Sex and Race-Related DNA Methylation Changes in Hepatocellular Carcinoma
Abstract
1. Health Disparity Is a Common Feature of Gastrointestinal Cancers
2. Liver Cancer, a Rising Malignancy with Sex and Race Disparities
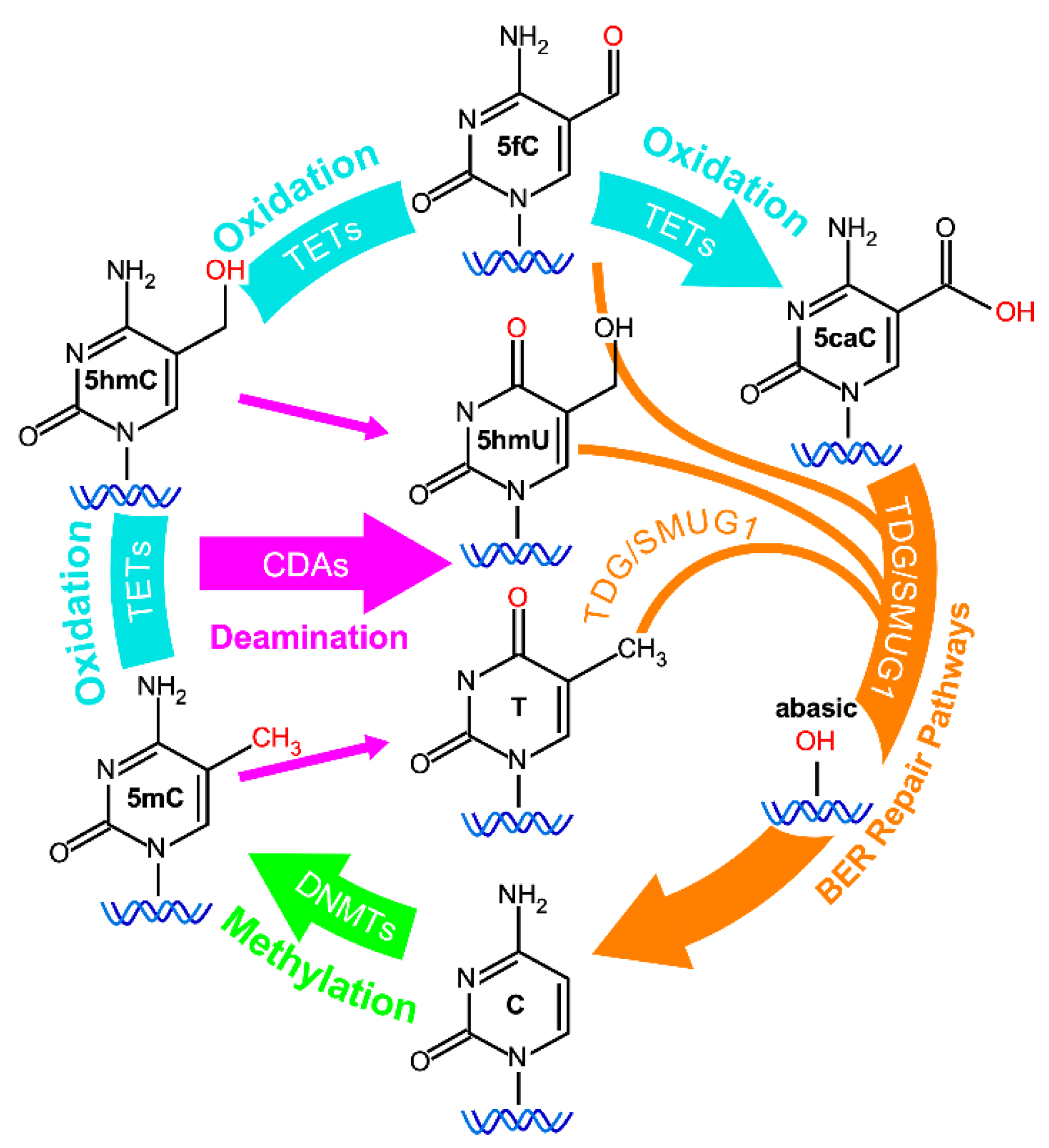
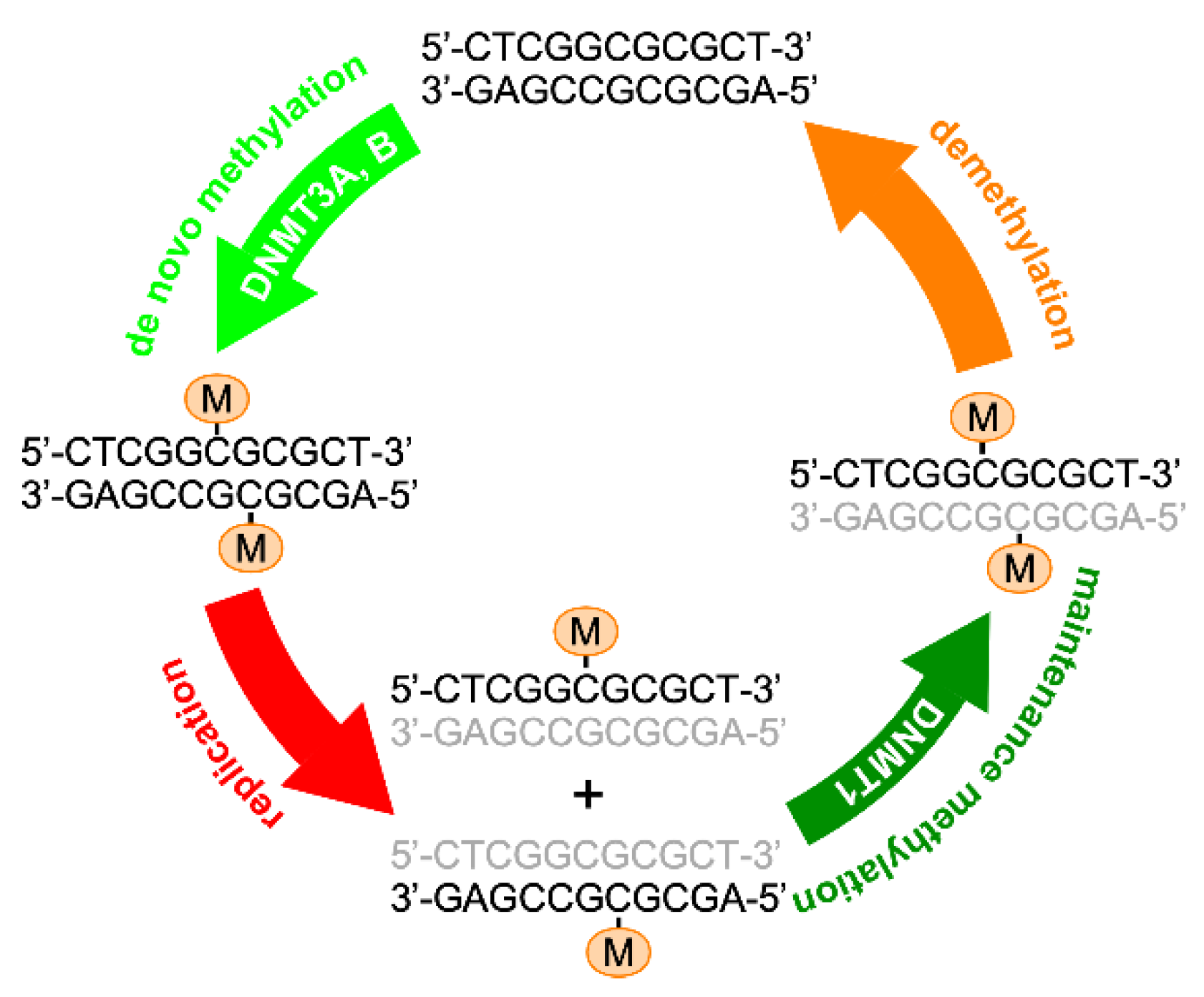
3. Epigenetic Changes: An Early Event in Oncogenesis
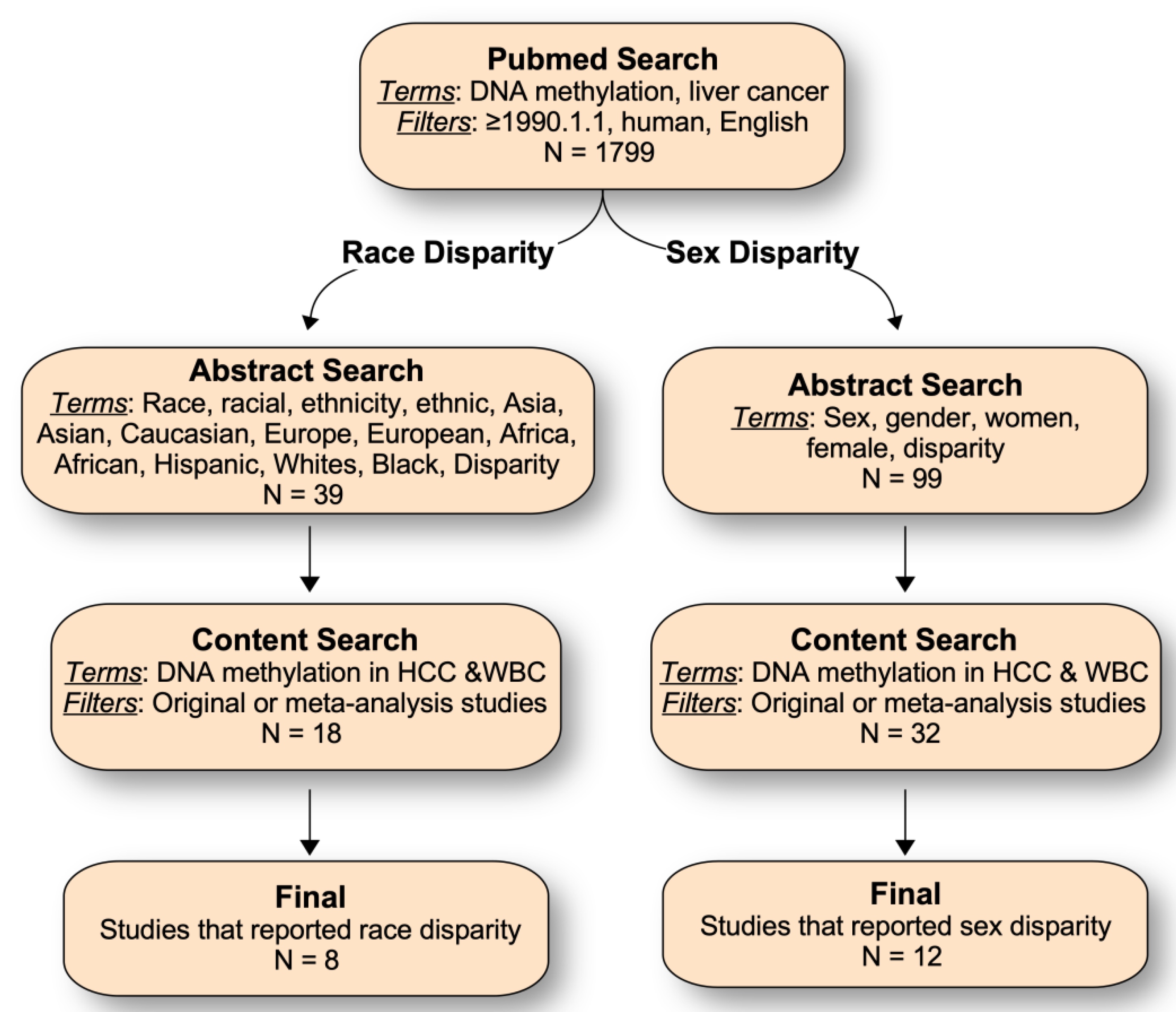
4. Literature Search on Sex and Race/Ethnic-Related DNA Methylation in HCC
5. Sex-Dependent DNA Methylation Events in HCC
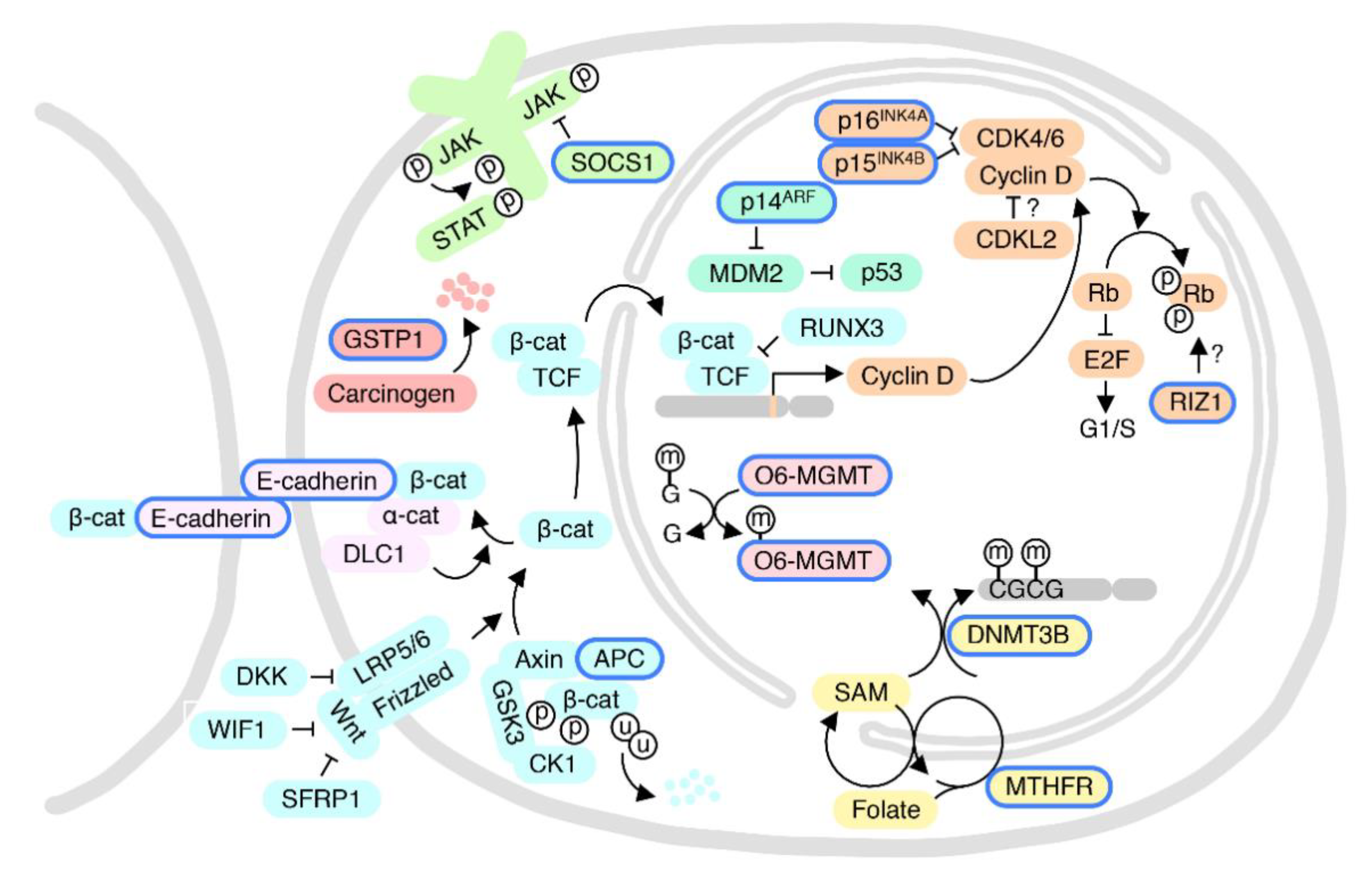
5.1. p16INK4A and CDKL2: The Cyclin-Dependent Kinase-RB Pathway
5.2. PPP2R1A and DNMT3B: Interaction between Methylation and Genetic Mutation
5.3. LINE1-Based Methylation Profiling
5.4. Tumor-Related Gene Panel
5.5. Genome-Wide Studies
5.6. Additional Sex-Related DNA Methylation Changes
5.7. Sex-Dependent DNA Methylation Changes in Premalignant Liver Conditions vs. HCC
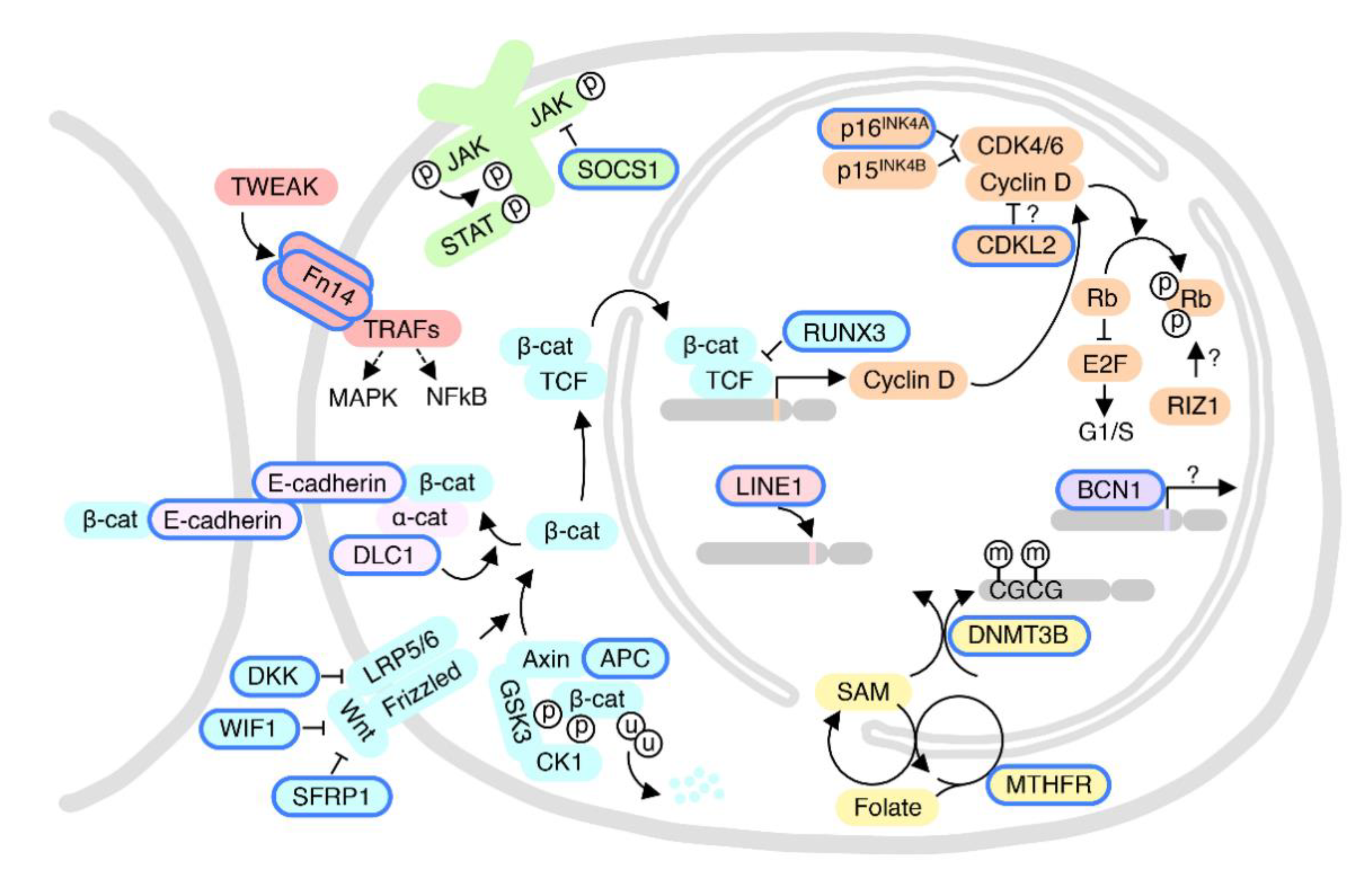
6. Race-Dependent DNA Methylation Events in HCC
6.1. Wnt Pathway and Cell-Cell Adhesion
6.2. The pRB and Cell Cycle Pathways
6.3. O6-MGMT DNA Repair Pathway
6.4. The GSTP1-Detoxification and SOCS1-JAK-STAT Pathways
6.5. Additional Race/Ethnic-Related DNA Methylation Changes
6.6. Race-Related DNA Methylation Changes in Premalignant Liver Conditions vs. HCC
7. Future Research Trends
7.1. Common Confounding Factors in Health Disparity Analyses
7.2. Basic and Translational Research
7.3. Clinical Importance
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.B.; Rim, S.H.; Joseph, D.A.; King, J.B.; Seeff, L.C.; Centers for Disease Control and Prevention. Colorectal cancer incidence and screening-United States, 2008 and 2010. MMWR Surveill Summ 2013, 62, 53–60. [Google Scholar]
- Mittal, S.; El-Serag, H.B. Epidemiology of hepatocellular carcinoma: Consider the population. J. Clin. Gastroenterol. 2013, 47, S2–S6. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Li, A.A.; Perumpail, B.J.; Gadiparthi, C.; Kim, W.; Cholankeril, G.; Glenn, J.S.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. Changing trends in etiology-based and ethnicity-based annual mortality rates of cirrhosis and hepatocellular carcinoma in the United States. Hepatology 2019, 69, 1064–1074. [Google Scholar] [CrossRef]
- Makarova-Rusher, O.V.; Altekruse, S.F.; McNeel, T.S.; Ulahannan, S.; Duffy, A.G.; Graubard, B.I.; Greten, T.F.; McGlynn, K.A. Population attributable fractions of risk factors for hepatocellular carcinoma in the United States. Cancer 2016, 122, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Ott, J.J.; Stevens, G.A.; Groeger, J.; Wiersma, S.T. Global epidemiology of hepatitis B virus infection: New estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012, 30, 2212–2219. [Google Scholar] [CrossRef]
- Ott, J.J.; Horn, J.; Krause, G.; Mikolajczyk, R.T. Time trends of chronic HBV infection over prior decades—A global analysis. J. Hepatol. 2017, 66, 48–54. [Google Scholar] [CrossRef]
- Islami, F.; Miller, K.D.; Siegel, R.L.; Fedewa, S.A.; Ward, E.M.; Jemal, A. Disparities in liver cancer occurrence in the United States by race/ethnicity and state. CA Cancer J. Clin. 2017, 67, 273–289. [Google Scholar] [CrossRef]
- Jones, P.A.; Gonzalgo, M.L. Altered DNA methylation and genome instability: A new pathway to cancer? Proc. Natl. Acad. Sci. USA 1997, 94, 2103–2105. [Google Scholar] [CrossRef]
