Heteronuclear single quantum correlation |
| HMQC-NOESY | Heteronuclear multiple quantum correlation-nuclear Overhauser effect spectroscopy |
| BHI | Brain heart infusion |
| CP | Cross-polarization |
| PISA | Polarity index at slant angle |
| SAMPI4 | Selective averaging via magic sandwich pulses using π/4 flip pulses |
| 14-O-PC | 1,2-di-O-tetradecyl-sn-glycero−3-phosphocholine |
| DMPG | 1,2-dimyristoyl-sn-glycero−3-phospho-(1′-rac-glycerol) |
| 6-O-PC | 1,2-di-O-hexyl-sn-glycero−3-phosphocholine |
| OD | Optical density |
| IPTG | Isopropyl β-d−1-thiogalactopyranoside |
| ACN | Acetonitrile |
| TFA | Trifluoroacetic acid |
| PISEMA | Polarization inversion spin exchange at the magic angle |
References
- Mohamed, M.F.; Brezden, A.; Mohammad, H.; Chmielewski, J.; Seleem, M.N. A short D-enantiomeric antimicrobial peptide with potent immunomodulatory and antibiofilm activity against multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Courvalin, P. Why is antibiotic resistance a deadly emerging disease? Clin. Microbiol. Infect. 2016, 22, 405–407. [Google Scholar] [CrossRef]
- Bell, M. Antibiotic misuse: A global crisis. JAMA Intern. Med. 2014, 174, 1920–1921. [Google Scholar] [CrossRef]
- Roche, M.; Bornet, C.; Monges, P.; Stein, A.; Gensollen, S.; Seng, P. Misuse of antibiotics reserved for hospital settings in outpatients: A prospective clinical audit in a university hospital in Southern France. Int. J. Antimicrob. Agents 2016, 48, 96–100. [Google Scholar] [CrossRef]
- Walsh, C.; Wright, G. Introduction: Antibiotic resistance. Chem. Rev. 2015, 105, 391–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski, L.d.S.; Silva-Pereira, I.; Kyaw, C.M. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front. Microbiol. 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rios, A.C.; Moutinho, C.G.; Pinto, F.C.; Del Fiol, F.S.; Jozala, A.; Chaud, M.V.; Vila, M.M.; Teixeira, J.A.; Balcão, V.M. Alternatives to overcoming bacterial resistances: State-of-the-art. Microbiol. Res. 2016, 191, 51–80. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Multidrug Resistance in Bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [Green Version]
- El-Shazly, D.A.; Nasef, S.A.; Mahmoud, F.F.; Jonas, D. Expanded spectrum β–lactamase producing Escherichia coli isolated from chickens with colibacillosis in Egypt. Poult. Sci. 2017, 96, 2375–2384. [Google Scholar] [CrossRef]
- Saido-Sakanaka, H.; Ishibashi, J.; Momotani, E.; Amano, F.; Yamakawa, M. In vitro and in vivo activity of antibacterial peptides synthesized based on the insect defensin. Peptides 2004, 25, 19–27. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Bryan, L.; Silphadaung, U.; Noga, E.; Wade, D.; Rollins-Smith, L. Inactivation of viruses infecting ectothermic animals by amphibian and piscine antibacterial peptides. Virology 2004, 323, 268–275. [Google Scholar] [CrossRef] [Green Version]
- Sierra, J.M.; Fusté, E.; Rabanal, F.; Vinuesa, T.; Viñas, M. An overview of antimicrobial peptides and the latest advances in their development, Expert Opinion on Biological Therapy. Expert Opin. Biol. Ther. 2017, 17, 663–676. [Google Scholar] [CrossRef]
- Chen, C.H.; Lu, T.K. Development and Challenges of Antimicrobial Peptides for Therapeutic Applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [Green Version]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef]
