Telomere Length and Male Fertility
Abstract
:1. Introduction
2. Results
2.1. Study Subjects Included in the Analyses
2.2. Association between STL and Sperm Parameters
2.3. Association between Teloscore and Sperm Parameters
2.4. Association between SNPs, Teloscore, and STL
2.5. Association between Individual SNPs and Sperm Parameters
2.6. Sperm ABCD Scores Association Analysis
2.7. Functional Effects of the SNPs
3. Discussion
4. Materials and Methods
4.1. Study Design and Populations
4.2. Characterization of Sperm Parameters
4.3. DNA Extraction
4.4. q-PCR Measurement of Sperm Telomere Length
4.5. SNP Selection and Genotyping
4.6. Genotyping and Quality Control
4.7. Teloscore Computation
4.8. Statistical Analysis
4.9. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). International Classification of Diseases, 11th Revision (ICD-11); WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A Unique View on Male Infertility around the Globe. Reprod. Biol. Endocrinol. 2015, 13, 37. [Google Scholar] [CrossRef] [Green Version]
- Tournaye, H.; Krausz, C.; Oates, R.D. Novel Concepts in the Aetiology of Male Reproductive Impairment. lancet. Diabetes Endocrinol. 2017, 5, 544–553. [Google Scholar] [CrossRef]
- World Health Organization, Department of Reproductive Health and Research. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; WHO Press: Geneva, Switzerland, 2010. [Google Scholar]
- Krausz, C. Male Infertility: Pathogenesis and Clinical Diagnosis. Clin. Endocrinol. Metab. 2011, 25, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, M.L.; Li, S.; Behr, B.; Pera, R.R.; Cullen, M.R. Relationship between Semen Production and Medical Comorbidity. Fertil. Steril. 2015, 103, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Salonia, A.; Matloob, R.; Gallina, A.; Abdollah, F.; Saccà, A.; Briganti, A.; Suardi, N.; Colombo, R.; Rocchini, L.; Guazzoni, G.; et al. Are Infertile Men Less Healthy than Fertile Men? Results of a Prospective Case-Control Survey. Eur. Urol. 2009, 56, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.K.; Swan, S.; Jørgensen, N.; Toppari, J.; Redmon, B.; Punab, M.; Drobnis, E.Z.; Haugen, T.B.; Zilaitiene, B.; Sparks, A.E.; et al. Alcohol and Male Reproductive Health: A Cross-Sectional Study of 8344 Healthy Men from Europe and the USA. Hum. Reprod. 2014, 29, 1801–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Lin, H.; Li, Y.; Cao, J. Association between Socio-Psycho-Behavioral Factors and Male Semen Quality: Systematic Review and Meta-Analyses. Fertil. Steril. 2011, 95, 116–123. [Google Scholar] [CrossRef]
- Cerván-Martín; Castilla; Palomino-Morales; Carmona Genetic Landscape of Nonobstructive Azoospermia and New Perspectives for the Clinic. J. Clin. Med. 2020, 9, 300. [CrossRef] [PubMed] [Green Version]
- Oud, M.S.; Volozonoka, L.; Smits, R.M.; Vissers, L.E.L.M.; Ramos, L.; Veltman, J.A. A Systematic Review and Standardized Clinical Validity Assessment of Male Infertility Genes. Hum. Reprod. 2019, 34, 932–941. [Google Scholar] [CrossRef] [Green Version]
- Ferlin, A.; Dipresa, S.; Delbarba, A.; Maffezzoni, F.; Porcelli, T.; Cappelli, C.; Foresta, C. Contemporary Genetics-Based Diagnostics of Male Infertility. Expert Rev. Mol. Diagn. 2019, 19, 623–633. [Google Scholar] [CrossRef]
- Krausz, C.; Riera-Escamilla, A. Genetics of Male Infertility. Nat. Rev. Urol. 2018, 15, 369–384. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn O’Brien, K.L.; Varghese, A.C.; Agarwal, A. The Genetic Causes of Male Factor Infertility: A Review. Fertil. Steril. 2010, 93, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Aston, K.I.; Carrell, D.T. Genome-Wide Study of Single-Nucleotide Polymorphisms Associated With Azoospermia and Severe Oligozoospermia. J. Androl. 2009, 30, 711–725. [Google Scholar] [CrossRef]
- Aston, K.I.; Krausz, C.; Laface, I.; Ruiz-Castane, E.; Carrell, D.T.; Ruiz-Castané, E.; Carrell, D.T. Evaluation of 172 Candidate Polymorphisms for Association with Oligozoospermia or Azoospermia in a Large Cohort of Men of European Descent. Hum. Reprod. 2010, 25, 1383–1397. [Google Scholar] [CrossRef] [Green Version]
- Kosova, G.; Scott, N.M.; Niederberger, C.; Prins, G.S.; Ober, C. Genome-Wide Association Study Identifies Candidate Genes for Male Fertility Traits in Humans. Am. J. Hum. Genet. 2012, 90, 950–961. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Tajima, A.; Kiguchi, M.; Kogusuri, S.; Fujii, A.; Sato, T.; Nozawa, S.; Yoshiike, M.; Mieno, M.; Kojo, K.; et al. Genome-Wide Association Study of Semen Volume, Sperm Concentration, Testis Size, and Plasma Inhibin B Levels. J. Hum. Genet. 2020, 65, 683–691. [Google Scholar] [CrossRef]