- Umbricht, C.B.; Evron, E.; Gabrielson, E.; Ferguson, A.; Marks, J.; Sukumar, S. Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene 2001, 20, 3348–3353. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Hong, S.; Bhagwat, A.S.; Zhang, X.; Cheng, X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: Its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012, 40, 10203–10214. [Google Scholar] [CrossRef]
- Cortellino, S.; Xu, J.; Sannai, M.; Moore, R.; Caretti, E.; Cigliano, A.; Le Coz, M.; Devarajan, K.; Wessels, A.; Soprano, D. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011, 146, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Bronner, C.; Alhosin, M.; Hamiche, A.; Mousli, M. Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns. Genes 2019, 10, 65. [Google Scholar] [CrossRef]
- Gujar, H.; Weisenberger, D.J.; Liang, G. The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes 2019, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Gillette, T.G.; Hill, J.A. Readers, writers, and erasers: Chromatin as the whiteboard of heart disease. Circ. Res. 2015, 116, 1245–1253. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Archer, S.Y.; Hodin, R.A. Histone acetylation and cancer. Curr. Opin. Genet. Dev. 1999, 9, 171–174. [Google Scholar] [CrossRef]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Filip, M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox. Res. 2015, 27, 172–197. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal. Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Antequera, F.; Bird, A. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. USA 1993, 90, 11995–11999. [Google Scholar] [CrossRef]
- Baba, Y.; Yagi, T.; Sawayama, H.; Hiyoshi, Y.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Baba, H. Long interspersed Element-1 methylation level as a prognostic biomarker in gastrointestinal cancers. Digestion 2018, 97, 26–30. [Google Scholar] [CrossRef]
- Liu, H.; Liu, X.W.; Dong, G.; Zhou, J.; Liu, Y.; Gao, Y.; Liu, X.Y.; Gu, L.; Sun, Z.; Deng, D. P16 Methylation as an early predictor for cancer development from oral epithelial dysplasia: A double-blind multicentre prospective study. EBioMedicine 2015, 2, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Wang, Z.W.; Cang, H.; Chen, Y.Y.; Zhao, R.; Yu, B.M.; Tang, X.M. P16 gene methylation in colorectal cancers associated with Duke’s staging. World J. Gastroenterol. 2001, 7, 722–725. [Google Scholar] [CrossRef]
- Ye, X.; Mo, M.; Xu, S.; Yang, Q.; Wu, M.; Zhang, J.; Chen, B.; Li, J.; Zhong, Y.; Huang, Q. The hypermethylation of p16 gene exon 1 and exon 2: Potential biomarkers for colorectal cancer and are associated with cancer pathological staging. BMC Cancer 2018, 18, 1023. [Google Scholar] [CrossRef]
- Stirzaker, C.; Millar, D.S.; Paul, C.L.; Warnecke, P.M.; Harrison, J.; Vincent, P.C.; Frommer, M.; Clark, S.J. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997, 57, 2229–2237. [Google Scholar] [PubMed]
- Herman, J.G.; Latif, F.; Weng, Y.; Lerman, M.I.; Zbar, B.; Liu, S.; Samid, D.; Duan, D.S.; Gnarra, J.R.; Linehan, W.M. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. USA 1994, 91, 9700–9704. [Google Scholar] [CrossRef] [PubMed]
- Maryam, M.; Idrees, M. Study of promoter hypomethylation profiles of RAS oncogenes in hepatocellular carcinoma derived from hepatitis C virus genotype 3a in Pakistani population. J. Med. Virol. 2018, 90, 1516–1523. [Google Scholar] [CrossRef]
