tRNA-Dependent Import of a Transit Sequence-Less Aminoacyl-tRNA Synthetase (LeuRS2) into the Mitochondria of Arabidopsis
Abstract
:1. Introduction
2. Results
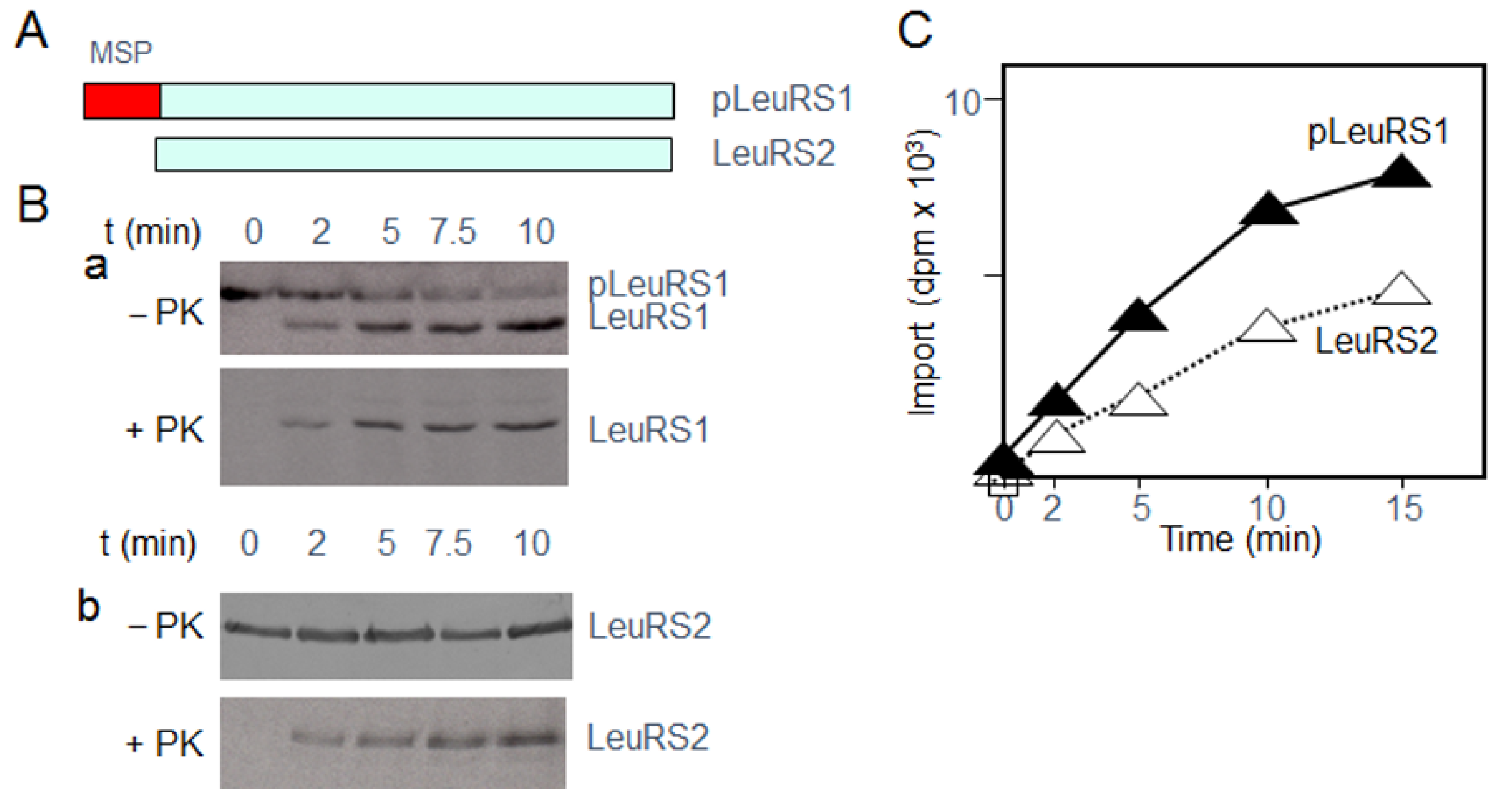
2.1. Identification of Proteins Interacting with LeuRS2 during Its Uptake by Isolated Arabidopsis Mitochondria
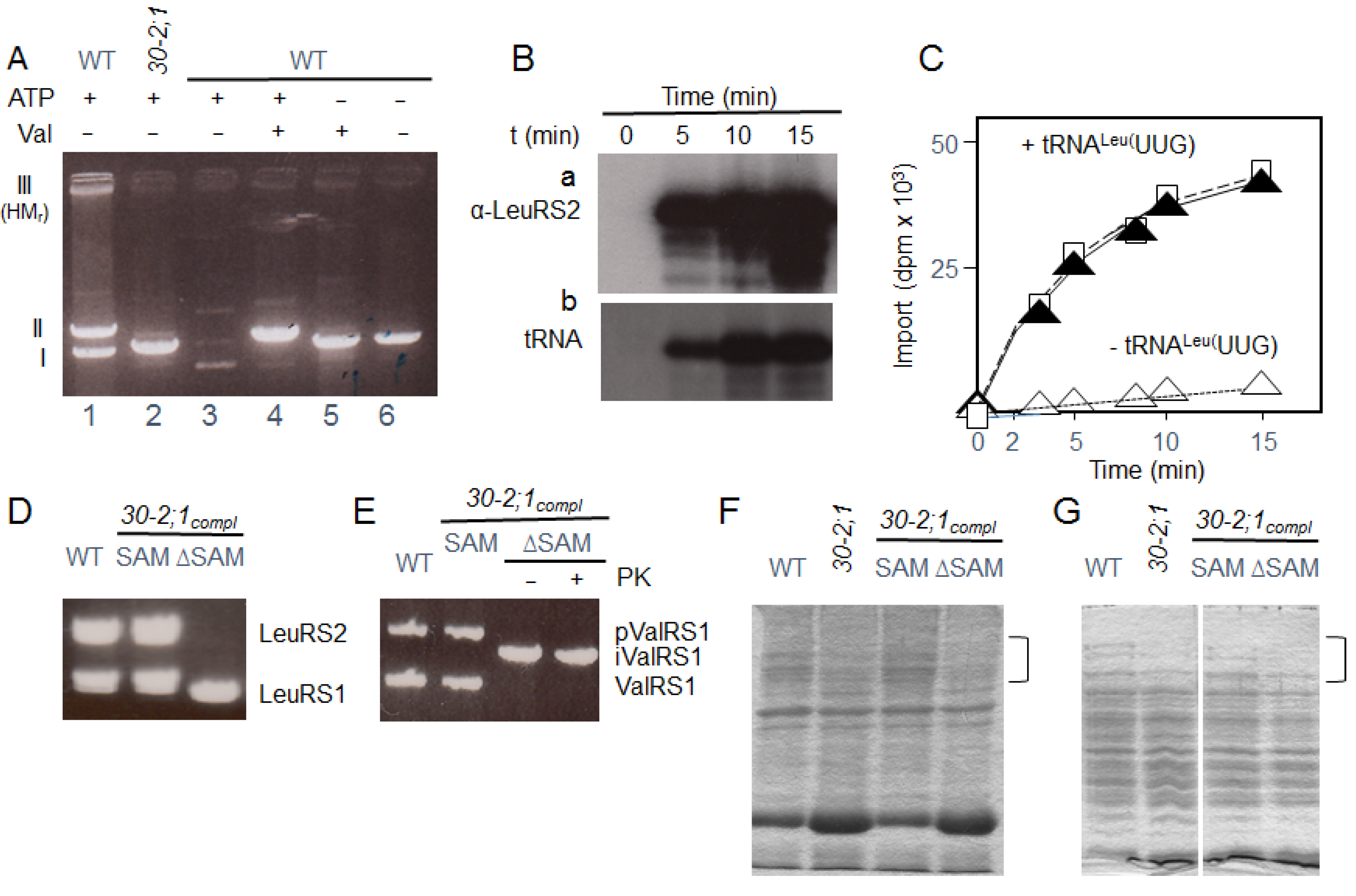
2.2. Import of LeuRS2 into Mutant Mitochondria Deprived of HP30-2
2.3. Evidence for a tRNA Requirement of LeuRS2 Import
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. tRNA Isolation and Uptake Assays
4.3. Organelle Isolation, Protein Synthesis and Protein Import Assays
4.4. Isolation of Translocation Intermediates of LeuRS2
4.5. Protein Analyses
4.6. Cell Death Measurements
4.7. Structural Modelling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AaRS | aminoacyl tRNA synthetase |
| MSP | mitochondrial signal peptide |
| PRAT | preprotein and amino acid transporter |
| SAM | sterile alpha motif |
| TIM | translocase of the inner mitochondrial membrane |
| TRBD | tRNA binding domain |
References
- Pang, Y.L.; Poruri, K.; Martinis, S.A. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscip. Rev. RNA 2014, 5, 461–480. [Google Scholar] [CrossRef] [Green Version]
- Duchêne, A.M.; Pujol, C.; Maréchal-Drouard, L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr. Genet. 2009, 55, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Tolkunova, E.; Park, H.; Xia, J.; King, M.P.; Davidson, E. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual alternative splicing of the primary transcript. J. Biol. Chem. 2000, 275, 35063–35069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rettig, J.; Wang, Y.; Schneider, A.; Ochsenreiter, T. Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucl. Acids Res. 2012, 40, 1299–1306. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.-L.; Yeh, L.S.; Chen, N.K.; Ripmaster, T.; Schimmel, P.; Wang, C.C. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 2004, 279, 49656–49663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, K.J.; Lin, G.; Men, L.C.; Wang, C.C. Redundancy of non-AUG initiators. A clever mechanism to enhance the efficiency of translation in yeast. J. Biol. Chem. 2006, 281, 7775–7783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatton, B.; Walter, P.; Ebel, J.P.; Lacroute, F.; Fasiolo, F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J. Biol. Chem. 1988, 263, 52–57. [Google Scholar] [CrossRef]