- Sun, E.; Belanger, C.R.; Haney, E.F.; Hancock, R.E.W. Host Defense (Antimicrobial) Peptides. Pept. Appl. Biomed. Biotechnol. Bioeng. 2018, 253–285. [Google Scholar] [CrossRef]
- Khurshid, Z.; Zafar, M.S.; Naseem, M.; Khan, R.S.; Najeeb, S. Human Oral Defensins Antimicrobial Peptides: A Future Promising Antimicrobial Drug. Curr. Pharm. Des. 2018, 24, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.A.E.; Hammami, R. Recent insights into structure–function relationships of antimicrobial peptides. J. Food Biochem. 2018, 43, e12546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Rozek, A. Role of membranes in the activities of antibacterial cationic peptides. FEMS Microbiol. Lett. 2002, 206, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Deeth, H.; Bansal, N. Whey Proteins. Whey Proteins 2019, 1–50. [Google Scholar] [CrossRef]
- Pedersen, L.R.L.; Hansted, J.G.; Nielsen, S.B.; Petersen, T.E.; Sørensen, U.S.; Otzen, D.; Sørensen, E.S. Proteolytic activation of proteose peptone component 3 by release of a C-terminal peptide with antibacterial properties. J. Dairy Sci. 2012, 95, 2819–2829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, T.J.; Kim, J.S.; Choi, S.S.; Kim, Y. Cloning, expression, isotope labeling, purification, and characterization of bovine antibacterial peptide, lactophoricin in Escherichia coli. Protein Expr. Purif. 2009, 65, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Gor’kov, L.; Chekmenev, E.Y.; Li, C.; Cotten, M.; Buffy, J.J.; Traaseth, N.J.; Veglia, G.; Brey, W.W. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J. Magn. Reson. 2007, 185, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Campagna, S.; Mathot, A.G.; Fleury, Y.; Girardet, J.M.; Gaillard, J.L. Antibacterial Activity of Lactophoricin, a Synthetic 23-Residues Peptide Derived from the Sequence of Bovine Milk Component-3 of Proteose Peptone. J. Dairy Sci. 2004, 87, 1621–1626. [Google Scholar] [CrossRef] [Green Version]
- Becucci, L.; Aloisi, G.; Scaloni, A.; Guidelli, R. Channel-forming activity of lactophoricins I and II in mercury-supported tethered bilayer lipid membranes. J. Electroanal. Chem. 2018, 819, 65–72. [Google Scholar] [CrossRef]
- Campagna, S.; Vitoux, B.; Humbert, G.; Girardet, J.M.; Linden, G.; Haertle, T.; Gaillard, J.L. Conformational studies of a synthetic peptide from the putative lipidbinding domain of bovine milk component PP3. J. Dairy Sci. 1998, 81, 3139–3148. [Google Scholar] [CrossRef]
- Campagna, S.; Cosette, P.; Molle, G.; Gaillard, J.L. Evidence for membrane affinity of the C-terminal domain of bovine milk PP3 component. Biochim. Biophys. Acta Biomembr. 2001, 1513, 217–222. [Google Scholar] [CrossRef] [Green Version]
- Park, T.J.; Kim, J.S.; Ahn, H.C.; Kim, Y. Solution and Solid-State NMR Structural Studies of Antibacterial Peptides LPcin-I and LPcin-II. Biophys. J. 2011, 101, 1193–1201. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.S.; Jeong, J.H.; Kim, Y. Design, Characterization, and Antibacterial Activity of a Novel Antibacterial Peptide Derived from Bovine Lactophoricin. J. Microbiol. Biotechnol. 2017, 27, 759–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; Jeong, J.H.; Kim, Y. Design and Engineering of Antibacterial Peptides Based on LPcin-YK3, an Antibacterial Peptide Derivative from Bovine Milk. J. Microbiol. Biotechnol. 2018, 28, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.-H. Tryptophan-Rich and Proline-Rich Antimicrobial Peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef] [Green Version]
