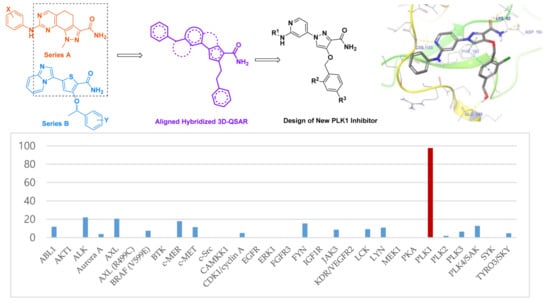
3.2. Chemistry
3.2.1. General Chemical Methods
All chemicals were of reagent grade and were purchased from Aldrich (USA). Separation of the compounds by column chromatography was carried out with silica gel 60 (200–300 mesh ASTM, E. Merck, Germany). The quantity of silica gel used was 50–100 times the weight charged on the column. Thin layer chromatography (TLC) was run on silica gel-coated aluminum sheets (silica gel 60 GF254, E. Merck, Germany) and visualized under ultraviolet (UV) light (254 nm). Both 1H NMR and 13C NMR spectra were recorded on a Brucker model digital AVANCE III 400 MHz spectrometer at 25 °C using tetramethylsilane (TMS) as an internal standard. High-resolution MS (HR/MS) experiments were conducted with Q-TOF/Mass spectrometer 6530 (Agilent Technologies, Santa Clara, CA, USA) operated in positive-ion electrospray mode.
3.2.2. Synthesis of Ethyl 1-(2-fluoropyridin-4-yl)-4-hydroxy-1H-pyrazole-3-carboxylate (3)
To a solution of 2-fluoropyridin-4-amine (0.776 mmol) in 1.35 mL of H2O and 0.206 mL (2.33 mmol) of 35% HCl at 0 °C, a solution of NaNO2 (0.776 mmol) and 0.2 mL of H2O was slowly added and reacted for 30 min. Then, a solution of ethyl 4-chloro-3-oxobutanoate (0.776 mmol) and 0.388 mL of EtOH was added to the reaction vessel, and NaOAc (4.66 mmol) was added, producing a solid. After stirring for 2 h, the organic layer was extracted with ethyl acetate and dried with anhydrous sodium sulphate (Na2SO4), and the solvent was evaporated to obtain compound 2. Subsequently, compound 2 was dissolved in 0.776 mL of THF at 0 °C, and potassium t-butoxide (0.854 mmol) was added, followed by stirring for 1 h. After completion of the reaction, saturated NH4Cl solution was added, the organic layer was extracted with ethyl acetate and dried with anhydrous sodium sulphate (Na2SO4), and the solvent was evaporated, followed by column chromatography and purification under EA:Hex (1:3) conditions to obtain compound 3 (74%). 1H NMR (400 MHz, DMSO-d6)) δ 9.58 (s, 1H), 8.32 (d, J = 5.7 Hz, 1H), 8.28 (d, J = 1.0 Hz, 1H), 7.84 (dd, J = 5.7, 1.3 Hz, 1H), 7.64 (d, J = 1.6 Hz, 1H), 4.32 (q, J = 7.1 Hz, 2H), 1.31 (t, J = 7.1 Hz, 3H).); HRMS (ESI+) m/z calculated for C11H11FN3O3 [M+H]+: 252.0779, found 252.0778.
3.2.3. General Procedure A (4a–4c)
Compound 3 (0.0199 mmol) was dissolved in 0.995 mL of DMF at 0 °C, and NaH (0.0239 mmol) and the appropriate benzyl bromide (0.0199 mmol) were added, followed by stirring for 1 h. After completion of the reaction, the reaction mixture was worked up 6 times with ethyl acetate and brine. The organic layer was dried with anhydrous sodium sulphate (Na2SO4), and the solvent was evaporated to give compound 4.