- Chiu, W.C.; Chang, C.P.; Wang, C.C. Evolutionary basis of converting a bacterial tRNA synthetase into a yeast cytoplasmic or mitochondrial enzyme. J. Biol. Chem. 2009, 284, 23954–23960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duchêne, A.M.; Giritch, A.; Hoffman, B.; Cognat, V.; Lancelin, D.; Zaepfel, M.; Peeters, N.; Laurence, M.; Small, I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 16484–16489. [Google Scholar] [CrossRef] [Green Version]
- Brandão, M.M.; Silva-Filho, M.C. Evolutionary history of Arabidopsis thaliana aminoacyl-tRNA synthetase dual-targeted proteins. Mol. Biol. Evol. 2011, 28, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Verner, K.; Schatz, G. Import of an incompletely folded precursor protein into isolated mitochondria requires an energized inner membrane, but no added ATP. EMBO J. 1987, 6, 2449–2456. [Google Scholar] [CrossRef]
- Pfanner, R.; Neupert, W. High-affinity binding sites involved in the import of porin into mitochondria. EMBO J. 1987, 6, 2635–2642. [Google Scholar]
- Wiedemann, N.; Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017, 20, 685–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schleyer, M.; Neupert, W. Transport of proteins into mitochondria: Translocational intermediates spanning contact sites between outer and inner membranes. Cell 1985, 43, 339–350. [Google Scholar] [CrossRef]
- Schnell, D.J.; Kessler, F.; Blobel, G. Isolation of components of the chloroplast protein import machinery. Science 1994, 266, 1007–10012. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Salvi, D.; Brugière, S.; Miras, S.; Kowalski, S.; Louwagie, M.; Garin, J.; Joyard, J.; Rolland, N. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell Proteom. 2003, 25, 325–345. [Google Scholar] [CrossRef] [Green Version]