- Booth, V.; Warschawski, D.E.; Santisteban, N.P.; Laadhari, M.; Marcotte, I. Recent progress on the application of 2H solid-state NMR to probe the interaction of antimicrobial peptides with intact bacteria. BBA Proteins Proteom. 2017, 1865, 1500–1511. [Google Scholar] [CrossRef]
- Lu, G.J.; Son, W.S.; Opella, S.J. A general assignment method for oriented sample (OS) solid-state NMR of proteins based on the correlation of resonances through heteronuclear dipolar couplings in samples aligned parallel and perpendicular to the magnetic field. J. Magn. Reson. 2011, 209, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.S.; Jeong, J.H.; Kim, K.S.; Kim, Y. Optimized Expression and Characterization of Antibacterial Peptides, LPcin Analogs. Bull. Korean Chem. Soc. 2015, 36, 1148–1154. [Google Scholar] [CrossRef]
- Jeong, J.H.; Kim, J.S.; Choi, S.S.; Kim, Y. NMR Structural Studies of Antibacterial Peptides: LPcin Analogs. Biophys. J. 2016, 110, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.H.; Kim, M.; Kim, Y. NMR structural studies and mechanism of action of Lactophoricin analogs as antibacterial peptides. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183469. [Google Scholar] [CrossRef]
- Jeong, J.H.; Kim, M.; Son, J.; Kim, Y. 1H-31P home-built solid-state NMR probe with a scroll coil for 400-MHz NB magnet for biological lossy sample. J. Anal. Sci. Technol. 2020, 11, 1–6. [Google Scholar] [CrossRef]
- De Angelis, A.A.; Grant, C.V.; Baxter, M.K.; MaGavin, J.A.; Opella, S.J.; Cotton, M.L. Amphipathic antibacterial piscidin in magnetically aligned lipid bilayers. Biophys. J. 2011, 101, 1086–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, É.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta BBA Biomembr. 2011, 1808, 1957–1974. [Google Scholar] [CrossRef] [Green Version]
- Stanberg, E.; Ulrich, A.S. NMR methods for studying membrane-active antibacterial peptides. Concepts Magn. Reson. Part A 2004, 23A, 89–120. [Google Scholar]
- Triba, M.N.; Warschawski, D.E.; Devaux, P.F. Reinvestigation by phosphorus NMR of lipid distribution in bicelles. Biophys. J. 2005, 88, 1887–1901. [Google Scholar] [CrossRef] [Green Version]
- Grélard, A.; Loudet, C.; Diller, A.; Dufourc, E.J. NMR spectroscopy of lipid bilayers. Methods Mol. Biol. 2010, 654, 341–359. [Google Scholar] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antibacterial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.K.; Song, J.W.; Gong, F.; Li, S.B.; Chang, H.Y.; Xie, H.M.; Gao, H.W.; Tan, Y.X.; Ji, S.P. Design of an α-helical antibacterial peptide with improved cell-selective and potent anti-biofilm activity. Sci. Rep. 2016, 6, 27394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wu, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Zhou, M.; Duan, J.; Bininda-Emonds, O.R.P.; Chen, T.; et al. Targeted Modification of a Novel Amphibian Antimicrobial Peptide from Phyllomedusa tarsius to Enhance Its Activity against MRSA and Microbial Biofilm. Front. Microbiol. 