4a as yellow solid (98%): General procedure A was followed, using benzyl bromide and 31H NMR (400 MHz, DMSO-d6): 1H NMR (400 MHz, DMSO) δ 8.81 (s, 1H), 8.37 (d, J = 5.7 Hz, 1H), 7.86 (d, J = 5.7 Hz, 1H), 7.65 (d, J = 1.7 Hz, 1H), 7.52–7.46 (m, 2H), 7.46–7.40 (m, 2H), 7.38 (dd, J = 5.0, 3.6 Hz, 1H), 5.12 (s, 2H), 4.32 (d, J = 7.1 Hz, 2H), 1.31 (t, J = 7.1 Hz, 3H).; HRMS (ESI+) m/z calculated for C18H17FN3O3 [M+H]+: 342.1248, found 342.1262.
4b (yellow solid, 92%): General procedure A was followed, using 1-(bromomethyl)-2-(trifluoromethyl)benzene and 3.1H NMR (400 MHz, DMSO-d6) δ 8.89 (s, 1H), 8.38 (d, J = 5.7 Hz, 1H), 7.91 (dd, J = 12.4, 6.8 Hz, 2H), 7.80 (dd, J = 17.3, 7.9 Hz, 2H), 7.70 (d, J = 1.7 Hz, 1H), 7.62 (t, J = 7.8 Hz, 1H), 5.26 (s, 2H), 4.33 (q, J = 7.1 Hz, 2H), 1.30 (t, J = 7.1 Hz, 3H). HRMS (ESI+) m/z calculated for C19H16F4N3O3 [M+H]+ 410.1122, found 410.1111.
4c (yellow solid, 56%): General procedure A was followed, using ((4-(bromomethyl)-3-chlorobenzyl)oxy)(tert-butyl)dimethylsilane and 3. 1H NMR (400 MHz, DMSO-d6) δ 8.88 (s, 1H), 8.37 (d, J = 5.7 Hz, 1H), 7.88 (d, J = 5.7 Hz, 1H), 7.67 (d, J = 8.1 Hz, 2H), 7.44 (s, 1H), 7.35 (d, J = 7.9 Hz, 1H), 5.16 (s, 2H), 4.75 (s, 2H), 4.32 (t, J = 7.1 Hz, 2H), 1.31 (t, J = 7.1 Hz, 3H), 0.91 (s, 9H), 0.09 (s, 6H). HRMS (ESI+) m/z calculated for C25H32ClFN3O4Si [M+H]+: 520.1829, found 520.1789.
3.2.4. General Procedure B (5a–7a)
Compound 4 (0.0293 mmol) was dissolved in 0.293 mL of DMSO, and the appropriate amine (0.0586 mmol) and TEA (0.0586 mmol) were added, followed by stirring at 100 °C for 24 h. After completion of the reaction, the reaction mixture was cooled to room temperature, and work up was performed 6 times with ethyl acetate and washed with brine. The organic layer was dried with anhydrous sodium sulphate (Na2SO4), and the solvent was evaporated, followed by column chromatography and purification under EA:Hex (1:1) conditions to obtain a compound.
5a (38%): General procedure B was followed, using tetrahydro-2H-pyran-4-amine and 4. 1H NMR (400 MHz, CDCl3) δ 8.01 (d, J = 6.0 Hz, 1H), 7.55 (s, 1H), 7.45 (dd, J = 7.8, 1.1 Hz, 2H), 7.42–7.34 (m, 3H), 6.88 (d, J = 1.7 Hz, 1H), 6.76 (dd, J = 6.0, 2.0 Hz, 1H), 5.94 (s, 1H), 5.12 (s, 2H), 4.46 (q, J = 7.1 Hz, 2H), 4.01 (dt, J = 12.2, 3.9 Hz, 2H), 3.92 (s, 1H), 3.57 (td, J = 11.8, 2.3 Hz, 2H), 2.02 (s, 2H), 1.65–1.56 (m, 2H), 1.43 (t, J = 7.1 Hz, 3H).; HRMS (ESI+) m/z calculated for C23H27N4O4 [M+H]+: 423.2027, found 423.2129.