- Sheaffer, K.L.; Elliott, E.N.; Kaestner, K.H. DNA hypomethylation contributes to genomic instability and intestinal cancer initiation. Cancer Prev. Res. 2016, 9, 534–546. [Google Scholar] [CrossRef]
- Abadie, V.; Lyonnet, S.; Maurin, N.; Berthelon, M.; Caillaud, C.; Giraud, F.; Mattei, J.F.; Rey, J.; Rey, F.; Munnich, A. CpG dinucleotides are mutation hot spots in phenylketonuria. Genomics 1989, 5, 936–939. [Google Scholar] [CrossRef]
- Rideout, W.M., 3rd; Coetzee, G.A.; Olumi, A.F.; Jones, P.A. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science 1990, 249, 1288–1290. [Google Scholar] [CrossRef]
- Mancini, D.; Singh, S.; Ainsworth, P.; Rodenhiser, D. Constitutively methylated CpG dinucleotides as mutation hot spots in the retinoblastoma gene (RB1). Am. J. Hum. Genet. 1997, 61, 80–87. [Google Scholar] [CrossRef]
- O’Neill, J.P.; Finette, B.A. Transition mutations at CpG dinucleotides are the most frequent in vivo spontaneous single-based substitution mutation in the human HPRT gene. Environ. Mol. Mutagen. 1998, 32, 188–191. [Google Scholar] [CrossRef]
- Buzin, C.H.; Feng, J.; Yan, J.; Scaringe, W.; Liu, Q.; den Dunnen, J.; Mendell, J.R.; Sommer, S.S. Mutation rates in the dystrophin gene: A hotspot of mutation at a CpG dinucleotide. Hum. Mutat. 2005, 25, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Denissenko, M.F.; Chen, J.X.; Tang, M.S.; Pfeifer, G.P. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc. Natl. Acad. Sci. USA 1997, 94, 3893–3898. [Google Scholar] [CrossRef]
- Liao, Y.J.; Liu, S.P.; Lee, C.M.; Yen, C.H.; Chuang, P.C.; Chen, C.Y.; Tsai, T.F.; Huang, S.F.; Lee, Y.H.; Chen, Y.M. Characterization of a glycine N-methyltransferase gene knockout mouse model for hepatocellular carcinoma: Implications of the gender disparity in liver cancer susceptibility. Int. J. Cancer 2009, 124, 816–826. [Google Scholar] [CrossRef]
- Ezzikouri, S.; El Feydi, A.E.; Benazzouz, M.; Afifi, R.; El Kihal, L.; Hassar, M.; Akil, A.; Pineau, P.; Benjelloun, S. Single nucleotide polymorphism in DNMT3B promoter and its association with hepatocellular carcinoma in a Moroccan population. Infect. Genet. Evol. 2009, 9, 877–881. [Google Scholar] [CrossRef]
- Chen, H.F.; Mai, J.R.; Wan, J.X.; Gao, Y.F.; Lin, L.N.; Wang, S.Z.; Chen, Y.X.; Zhang, C.Z.; Zhang, Y.J.; Xia, B. Role of a novel functional variant in the PPP2R1A promoter on the regulation of PP2A-Aalpha and the risk of hepatocellular carcinoma. PLoS ONE 2013, 8, e59574. [Google Scholar] [CrossRef]
- Banelli, B.; Brigati, C.; Di Vinci, A.; Casciano, I.; Forlani, A.; Borzi, L.; Allemanni, G.; Romani, M. A pyrosequencing assay for the quantitative methylation analysis of the PCDHB gene cluster, the major factor in neuroblastoma methylator phenotype. Lab. Investig. 2012, 92, 458–465. [Google Scholar] [CrossRef]
- Shen, J.; Wang, S.; Zhang, Y.J.; Wu, H.C.; Kibriya, M.G.; Jasmine, F.; Ahsan, H.; Wu, D.P.; Siegel, A.B.; Remotti, H. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics 2013, 8, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Varghese, R.S.; Zhou, Y.; Chen, Y.; Barefoot, M.E.; Tadesse, M.; Ressom, H.W. Epigenetic changes associated with mechanisms of disparities in hepatocellular carcinoma. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 5320–5325. [Google Scholar] [CrossRef]