- Carrie, C.; Murcha, M.W.; Whelan, J. An in silico analysis of the mitochondrial import apparatus of plants. BMC Plant. Biol. 2010, 16, 249. [Google Scholar] [CrossRef] [Green Version]
- Duncan, O.; Murcha, M.W.; Whelan, J. Unique components of the plant mitochondrial protein import apparatus. Biochim. Biophys. Acta 2013, 1833, 304–313. [Google Scholar] [CrossRef] [Green Version]
- Habeeb, A.F. Reaction of protein sulfhydryl groups with Ellman’s reagent. Methods Enzymol. 1972, 25, 457–464. [Google Scholar]
- Kim, C.A.; Bowie, J.U. SAM domains: Uniform structure, diversity of function. Trends Biochem. Sci. 2003, 28, 625–628. [Google Scholar] [CrossRef]
- Aviv, T.; Lin, Z.; Lau, S.; Rendl, L.M.; Sicheri, F.; Smibert, C.A. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 2003, 10, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 2011, 80, 1033–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirande, M. The ins and outs of tRNA transport. EMBO Rep. 2007, 8, 547–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adhya, S. Leishmania mitochondrial tRNA importers. Int. J. Biochem. Cell Biol. 2008, 40, 2681–2685. [Google Scholar] [CrossRef] [PubMed]
- Tarassov, I.A.; Entelis, N.S. Mitochondrially-imported cytoplasmic lysine tRNA (CUU) of Saccharomyces cerevisiae: In vivo and in vitro targeting systems. Nucl. Acids Res. 1992, 20, 1277–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowher, A.; Smirnov, A.; Tarassov, I.; Entelis, N. Induced tRNA import into human mitochondria: Implication of a host aminoacyl-tRNA-synthetase. PLoS ONE 2013, 8, e66228. [Google Scholar] [CrossRef]
- Rubio, M.A.; Rinehart, J.J.; Krett, B.; Duvezin-Caubet, S.; Reichert, A.S.; Söll, D.; Alfonzo, J.D. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Natl. Acad. Sci. USA 2008, 105, 9186–9191. [Google Scholar] [CrossRef] [Green Version]
- Small, I.; Laurence, M.; Masson, J.; Pelletier, G.; Cosset, A.; Weil, J.H.; Dietrich, A. In vivo import of a normal or mutagenized heterologous transfer RNA into the mitochondria of transgenic plants: Towards novel ways of influencing mitochondrial gene expression? EMBO J. 1992, 11, 1291–1296. [Google Scholar] [CrossRef]