6a (37%): General procedure B was followed, using piperidin-4-amine and 4. 1H NMR (400 MHz, DMSO-d6) δ 8.60 (s, 1H), 8.10 (d, J = 6.1 Hz, 1H), 7.48 (d, J = 6.9 Hz, 2H), 7.44–7.34 (m, 3H), 7.11 (d, J = 30.0 Hz, 2H), 5.10 (d, J = 8.8 Hz, 2H), 4.31 (q, J = 7.1 Hz, 2H), 3.82 (s, 1H), 3.62 (s, 2H), 1.91 (s, 2H), 1.78 (d, J = 13.1 Hz, 2H), 1.65 (s, 2H), 1.38 (s, 9H), 1.29 (d, J = 5.3 Hz, 3H).; HRMS (ESI+) m/z calculated for C28H36N5O5 [M+H]+: 522.2711, found 522.2722.
7a (53%): General procedure B was followed, using pyrrolidin-3-amine and 4. 1H NMR (400 MHz, CDCl3) δ 8.06 (s, 1H), 7.55 (s, 1H), 7.47-7.43 (m, 2H), 7.42−7.33 (m, 3H), 6.82 (d, J = 12.8 Hz, 2H), 5.11 (s, 2H), 4.46 (q, J = 6.8 Hz, 2H), 4.42-4.34 (m, 1H), 3.73 (dd, J = 11.2, 6.0 Hz, 1H), 3.31 (s, 1H), 3.22 (s, 1H), 2.28–2.19 (m, 1H), 1.89 (d, J = 9.7 Hz, 2H), 1.46 (s, 9H), 1.42 (d, J = 7.1 Hz, 3H), 1.25-1.25 (m, 1H).; HRMS (ESI+) m/z calculated for C27H34N5O3 [M+H]+: 508.2554, found 508.2535.
3.2.5. General Procedure C (8a–10a, 13a–13b, 14a–14b)
After adding 0.47 mL of 7N NH3 in MeOH to the appropriate ethyl 4-1H-pyrazole-3-carboxylate (0.0111 mmol), the mixture was stirred at 60 °C for 16 h. After completion of the reaction, the reaction mixture was cooled to room temperature and concentrated in vacuo. Column chromatography was performed and purified under MC:MeOH (20:1) conditions to obtain corresponding 4-1H-pyrazole-3-carboxamide.
8a as white solid (53%): General procedure C was followed, using
5a. 1H NMR (400 MHz, DMSO-d
6) δ 8.44 (s, 1H), 8.04 (d,
J = 5.7 Hz, 1H), 7.53–7.45 (m, 3H), 7.45–7.39 (m, 2H), 7.38–7.33 (m, 1H), 7.25 (s, 1H), 6.95 (dd,
J = 5.7, 2.0 Hz, 1H), 6.90 (d,
J = 1.7 Hz, 1H), 6.75 (d,
J = 7.7 Hz, 1H), 5.11 (s, 2H), 3.95 (d,
J = 7.8 Hz, 1H), 3.86 (dd,
J = 8.0, 3.3 Hz, 2H), 3.41 (td,
J = 11.4, 2.1 Hz, 2H), 1.87 (d,
J = 10.3 Hz, 2H), 1.49–1.40 (m, 2H);
13C NMR (101 MHz, DMSO-d
6) δ 161.7, 159.2, 149.3, 146.2, 145.5, 136.2, 128.5, 128.2, 128.0, 114.3, 101.1, 95.3, 73.5, 66.0, 46.4, 32.7 (
Supplementary S5); mp: 78.5–80.5 °C;
HRMS (ESI+) m/z calculated for C
21H
24N
5O
3 [M+H]+: 394.1874, found 394.1853.
9a (white solid, 30%): General procedure C was followed, using 6a. 1H NMR (400 MHz, DMSO-d6) δ 8.82 (s, 2H), 8.50 (s, 1H), 8.11 (d, J = 5.7 Hz, 1H), 7.52-7.48 (m, 2H), 7.44–7.35 (m, 3H), 7.08 (dd, J = 5.8, 2.0 Hz, 1H), 6.97 (d, J = 1.7 Hz, 1H), 5.12 (s, 2H), 4.49–4.41 (m, 1H), 3.51–3.42 (m, 1H), 3.28-3.24 (m, 1H), 3.09 (dt, J = 11.0, 4.3 Hz, 1H), 2.23 (dt, J = 20.9, 7.3 Hz, 2H), 1.93 (dd, J = 15.9, 9.5 Hz, 2H); 13C NMR (101 MHz, DMSO-d6) δ 161.6, 159.1, 149.2, 141.1, 145.5, 137.3, 136.7, 133.5, 127.3, 124.4, 121.3, 99.3, 88.9, 70.5, 36.4, 33.6; mp: 58.5–60.5 °C; HRMS (ESI+) calculated for C21H25N6O2 [M+H]+: 393.2034, found 393.2005.