- Dedeurwaerder, S.; Defrance, M.; Calonne, E.; Denis, H.; Sotiriou, C.; Fuks, F. Evaluation of the infinium methylation 450K technology. Epigenomics 2011, 3, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Wong, I.H.; Lo, Y.M.; Yeo, W.; Lau, W.Y.; Johnson, P.J. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin. Cancer Res. 2000, 6, 3516–3521. [Google Scholar]
- Tsutsui, M.; Iizuka, N.; Moribe, T.; Miura, T.; Kimura, N.; Tamatsukuri, S.; Ishitsuka, H.; Fujita, Y.; Hamamoto, Y.; Tsunedomi, R. Methylated cyclin D2 gene circulating in the blood as a prognosis predictor of hepatocellular carcinoma. Clin. Chim. Acta 2010, 411, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Goel, A. Genetic and epigenetic signatures in human hepatocellular carcinoma: A systematic review. Curr. Genom. 2011, 12, 130–137. [Google Scholar] [CrossRef]
- Zang, J.J.; Xie, F.; Xu, J.F.; Qin, Y.Y.; Shen, R.X.; Yang, J.M.; He, J. P16 gene hypermethylation and hepatocellular carcinoma: A systematic review and meta-analysis. World J. Gastroenterol. 2011, 17, 3043–3048. [Google Scholar] [CrossRef]
- Li, X.; Hui, A.M.; Sun, L.; Hasegawa, K.; Torzilli, G.; Minagawa, M.; Takayama, T.; Makuuchi, M. p16INK4A hypermethylation is associated with hepatitis virus infection, age, and gender in hepatocellular carcinoma. Clin. Cancer Res. 2004, 10, 7484–7489. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, J.; Xu, C.; Liu, S.; Jiang, S.; Xu, Q.; Chen, X.; Zhuang, H.; Lu, F. Quantitative methylation analysis reveals gender and age differences in p16INK4a hypermethylation in hepatitis B virus-related hepatocellular carcinoma. Liver Int. 2012, 32, 420–428. [Google Scholar] [CrossRef]
- Wiencke, J.K.; Zheng, S.; Lafuente, A.; Lafuente, M.J.; Grudzen, C.; Wrensch, M.R.; Miike, R.; Ballesta, A.; Trias, M. Aberrant methylation of p16INK4a in anatomic and gender-specific subtypes of sporadic colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 1999, 8, 501–506. [Google Scholar]
- Li, P.; Zhang, X.; Gu, L.; Zhou, J.; Deng, D. P16 methylation increases the sensitivity of cancer cells to the CDK4/6 inhibitor palbociclib. PLoS ONE 2019, 14, e0223084. [Google Scholar] [CrossRef]
- Zhou, Y.; Qiu, X.P.; Li, Z.H.; Zhang, S.; Rong, Y.; Yang, G.H.; Fang, Z. Clinical significance of aberrant cyclin-dependent kinase-like 2 methylation in hepatocellular carcinoma. Gene 2019, 683, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Yang, S.; Liao, Y.; Hu, Z.; Long, H.; Zeng, Y.; Wang, X.; Qiu, C.; Xu, A.; Lin, J. Decreased CDKL2 expression is correlated with the progression and poor prognosis of glioma. Pathol. Res. Pract. 2020, 216, 152920. [Google Scholar] [CrossRef]
- Khalid, M.; Bojang, P., Jr.; Hassanin, A.A.I.; Bowers, E.C.; Reyes-Reyes, E.M.; Ramos, I.N.; Ramos, K.S. Line-1: Implications in the etiology of cancer, clinical applications, and pharmacologic targets. Mutat. Res. 2018, 778, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Carnell, A.N.; Goodman, J.I. The long (LINEs) and the short (SINEs) of it: Altered methylation as a precursor to toxicity. Toxicol. Sci. 2003, 75, 229–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, C.; Utsunomiya, T.; Ikemoto, T.; Yamada, S.; Morine, Y.; Imura, S.; Arakawa, Y.; Takasu, C.; Ishikawa, D.; Imoto, I. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann. Surg. Oncol. 2014, 21, S729–S735. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Kim, B.H.; Cho, N.Y.; Yoo, E.J.; Choi, M.; Shin, S.H.; Jang, J.J.; Suh, K.S.; Kim, Y.S.; Kang, G.H. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin. Cancer Res. 2009, 15, 812–820. [Google Scholar] [CrossRef]