2017, 8, 628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKay, M.J.; Martfeld, A.N.; De Angelis, A.A.; Opella, S.J.; Greathouse, D.V.; Koeppe, R.E. Control of Transmembrane Helix Dynamics by Interfacial Tryptophan Residues. Biophys. J. 2018, 114, 2617–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foloppe, N.; MacKerell, A.D. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000, 21, 86–104. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Banavali, N.K. All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 2000, 21, 105–120. [Google Scholar] [CrossRef]
- Im, W.; Lee, M.S.; Brooks, C.L., III. Generalized born model with simple smoothing function. J. Comput. Chem. 2003, 15, 1691–1702. [Google Scholar] [CrossRef]


| Number | Peptide | Sequence | Net Charge at pH 7.0 | Antimicrobial Activity | |
|---|---|---|---|---|---|
| Gram-Positive | Gram-Negative | ||||
| LPcin-1 | NTVKE TIKYL KSLFS HAFEV VKT | + 2.1 | + | ++ | |
| 1 | LP1-C2 | NTVKE TIKYL KSLFS HAFEV V | + 1.1 | – | + |
| 2 | LP1-C4 | NTVKE TIKYL KSLFS HAFE | + 1.1 | + | ++ |
| 3 | LP1-C6 | NTVKE TIKYL KSLFS HA | + 2.1 | + | +++ |
| 4 | LP1-C8 (YK1) | NTVKE TIKYL KSLFS | + 2.0 | + | +++ |
| 5 | LP1-C10 | NTVKE TIKYL KSL | + 2.0 | – | – |
| 6 | LP1-T6W | NTVKE WIKYL KSLFS HAFEV VKT | + 2.1 | + | ++ |
| 7 | LP1-T2W | NWVKE TIKYL KSLFS HAFEV VKT | + 2.1 | ++ | ++ |
| 8 | LP1-T2,6W | NWVKE WIKYL KSLFS HAFEV VKT | + 2.1 | ++ | +++ |
| 9 | LP1-T2K | NKVKE TIKYL KSLFS HAFEV VKT | + 3.1 | + | ++ |
| 10 | LP1-T2K,T6W (YK2) | NKVKE WIKYL KSLFS HAFEV VKT | + 3.1 | ++ | +++ |
| 11 | LP1-T2K,T6W-C8 (YK3) | NKVKE WIKYL KSLFS | + 3.0 | ++ | +++ |
| 12 | YK4 | NKVKE WWKWL KSLFS | + 3.0 | ++ | – |
| 13 | YK5 | NKVKE WIKYL KSLFK | + 4.0 | ++ | +++ |
| 14 | YK6 | NKVKE WIKYL KSKFS | + 4.0 | ++ | ++ |
| 15 | YK7 | NKVKE WWKWL KSLFK | + 4.0 | ++ | +++ |
| 16 | YK8 | NKVKE WIKYL KSKFK | + 5.0 | ++ | +++ |
| 17 | YK9 | NKVKE WWKWL KSL | + 3.0 | ++ | ++ |
| 18 | YK10 | NKVKE WIKYL KKL | + 4.0 | + | ++ |
| 19 | YK11 | NKVKE WWKWL KKL | + 4.0 | ++ | ++ |
| 20 | YK12 | NKVKE WWKWL K | + 3.0 | + | + |
| Cell Lines | VERO | NIH 3T3 | L929 | HFL-1 | CHO-K1 | |
|---|---|---|---|---|---|---|
| Peptides (μΜ) | ||||||
| YK5 IC50 (μM) | >100 | >100 | >100 | >100 | 57.0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Son, J.; Kim, Y. Structural and Mechanismic Studies of Lactophoricin Analog, Novel Antibacterial Peptide. Int. J. Mol. Sci. 2021, 22, 3734. https://doi.org/10.3390/ijms22073734
Kim M, Son J, Kim Y. Structural and Mechanismic Studies of Lactophoricin Analog, Novel Antibacterial Peptide. International Journal of Molecular Sciences. 2021; 22(7):3734. https://doi.org/10.3390/ijms22073734
Chicago/Turabian StyleKim, Minseon, Jinyoung Son, and Yongae Kim. 2021. "Structural and Mechanismic Studies of Lactophoricin Analog, Novel Antibacterial Peptide" International Journal of Molecular Sciences 22, no. 7: 3734. https://doi.org/10.3390/ijms22073734
APA StyleKim, M., Son, J., & Kim, Y. (2021). Structural and Mechanismic Studies of Lactophoricin Analog, Novel Antibacterial Peptide. International Journal of Molecular Sciences, 22(7), 3734. https://doi.org/10.3390/ijms22073734