10a (yellow solid, 57%): General procedure C was followed, using 7a. 1H NMR (400 MHz, DMSO-d6) δ 8.48−8.43 (m, 1H), 8.06 (dd, J = 5.5, 3.6 Hz, 1H), 7.53−7.48 (m, 2H), 7.43–7.33 (m, 3H), 6.97 (d, J = 12.6 Hz, 1H), 6.85 (s, 1H), 5.11 (d, J = 5.8 Hz, 2H), 4.81−4.73 (m, 1H), 4.04 (s, 1H), 3.09 (s, 1H), 2.78 (s, 1H), 1.95 (d, J = 13.7 Hz, 2H), 1.47 (s, 2H), 1.30−1.25 (m, 2H); 13C NMR (101 MHz, DMSO-d6) δ 161.6, 159.1, 157.9, 149.4, 146.3, 145.5, 136.3; mp: 59.0–61.0 °C; HRMS (ESI+) calculated for C20H23N6O2 [M+H]+: 379.1877, found 379.1848.
13a (brown solid, 73%): General procedure C was followed, using
11a. 1H NMR (400 MHz, DMSO-d
6) δ 9.27 (s, 1H), 8.53 (s, 1H), 8.24 (d,
J = 5.7 Hz, 1H), 7.71–7.65 (m, 2H), 7.52 (dd,
J = 6.9, 5.3 Hz, 3H), 7.45−7.40 (m, 2H), 7.39−7.34 (m, 1H), 7.31 (d,
J = 1.6 Hz, 1H), 7.28 (dd,
J = 8.5, 7.5 Hz, 3H), 7.22 (dd,
J = 5.7, 2.0 Hz, 1H), 6.92 (t,
J = 7.3 Hz, 1H), 5.14 (s, 2H) (
Supplementary S4);
13C NMR (101 MHz, DMSO-d
6) δ 161.6, 157.1, 149.2, 146.3, 145.6, 141.4, 136.7, 136.2, 128.6, 128.2, 128.0, 120.8, 118.3, 114.3, 103.5, 97.8, 73.5 (
Supplementary S5);
mp: 191 °C–193 °C;
HRMS (ESI+) m/z calculated for C
22H
20N
5O
2 [M+H]
+: 386.1612, found 386.1450.
13b (45%): General procedure C was followed, using 11b. 1H NMR (400 MHz, DMSO-d6) δ 9.28 (s, 1H), 8.60 (s, 1H), 8.25 (d, J = 5.7 Hz, 1H), 7.89 (d, J = 7.6 Hz, 1H), 7.82 (d, J = 7.8 Hz, 1H), 7.77 (t, J = 7.6 Hz, 1H), 7.71–7.66 (m, 2H), 7.62 (t, J = 7.6 Hz, 1H), 7.53 (s, 1H), 7.32 (d, J = 1.6 Hz, 1H), 7.31−7.24 (m, 4H), 6.92 (t, J = 7.3 Hz, 1H), 5.28 (s, 2H); 13C NMR (101 MHz, DMSO-d6) δ 161.6 (s), 157.1 (d, J = 9.1 Hz), 149.1 (s), 146.3 (s), 145.4 (s), 141.4 (s), 136.7 (s), 134.2 (s), 132.9 (s), 130.5 (s), 129.0 (s), 128.6 (s), 126.9 (s), 126.1 (d, J = 5.5 Hz), 120.8 (s), 118.3 (d, J = 10.3 Hz), 114.7 (s), 103.6 (s), 97.9 (s), 70.2 (s); mp: 188.5–190.5 °C; HRMS (ESI+) m/z calculated for C23H19F3N5O2 [M+H]+: 454.1485, found 454.1450.