- Stern, L.L.; Mason, J.B.; Selhub, J.; Choi, S.W. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol. Biomarkers Prev. 2000, 9, 849–853. [Google Scholar] [PubMed]
- Qiao, K.; Zhang, S.; Huo, Z.; Hou, W.; Dai, Q.; Su, R.; Wang, F. LINE-1 methylation in chronic HBV infected patients: Association with HCC, gender and MTHFR C677T polymorphism. Clin. Lab. 2020, 66, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.B.; Zhang, Y.X.; Zhou, S.H.; Shi, M.X.; Cai, J.; Liu, Y.; Chen, K.P.; Qiang, F.L. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J. Gastroenterol. 2011, 17, 4718–4724. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Xie, X.; Chen, Z.; Wu, L.; Yu, Z.; Guo, X.; Chen, G. Association of TNFRSF12A methylation with prognosis in hepatocellular carcinoma with history of alcohol consumption. Front. Genet. 2019, 10, 1299. [Google Scholar] [CrossRef]
- Yoshida, T.; Ogata, H.; Kamio, M.; Joo, A.; Shiraishi, H.; Tokunaga, Y.; Sata, M.; Nagai, H.; Yoshimura, A. SOCS1 is a suppressor of liver fibrosis and hepatitis-induced carcinogenesis. J. Exp. Med. 2004, 199, 1701–1707. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Cheng, J.; Ding, S.; Li, M.; Sun, S.; Zhang, L.; Liu, S.; Chen, X.; Zhuang, H. An integrated analysis of SOCS1 down-regulation in HBV infection-related hepatocellular carcinoma. J. Viral. Hepat. 2014, 21, 264–271. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, X.; Liu, Y.; Lu, F.; Chen, X. Decreased expression of BNC1 and BNC2 is associated with genetic or epigenetic regulation in hepatocellular carcinoma. Int. J. Mol. Sci. 2016, 17, 153. [Google Scholar] [CrossRef]
- Su, F.H.; Chen, J.D.; Cheng, S.H.; Lin, C.H.; Liu, Y.H.; Chu, F.Y. Seroprevalence of Hepatitis-B infection amongst Taiwanese university students 18 years following the commencement of a national Hepatitis-B vaccination program. J. Med. Virol. 2007, 79, 138–143. [Google Scholar] [CrossRef]
- Chen, P.; Zheng, Y.; Cai, Y.; Zou, P.; Li, N.; Peng, C.; Gao, H.; Liu, J.; Chen, Y.; Tong, Z. Prevalence of hepatitis C virus infection in a surgical population of Southeast China: A large-scale multicenter study. Can. J. Gastroenterol. Hepatol. 2020, 2020, 8219536. [Google Scholar] [CrossRef] [PubMed]
- Rongrui, L.; Na, H.; Zongfang, L.; Fanpu, J.; Shiwen, J. Epigenetic mechanism involved in the HBV/HCV-related hepatocellular carcinoma tumorigenesis. Curr. Pharm. Des. 2014, 20, 1715–1725. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Ou, J.H. Genetic and epigenetic alterations in hepatitis B virus-associated hepatocellular carcinoma. Virol. Sin. 2015, 30, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Kan, Z.; Zheng, H.; Liu, X.; Li, S.; Barber, T.D.; Gong, Z.; Gao, H.; Hao, K.; Willard, M.D.; Xu, J. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013, 23, 1422–1433. [Google Scholar] [CrossRef]
- Bengochea, A.; de Souza, M.M.; Lefrancois, L.; Le Roux, E.; Galy, O.; Chemin, I.; Kim, M.; Wands, J.R.; Trepo, C.; Hainaut, P. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br. J. Cancer 2008, 99, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, A.M.; Fuentes, D.; Morshid, A.I.; Burke, M.R.; Kaseb, A.O.; Hassan, M.; Hazle, J.D.; Elsayes, K.M. Role of Wnt/beta-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J. Hepatocell. Carcinoma 2018, 5, 61–73. [Google Scholar] [CrossRef]