- Dietrich, A.; Marechal-Drouard, L.; Carneiro, V.; Cosset, A.; Small, I. A single base change prevents import of cytosolic alanyl-tRNA into mitochondria in transgenic plants. Plant. J. 1996, 10, 913–918. [Google Scholar] [CrossRef] [Green Version]
- Delage, L.; Dietrich, A.; Cosset, A.; Marechal-Drouard, L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell. Biol. 2003, 23, 4000–4012. [Google Scholar] [CrossRef] [Green Version]
- Murcha, M.W.; Kubiszewski-Jakubiak, S.; Teixeira, P.F.; Gügel, I.L.; Kmiec, B.; Narsai, R.; Ivanova, A.; Megel, C.; Schock, A.; Kraus, S.; et al. Plant-Specific Preprotein and Amino Acid Transporter Proteins Are Required for tRNA Import into Mitochondria. Plant. Physiol. 2016, 172, 2471–2490. [Google Scholar] [CrossRef] [Green Version]
- Rossig, C.; Gray, J.; Valdes, O.; Rustgi, S.; von Wettstein, D.; Reinbothe, C.; Reinbothe, S. HP30-2, a mitochondrial PRAT protein for import of signal sequence-less precursor proteins in Arabidopsis thaliana. J. Integr. Plant. Biol. 2017, 59, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; Marie, A.; Delage, L.; Nilsson, S.; Zaepfel, M.; Glaser, E.; Marechal, L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 18362–18367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diodato, D.; Ghezzi, D.; Tiranti, V. The Mitochondrial Aminoacyl tRNA Synthetases: Genes and Syndromes. Int. J. Cell Biol. 2014, 2014, 787956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giordano, C.; Morea, V.; Perli, E.; d’Amati, G. The phenotypic expression of mitochondrial tRNA-mutations can be modulated by either mitochondrial leucyl-tRNA synthetase or the C-terminal domain thereof. Front. Genet. 2015, 6, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perli, E.; Fiorillo, A.; Giordano, C.; Pisano, A.; Montanari, A.; Grazioli, P.; Campese, A.F.; Di Micco, P.; Tuppen, H.A.; Genovese, I.; et al. Short peptides from leucyl-tRNA synthetase rescue disease-causing mitochondrial tRNA point mutations. Hum. Mol. Genet. 2016, 1, 903–915. [Google Scholar] [CrossRef] [Green Version]
- Fukunaga, R.; Yokoyama, S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005, 12, 915–922. [Google Scholar] [CrossRef]