14a (yellow solid 73%): General procedure C was followed, using
12a. 1H NMR (400 MHz, DMSO-d
6) δ 8.47 (s, 1H), 8.26 (s, 1H), 8.23-8.19 (m, 2H), 7.55–7.46 (m, 4H), 7.45–7.39 (m, 2H), 7.38 (dd,
J = 5.0, 3.6 Hz, 1H), 7.28 (s, 1H), 7.22 (dd,
J = 5.7, 1.9 Hz, 1H), 7.03 (dd,
J = 8.0, 1.6 Hz, 1H), 6.97 (td,
J = 7.7, 1.8 Hz, 1H), 6.93 (dd,
J = 7.7, 1.7 Hz, 1H), 5.13 (s, 2H), 3.85 (s, 3H) (
Supplementary S4);
13C NMR (101 MHz, DMSO-d
6) δ 161.7, 157.3, 149.0, 146.3, 145.6, 136.3, 129.8, 128.5, 128.2, 128.0, 121.9, 120.4, 120.0, 114.2, 110.9, 103.5, 98.2, 73.5, 55.7 (
Supplementary S5);
mp: 197.0–199.5 °C;
HRMS (ESI+) m/z calculated for C
23H
22N
5O
3 [M+H]
+: 416.1717, found 416.1683.
14b (white solid 67%): General procedure C was followed, using 12b. 1H NMR (400 MHz, DMSO-d6) δ 8.53 (s, 1H), 8.24 (dd, J = 8.6, 2.6 Hz, 2H), 8.21 (d, J = 5.7 Hz, 1H), 7.89 (d, J = 7.7 Hz, 1H), 7.82 (d, J = 7.8 Hz, 1H), 7.77 (t, J = 7.6 Hz, 1H), 7.62 (t, J = 7.7 Hz, 1H), 7.51 (d, J = 5.7 Hz, 1H), 7.49 (d, J = 1.8 Hz, 1H), 7.30 (s, 1H), 7.26 (dd, J = 5.7, 1.9 Hz, 1H), 7.03 (dd, J = 8.0, 1.6 Hz, 1H), 6.97 (td, J = 7.7, 1.8 Hz, 1H), 6.92 (td, J = 7.6, 1.7 Hz, 1H), 5.28 (s, 2H), 3.86 (s, 3H); 13C NMR (101 MHz, DMSO-d6) δ 161.7 (s), 157.2 (d, J = 10.4 Hz), 148.9 (d, J = 12.6 Hz), 146.3 (s), 145.4 (s), 136.5 (s), 134.2 (s), 132.9 (s), 130.5 (s), 129.8 (d, J = 7.2 Hz), 128.9 (s), 121.9 (s), 120.4 (s), 119.9 (d, J = 15.6 Hz), 114.6 (s), 110.9 (s), 103.7 (s), 98.4 (s), 70.2 (s), 55.7 (s). mp: 178.5–180.5 °C; HRMS (ESI+) m/z calculated for C24H21F3N5O3 [M+H]+: 484.1591, found 484.1546.
3.2.6. General Procedure D (11a–11c, 12a–12c)
Compound 4 (0.0293 mmol) was dissolved in the appropriate aniline (0.134 mL, 50 eq) and stirred at 120° C for 18 h. After completion of the reaction, the reaction mixture was cooled to room temperature, and work up was performed with ethyl acetate and water. The organic layer was dried with anhydrous sodium sulphate (Na2SO4) and the solvent was evaporated, followed by column chromatography and purification under EA:Hex (1:3.5) conditions to obtain product.
11a (62%): General procedure D was followed, using aniline and 4a 1H NMR (400 MHz, DMSO-d6) δ 9.36 (s, 1H), 8.62 (s, 1H), 8.26 (d, J = 5.7 Hz, 1H), 7.70 (dd, J = 8.6, 1.0 Hz, 2H), 7.52–7.47 (m, 2H), 7.45–7.40 (m, 2H), 7.39–7.34 (m, 2H), 7.30–7.25 (m, 2H), 7.22 (dd, J = 5.8, 2.0 Hz, 1H), 6.92 (t, J = 7.3 Hz, 1H), 5.13 (s, 2H), 4.32 (q, J = 7.1 Hz, 2H), 1.31 (t, J = 7.1 Hz, 3H). HRMS (ESI+) m/z calculated for C24H23N4O3 [M+H]+: 415.1765, found 415.1821.