- Hiltunen, M.O.; Alhonen, L.; Koistinaho, J.; Myohanen, S.; Paakkonen, M.; Marin, S.; Kosma, V.M.; Janne, J. Hypermethylation of the APC (adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. Int. J. Cancer 1997, 70, 644–648. [Google Scholar] [CrossRef]
- Huang, H.; Fujii, H.; Sankila, A.; Mahler-Araujo, B.M.; Matsuda, M.; Cathomas, G.; Ohgaki, H. Beta-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am. J. Pathol. 1999, 155, 1795–1801. [Google Scholar] [CrossRef]
- Liu, M.; Cui, L.H.; Li, C.C.; Zhang, L. Association of APC, GSTP1 and SOCS1 promoter methylation with the risk of hepatocellular carcinoma: A meta-analysis. Eur. J. Cancer Prev. 2015, 24, 470–483. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, J.; Ma, R.; Lin, H.; Liang, X.; Cai, X. Prognostic significance of E-cadherin expression in hepatocellular carcinoma: A meta-analysis. PLoS ONE 2014, 9, e103952. [Google Scholar] [CrossRef] [PubMed]
- Herath, N.I.; Walsh, M.D.; Kew, M.C.; Young, J.; Leggett, B.A.; Macdonald, G.A. Cadherin/catenin complex appears to be intact in hepatocellular carcinomas from Australia and South Africa. J. Gastroenterol. Hepatol. 2004, 19, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Herath, N.I.; Purdie, D.M.; Kew, M.C.; Smith, J.L.; Young, J.; Leggett, B.A.; MacDonald, G.A. Varying etiologies lead to different molecular changes in Australian and South African hepatocellular carcinomas. Int. J. Oncol. 2009, 35, 1081–1089. [Google Scholar] [CrossRef][Green Version]
- Li, C.C.; Yu, Z.; Cui, L.H.; Piao, J.M.; Liu, M. Role of P14 and MGMT gene methylation in hepatocellular carcinomas: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 6591–6596. [Google Scholar] [CrossRef][Green Version]
- Chiam, K.; Centenera, M.M.; Butler, L.M.; Tilley, W.D.; Bianco-Miotto, T. GSTP1 DNA methylation and expression status is indicative of 5-aza-2’-deoxycytidine efficacy in human prostate cancer cells. PLoS ONE 2011, 6, e25634. [Google Scholar] [CrossRef] [PubMed]
- Anzola, M.; Cuevas, N.; Lopez-Martinez, M.; Saiz, A.; Burgos, J.J.; de Pancorbo, M.M. No association between GSTP1 gene aberrant promoter methylation and prognosis in surgically resected hepatocellular carcinoma patients from the Basque Country (Northern Spain). Liver Int. 2003, 23, 249–254. [Google Scholar] [CrossRef]
- Tchou, J.C.; Lin, X.; Freije, D.; Isaacs, W.B.; Brooks, J.D.; Rashid, A.; De Marzo, A.M.; Kanai, Y.; Hirohashi, S.; Nelson, W.G. GSTP1 CpG island DNA hypermethylation in hepatocellular carcinomas. Int. J. Oncol. 2000, 16, 663–676. [Google Scholar] [CrossRef]
- Yuan, J.M.; Lu, S.C.; Van Den Berg, D.; Govindarajan, S.; Zhang, Z.Q.; Mato, J.M.; Yu, M.C. Genetic polymorphisms in the methylenetetrahydrofolate reductase and thymidylate synthase genes and risk of hepatocellular carcinoma. Hepatology 2007, 46, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, L.; Spitz, M.R.; Hong, W.K.; Mao, L.; Wei, Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002, 62, 4992–4995. [Google Scholar] [PubMed]