- Kovaleski, B.J.; Kennedy, R.; Hong, M.K.; Datta, S.A.; Kleiman, L.; Rein, A.; Musier-Forsyth, K. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J. Biol. Chem. 2006, 281, 19449–19456. [Google Scholar] [CrossRef] [Green Version]
- Servillo, L.; Balestrieri, C.; Quagliuolo, L.; Iorio, E.L.; Giovane, A. tRNA fluorescent labeling at 3’ end inducing an aminoacyl-tRNA-like behavior. Eur. J. Biochem. 1993, 213, 583–589. [Google Scholar] [CrossRef]
- Raina, M.; Ibba, M. tRNAs as regulators of biological processes. Front. Genet. 2014, 5, 171. [Google Scholar] [CrossRef] [Green Version]
- Abbott, J.A.; Francklyn, C.S.; Robey-Bond, S.M. Transfer RNA and human disease. Front. Genet. 2014, 5, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, C.M.; Krzesinski, P.R.; Greaser, M.L. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis 2003, 24, 1695–1702. [Google Scholar] [CrossRef] [PubMed]
- Towbin, M.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets; procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.Y. Manual micro-sequence analysis of polypeptides using dimethylaminobenzene isothiocyanate. Methods Enzymol. 1983, 91, 455–466. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reinbothe, S.; Rossig, C.; Gray, J.; Rustgi, S.; von Wettstein, D.; Reinbothe, C.; Rassow, J. tRNA-Dependent Import of a Transit Sequence-Less Aminoacyl-tRNA Synthetase (LeuRS2) into the Mitochondria of Arabidopsis. Int. J. Mol. Sci. 2021, 22, 3808. https://doi.org/10.3390/ijms22083808
Reinbothe S, Rossig C, Gray J, Rustgi S, von Wettstein D, Reinbothe C, Rassow J. tRNA-Dependent Import of a Transit Sequence-Less Aminoacyl-tRNA Synthetase (LeuRS2) into the Mitochondria of Arabidopsis. International Journal of Molecular Sciences. 2021; 22(8):3808. https://doi.org/10.3390/ijms22083808
Chicago/Turabian StyleReinbothe, Steffen, Claudia Rossig, John Gray, Sachin Rustgi, Diter von Wettstein, Christiane Reinbothe, and Joachim Rassow. 2021. "tRNA-Dependent Import of a Transit Sequence-Less Aminoacyl-tRNA Synthetase (LeuRS2) into the Mitochondria of Arabidopsis" International Journal of Molecular Sciences 22, no. 8: 3808. https://doi.org/10.3390/ijms22083808