11b (42%): General procedure D was followed, using aniline and 4b. 1H NMR (400 MHz, DMSO-d6) δ 9.37 (s, 1H), 8.71 (s, 1H), 8.27 (d, J = 5.7 Hz, 1H), 7.93 (d, J = 7.6 Hz, 1H), 7.79 (dd, J = 17.5, 7.9 Hz, 2H), 7.71 (dd, J = 8.6, 1.0 Hz, 2H), 7.61 (t, J = 7.6 Hz, 1H), 7.39 (d, J = 1.6 Hz, 1H), 7.31-7.25 (m, 3H), 6.92 (t, J = 7.3 Hz, 1H), 5.27 (s, 2H), 4.32 (q, J = 7.1 Hz, 2H), 1.30 (t, J = 7.1 Hz, 3H). HRMS (ESI+) m/z calculated for C25H22F3N4O3 [M+H]+: 483.1639, found 483.1793
11c (63%): General procedure D was followed, using aniline and 4c. 1H NMR (400 MHz, DMSO-d6) δ 9.36 (s, 1H), 8.71 (s, 1H), 8.27 (d, J = 5.7 Hz, 1H), 7.73−7.69 (m, 2H), 7.67 (d, J = 7.9 Hz, 1H), 7.44 (s, 1H), 7.39 (d, J = 1.7 Hz, 1H), 7.35 (d, J = 7.8 Hz, 1H), 7.31–7.24 (m, 3H), 6.92 (t, J = 7.3 Hz, 1H), 5.17 (s, 2H), 4.75 (s, 2H), 4.32 (q, J = 7.1 Hz, 2H), 1.31 (t, J = 7.1 Hz, 3H), 0.91 (s, 9H), 0.09 (s, 6H). HRMS (ESI+) m/z calculated for C31H38ClN4O4Si [M+H]+: 593.2345, found 593.2464.
12a (79%): General procedure D was followed, using 2-methoxyaniline and 4a. 1H NMR (400 MHz, DMSO-d6) δ 8.56 (s, 1H), 8.45 (s, 1H), 8.21 (dd, J = 6.7, 5.2 Hz, 2H), 7.54–7.47 (m, 3H), 7.45–7.39 (m, 2H), 7.36 (dt, J = 9.6, 4.3 Hz, 1H), 7.19 (dd, J = 5.7, 2.0 Hz, 1H), 7.03 (dd, J = 8.0, 1.6 Hz, 1H), 6.97 (td, J = 7.7, 1.7 Hz, 1H), 6.91 (td, J = 7.6, 1.6 Hz, 1H), 5.12 (s, 2H), 4.32 (q, J = 7.1 Hz, 2H), 3.85 (s, 3H), 1.31 (t, J = 7.1 Hz, 3H). HRMS (ESI+) m/z calculated for C25H25N4O4 [M+H]+: 445.1870, found 445.1954.
12b (48%): General procedure D was followed, using 2-methoxyaniline and 4b. 1H NMR (400 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.46 (s, 1H), 8.22 (dd, J = 5.5, 3.9 Hz, 2H), 7.93 (d, J = 7.7 Hz, 1H), 7.79 (dd, J = 17.2, 8.0 Hz, 2H), 7.61 (t, J = 7.6 Hz, 1H), 7.55 (dd, J = 4.9, 1.7 Hz, 1H), 7.24 (dd, J = 5.7, 2.0 Hz, 1H), 7.04–7.01 (m, 1H), 6.97 (td, J = 7.7, 1.8 Hz, 1H), 6.92 (td, J = 7.6, 1.6 Hz, 1H), 5.27 (s, 2H), 4.32 (q, J = 7.1 Hz, 2H), 3.85 (s, 3H), 1.30 (t, J = 7.1 Hz, 3H). HRMS (ESI+) m/z calculated for C26H24F3N4O4 [M+H]+: 513.1744, found 513.1765.