- Wu, Y.; Lin, J.S. DNA methyltransferase 3B promoter polymorphism and its susceptibility to primary hepatocellular carcinoma in the Chinese Han nationality population: A case-control study. World J. Gastroenterol. 2007, 13, 6082–6086. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.R. Epidemiology of hepatitis B in the United States. Hepatology 2009, 49, S28–S34. [Google Scholar] [CrossRef]
- McCormick, H.; Young, P.E.; Hur, S.S.J.; Booher, K.; Chung, H.; Cropley, J.E.; Giannoulatou, E.; Suter, C.M. Isogenic mice exhibit sexually-dimorphic DNA methylation patterns across multiple tissues. BMC Genom. 2017, 18, 966. [Google Scholar] [CrossRef]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef]
- Wu, H.C.; Yang, H.I.; Wang, Q.; Chen, C.J.; Santella, R.M. Plasma DNA methylation marker and hepatocellular carcinoma risk prediction model for the general population. Carcinogenesis 2017, 38, 1021–1028. [Google Scholar] [CrossRef]
- Gurioli, G.; Martignano, F.; Salvi, S.; Costantini, M.; Gunelli, R.; Casadio, V. GSTP1 methylation in cancer: A liquid biopsy biomarker? Clin. Chem. Lab. Med. 2018, 56, 702–717. [Google Scholar] [CrossRef]
- Oussalah, A.; Rischer, S.; Bensenane, M.; Conroy, G.; Filhine-Tresarrieu, P.; Debard, R.; Forest-Tramoy, D.; Josse, T.; Reinicke, D.; Garcia, M. Plasma mSEPT9: A Novel Circulating Cell-free DNA-Based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine 2018, 30, 138–147. [Google Scholar] [CrossRef]
- El-Bendary, M.; Nour, D.; Arafa, M.; Neamatallah, M. Methylation of tumour suppressor genes RUNX3, RASSF1A and E-Cadherin in HCV-related liver cirrhosis and hepatocellular carcinoma. Br. J. Biomed. Sci. 2020, 77, 35–40. [Google Scholar] [CrossRef] [PubMed]
| Colorectal | Liver | Gastric | Esophagus | Pancreatic | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Incidence | Mortality | Incidence | Mortality | Incidence | Mortality | Incidence | Mortality | Incidence | Mortality | ||
| Sex | M | 44.4 | 16.6 | 12.7 | 9.6 | 9.0 | 4.1 | 7.9 | 7.0 | 14.6 | 12.7 |
| F | 34.0 | 11.7 | 4.4 | 4.0 | 4.6 | 2.2 | 1.8 | 1.4 | 11.2 | 9.6 | |
| Race | NHW | 38.6 | 13.8 | 6.9 | 5.8 | 5.4 | 2.3 | 5.0 | 4.4 | 12.7 | 11.1 |
| NHB | 45.7 | 19.0 | 10.9 | 8.6 | 10.1 | 5.4 | 4.0 | 3.2 | 15.9 | 13.6 | |
| H/L | 34.1 | 11.1 | 13.4 | 9.3 | 9.6 | 5.0 | 2.7 | 2.0 | 11.2 | 8.5 | |
| A/PI | 30.0 | 9.5 | 12.7 | 9.0 | 10.3 | 5.1 | 2.1 | 1.6 | 9.3 | 7.5 | |
| AI/AN | 43.3 | 15.8 | 15.1 | 10.6 | 8.8 | 4.8 | 4.5 | 3.4 | 11.5 | 8.9 | |
| Reference | Genes | Results | Design and Method | |||
|---|---|---|---|---|---|---|
| Tissues | Etiology | Population | Method | |||
| Li X et al. [49] | CDKN2A (p16INK4A) | Hypermethylation (F > M) | Liver | Virus, Unknown | Japanese | MSP |
| Wang Y et al. [50] | CDKN2A (p16INK4A) | Hypermethylation (M > F) | Liver | Virus (HBV) | Chinese | MSRE-qPCR, qBS |
| Zhou Y et al. [53] | CDKL2 | Hypermethylation (F > M) | Liver | Virus, Unknown | Chinese | MSRE-qPCR, qBS |
| Zhu C et al. [57] | LINE1 | Hypomethylation (M > F) | Liver | Virus, Alcohol, Unknown | Japanese | qBS |
| Lee HS et al. [58] | HOXA1, CDKN1C, CRABP1, DLEC1, CDKN2A (p16INK4A), CCND2, CACNA1G, RUNX3, PTGS2, BCL2, GRIN2B, NEUROG1, GSTP1, SYK, SFRP1, CALCA, SOCS3, APC and TERT | Numbers of hypermethylated genes (F > M) | Liver | Virus (HBV) | Korean | qMSP |
| LINE1 | Hypomethylation (No sex difference) | Liver | MSRE-PCR | |||
| Liu JB et al. [61] | APC, WIF1, RUNX3, DLC1, SFRP1, DKK, CDH1 | ≥ 3 hypermethylated genes (M > F) | Liver | Virus, Unknown | Chinese | MSP |
| Shen W et al. [42] | 75 DMCs | Significant sex difference (site-dependent) | Liver | Virus, Alcoholic, Tobacco | American | BeadChip (450K) |
| Wang Y et al. [62] | TNFRSF12A | Association of hypomethylation and poor outcome (M > F) | Liver | Virus, Alcohol, Unknown | American | BeadChip (450K, 27K) |
| Zhang X et al. [64] | SOCS1 | Hypermethylation (M > F) | Liver | Virus (HBV) | Chinese | MSRE-qPCR |
| Wu Y et al. [65] | BNC1 | Hypermethylation (F > M) | Liver | Virus, Unknown | Chinese | MSRE-qPCR |
| Ezzikouri S et al. [39] | DNMT3B | Higher HCC risk in DNMT3B -149T carriers (F > M) | Blood | Virus, Unknown | Moroccan | PCR-RFLP |
| Qiao K et al. [60] | MTHFR | LINE1 hypomethylation in MTHFR 677T carriers (F > M) | Blood | Virus (HBV) | Chinese | gPCR, qMSP |
| Reference | Genes | Results | Design and Method | |||
|---|---|---|---|---|---|---|
| Tissue | Etiology | Population | Method | |||
| Liu M et al. [75] | APC, GSTP, SOCS1 | Association of hypermethylation and HCC risk (Asian > Whites) | Liver | Unavailable | Asian, American, European | MSP, qMSP |
| Herald NI et al. [77] | CDH1 | Hypermethylation (Caucasian/Asian > African Black) | Liver | Virus, Alcohol, Aflatoxin, Hemochromatosis, Allagille’s syndrom, Unknown | Australian, South African | MSP |
| Herald NI et al. [78] | CDKN2A (p16INK4A), CDKN2A (p14ARF), CDKN2B (p15INK4B), CDH1, MGMT, RIZ1 | Hypermethylation (Caucasian/Asian > African Black) | Liver | Virus, Alcohol, Aflatoxin, Hemochromatosis, Allagille’s syndrom, Unknown | Australian, South African | MSP |
| 5 CpG islands | Hypermethylation (Caucasian/Asian > African Black) | MSRE-qPCR | ||||
| Li CC et al. [79] | CDKN2A (p14ARF), MGMT | Association of hypermethylation and HCC risk (Chinese > Western) | Liver | Unavailable | Chinese, Japanese, Australian, South African, Spain, American, German | MSP |
| Anzola M et al. [81] | SOCS1 | Hypermethylation (Asian > European) | Liver | Virus, Alcohol, Tobacco, Unknown | Spain | MSP |
| Varghese RS et al. [43] | 40 DMDE genes in European American, 32 DMDE genes in African American | Only 5 genes overlapped in DMDE genetic profiles | Liver | Tobacco, Virus, Alcohol | American | BeadChip (850K) |
| Yuan JM et al. [83] | MTHFR | Frequency of MTHFR 677T allele carriers (non-Asian > Asian) | Blood | Tobacco, Alcohol, Virus | American, Chinese | gPCR |
| Ezzikouri S et al. [39] | DNMT3B | Frequency of DNMT3B -149T carriers (Chinese > Moroccan) | Blood | Virus, Unknown | Moroccan | PCR-RFLP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, W.; Siwko, S.; Tsai, R.Y.L. Sex and Race-Related DNA Methylation Changes in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 3820. https://doi.org/10.3390/ijms22083820
Ye W, Siwko S, Tsai RYL. Sex and Race-Related DNA Methylation Changes in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2021; 22(8):3820. https://doi.org/10.3390/ijms22083820
Chicago/Turabian StyleYe, Wenrui, Stefan Siwko, and Robert Y. L. Tsai. 2021. "Sex and Race-Related DNA Methylation Changes in Hepatocellular Carcinoma" International Journal of Molecular Sciences 22, no. 8: 3820. https://doi.org/10.3390/ijms22083820
APA StyleYe, W., Siwko, S., & Tsai, R. Y. L. (2021). Sex and Race-Related DNA Methylation Changes in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 22(8), 3820. https://doi.org/10.3390/ijms22083820