12c (81%): General procedure D was followed, using 2-methoxyaniline and 4c. 1H NMR (400 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.45 (s, 1H), 8.24−8.19 (m, 2H), 7.67 (d, J = 7.9 Hz, 1H), 7.54 (d, J = 1.8 Hz, 1H), 7.44 (s, 1H), 7.35 (d, J = 7.9 Hz, 1H), 7.23 (dd, J = 5.7, 2.0 Hz, 1H), 7.03 (dd, J = 8.0, 1.6 Hz, 1H), 6.97 (td, J = 7.7, 1.8 Hz, 1H), 6.94−6.89 (m, 1H), 5.17 (s, 2H), 4.75 (s, 2H), 4.32 (q, J = 7.1 Hz, 2H), 3.85 (s, 3H), 1.31 (t, J = 7.1 Hz, 3H), 0.91 (s, 9H), 0.09 (s, 6H). HRMS (ESI+) m/z calculated for C32H40ClN4O5Si [M+H]+: 623.2451, found 623.2416.
3.2.7. 4-((2-chloro-4-(hydroxymethyl)benzyl)oxy)-1-(2-(phenylamino)pyridin-4-yl)-1H-pyrazole-3-carboxamide (15)
After adding 1.45 mL of 7 N NH
3 in MeOH to
11c (0.0244 mmol), the mixture was stirred at 60 °C for 24 h. After confirmation of completion of the reaction, the reaction mixture was cooled to room temperature and concentrated in vacuo to obtain compound ethyl 4-((4-(((tert-butyldimethylsilyl)oxy)methyl)-2-chlorobenzyl)oxy)-1-(2-(phenylamino)pyridin-4-yl)-1H-pyrazole-3-carboxylate. Next, the product (0.0244 mmol) was dissolved in 0.244 mL of THF, and teterabutylammonium fluoride 1M in THF (0.0244 mmol) was slowly added. After the reaction, work up was performed with ethyl acetate and saturated NH
4Cl solution. The organic layer was dried with anhydrous sodium sulphate (Na
2SO
4), and the solvent was evaporated, followed by column chromatography and purification under MC:MeOH (40:1) conditions to obtain compound
15 (60%):
1H NMR (400 MHz, DMSO-d
6) δ 9.27 (s, 1H), 8.61 (s, 1H), 8.24 (d,
J = 5.7 Hz, 1H), 7.68 (dd,
J = 8.6, 1.0 Hz, 2H), 7.63 (d,
J = 7.9 Hz, 1H), 7.54 (s, 1H), 7.46 (s, 1H), 7.35−7.31 (m, 2H), 7.30−7.22 (m, 4H), 6.92 (t,
J = 7.3 Hz, 1H), 5.35 (t,
J = 5.8 Hz, 1H), 5.19 (s, 2H), 4.52 (d,
J = 5.8 Hz, 2H) (
Supplementary S4);
13C NMR (101 MHz, DMSO-d
6) δ 161.6, 157.2, 149.2, 146.3, 145.3, 141.4, 136.6, 132.5, 131.7, 130.2, 128.7, 127.0, 125.1, 124.2, 120.8, 118.3, 114.5, 103.6, 97.9, 70.9, 61.9 (
Supplementary S5);
mp: 179.5–180.5°C;
HRMS (ESI+) m/z calculated for C
23H
21ClN
5O
5 [M+H]
+: 450.1327, found 450.1294.
3.2.8. 4-((2-chloro-4-(hydroxymethyl)benzyl)oxy)-1-(2-((2-methoxyphenyl)amino) pyridin -4-yl)-1H-pyrazole-3-carboxamide (17)
Compound
17 was prepared by an analogous procedure as described for the preparation of
15. (67%):
1H NMR (400 MHz, DMSO-d
6) δ 8.54 (s, 1H), 8.27−8.19 (m, 3H), 7.63 (d,
J = 7.9 Hz, 1H), 7.53 (s, 1H), 7.49 (d,
J = 1.8 Hz, 1H), 7.46 (d,
J = 1.3 Hz, 1H), 7.33 (d,
J = 7.8 Hz, 1H), 7.30–7.23 (m, 2H), 7.03 (dd,
J = 7.9, 1.6 Hz, 1H), 6.99-6.94 (m, 1H), 6.92 (dd,
J = 7.5, 5.9 Hz, 1H), 5.35 (t,
J = 5.8 Hz, 1H), 5.19 (s, 2H), 4.52 (d,
J = 5.8 Hz, 2H), 3.86 (s, 3H) (
Supplementary S4).
13C NMR (101 MHz, DMSO-d
6) δ 161.6, 157.2, 149.2, 146.3, 145.4, 141.4, 137.2, 136.6, 132.4, 130.1, 129.4, 128.6, 120.8, 118.3, 118.3, 114.6, 103.6, 97.9, 70.9, 44.8, 29.0.
mp: 198.5-200.5 °C;
HRMS (ESI+) m/z calculated for C
24H
23ClN
5O
4 [M+H]
+: 480.1433, found 480.1406.
3.2.9. General Procedure E (16, 18)
After dissolving compound
15 (9.56 mmol) in 0.1 mL of MC, triethylamine (10.5 mmol) and methanesulfonyl chloride (14.3 mmol) were sequentially added dropwise at 0 °C and stirred for 3 h. After the reaction was complete, the organic layer was extracted using water and methylene chloride. The organic layer was dried with anhydrous sodium sulfate, the solvent was evaporated, and the next reaction proceeded. The obtained compound (0.00956 mmol) was dissolved in 0.1 mL of THF, and dimethylamine (19.1 mmol) and DIPEA (19.1 mmol) were added, followed by stirring at 60 °C for 18 h. After completion of the reaction, the mixture was cooled to room temperature, and work up was performed with ethyl acetate and water. The organic layer was dried with anhydrous sodium sulphate, and the solvent was evaporated, followed by column chromatography and purification under MC:MeOH (10:1) to obtain compound
16 as white solid (50%):
1H NMR (400 MHz, DMSO-d
6) δ 9.27 (s, 1H), 8.61 (s, 1H), 8.24 (d,
J = 5.7 Hz, 1H), 7.71−7.66 (m, 2H), 7.63 (d,
J = 7.8 Hz, 1H), 7.54 (s, 1H), 7.44 (s, 1H), 7.34-7.31 (m, 2H), 7.30–7.23 (m, 4H), 6.92 (t,
J = 7.3 Hz, 1H), 5.19 (s, 2H), 3.42 (s, 2H), 2.16 (s, 6H) (
Supplementary S4).
13C NMR (101 MHz, DMSO-d
6) δ 161.6, 157.2, 149.2, 146.3, 145.4, 141.4, 137.2, 136.6, 132.4, 130.1, 129.4, 128.6, 120.8, 118.3, 118.3, 114.6, 103.6, 97.9, 70.9, 44.8, 29.0;
mp: 138.5–140.5 °C;
HRMS (ESI+) m/z calculated for C
25H
26ClN
6O
2 [M+H]
+: 477.1800, found 477.1767.
18(. white solid, 39%): Compound
18 was prepared by the same procedure as described for the preparation of
16 using
17. 1H NMR (400 MHz, DMSO-d
6) δ 8.54 (s, 1H), 8.27–8.19 (m, 3H), 7.63 (d,
J = 7.8 Hz, 1H), 7.54–7.48 (m, 2H), 7.44 (d,
J = 1.3 Hz, 1H), 7.32 (d,
J = 7.8 Hz, 1H), 7.29 (s, 1H), 7.25 (dd,
J = 5.7, 1.9 Hz, 1H), 7.03 (dd,
J = 7.9, 1.6 Hz, 1H), 6.97 (td,
J = 7.7, 1.8 Hz, 1H), 6.92 (td,
J = 7.6, 1.7 Hz, 1H), 5.18 (s, 2H), 3.86 (s, 3H), 3.41 (s, 2H), 2.15 (s, 6H) (
Supplementary S4);
13C NMR (101 MHz, DMSO-d
6) δ 161.7, 157.2, 148.9, 145.9, 136.4, 132.4, 132.1, 130.1, 129.8, 129.3, 127.6, 121.9, 120.4, 119.9, 114.5, 110.9, 103.6, 98.3, 70.9, 62.2, 55.7, 44.9;
mp: 160–162 °C;
HRMS (ESI+) m/z calculated for C
26H
28ClN
6O
3 [M+H]
+: 507.1906, found 507.1881.