- Leslie, J.S.; Rawlins, L.E.; Chioza, B.A.; Olubodun, O.R.; Salter, C.G.; Fasham, J.; Jones, H.F.; Cross, H.E.; Lam, S.; Harlalka, G.V.; et al. MNS1 Variant Associated with Situs Inversus and Male Infertility. Eur. J. Hum. Genet. 2020, 28, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Mu, C.; Ma, J.; Dai, X.; Jiao, H. The Association of Four SNPs in DNA Mismatch Repair Genes with Idiopathic Male Infertility in Northwest China. Int. J. Immunogenet. 2019, 46, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Miyagawa, Y.; Tsujimura, A.; Wada, M. Genetic Polymorphisms within The Intronless ACTL7A and ACTL7B Genes Encoding Spermatogenesis-Specific Actin-Like Proteins in Japanese Males. Int. J. Fertil. Steril. 2019, 13, 245–249. [Google Scholar]
- Singh, V.; Bansal, S.K.; Sudhakar, D.V.S.; Neelabh; Chakraborty, A.; Trivedi, S.; Gupta, G.; Thangaraj, K.; Rajender, S.; Singh, K. SNPs in ERCC1, ERCC2, and XRCC1 Genes of the DNA Repair Pathway and Risk of Male Infertility in the Asian Populations: Association Study, Meta-Analysis, and Trial Sequential Analysis. J. Assist. Reprod. Genet. 2019, 36, 79–90. [Google Scholar] [CrossRef] [PubMed]
- García Rodríguez, A.; de la Casa, M.; Johnston, S.; Gosálvez, J.; Roy, R. Association of Polymorphisms in Genes Coding for Antioxidant Enzymes and Human Male Infertility. Ann. Hum. Genet. 2019, 83, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Gentiluomo, M.; Crifasi, L.; Luddi, A.; Locci, D.; Barale, R.; Piomboni, P.; Campa, D. Taste Receptor Polymorphisms and Male Infertility. Hum. Reprod. 2017, 32, 2324–2331. [Google Scholar] [CrossRef]
- Darmishonnejad, Z.; Zarei-Kheirabadi, F.; Tavalaee, M.; Zarei-Kheirabadi, M.; Zohrabi, D.; Nasr-Esfahani, M.H. Relationship between Sperm Telomere Length and Sperm Quality in Infertile Men. Andrologia 2020, 52, e13546. [Google Scholar] [CrossRef]
- Boniewska-Bernacka, E.; Pańczyszyn, A.; Cybulska, N. Telomeres as a Molecular Marker of Male Infertility. Hum. Fertil. 2019, 22, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, S.; Tavalaee, M.; Izadi, T.; Barikrow, N.; Zakeri, Z.; Lockshin, R.A.; Abbasi, H.; Nasr- Esfahani, M.H. Reduced Sperm Telomere Length in Individuals with Varicocele Is Associated with Reduced Genomic Integrity. Sci. Rep. 2019, 9, 1–7. [Google Scholar]
- Rocca, M.S.; Foresta, C.; Ferlin, A. Telomere Length: Lights and Shadows on Their Role in Human Reproduction. Biol. Reprod. 2018, 100, 305–317. [Google Scholar] [CrossRef]
- Balmori, C.; Varela, E. Should We Consider Telomere Length and Telomerase Activity in Male Factor Infertility? Curr. Opin. Obstet. Gynecol. 2018, 30, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Cariati, F.; Jaroudi, S.; Alfarawati, S.; Raberi, A.; Alviggi, C.; Pivonello, R.; Wells, D. Investigation of Sperm Telomere Length as a Potential Marker of Paternal Genome Integrity and Semen Quality. Reprod. Biomed. Online 2016, 33, 404–411. [Google Scholar] [CrossRef] [Green Version]
- Rocca, M.S.; Speltra, E.; Menegazzo, M.; Garolla, A.; Foresta, C.; Ferlin, A. Sperm Telomere Length as a Parameter of Sperm Quality in Normozoospermic Men. Hum. Reprod. 2016, 31, 1158–1163. [Google Scholar] [CrossRef]
- Thilagavathi, J.; Kumar, M.; Mishra, S.S.; Venkatesh, S.; Kumar, R.; Dada, R. Analysis of Sperm Telomere Length in Men with Idiopathic Infertility. Arch. Gynecol. Obstet. 2013, 287, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.; Hartshorne, G.M. Telomere Lengths in Human Pronuclei, Oocytes and Spermatozoa. Mol. Hum. Reprod. 2013, 19, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Ferlin, A.; Rampazzo, E.; Rocca, M.S.; Keppel, S.; Frigo, A.C.; De Rossi, A.; Foresta, C. In Young Men Sperm Telomere Length Is Related to Sperm Number and Parental Age. Hum. Reprod. 2013, 28, 3370–3376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achi, M.V.; Ravindranath, N.; Dym, M. Telomere Length in Male Germ Cells Is Inversely Correlated with Telomerase Activity1. Biol. Reprod. 2000, 63, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Vasilopoulos, E.; Fragkiadaki, P.; Kalliora, C.; Fragou, D.; Docea, A.O.; Vakonaki, E.; Tsoukalas, D.; Calina, D.; Buga, A.M.; Georgiadis, G.; et al. The Association of Female and Male Infertility with Telomere Length (Review). Int. J. Mol. Med. 2019, 44, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Montpetit, A.J.; Alhareeri, A.A.; Montpetit, M.; Starkweather, A.R.; Elmore, L.W.; Filler, K.; Mohanraj, L.; Burton, C.W.; Menzies, V.S.; Lyon, D.E.; et al. Telomere Length: A Review of Methods for Measurement. Nurs. Res. 2015, 63, 289–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, T.P.; Wright, W.E.; Shay, J.W. Comparison of Telomere Length Measurement Methods. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, e20160451. [Google Scholar] [CrossRef] [Green Version]
- Barrett, J.H.; Iles, M.M.; Dunning, A.M.; Pooley, K.A. Telomere Length and Common Disease: Study Design and Analytical Challenges. Hum. Genet. 2015, 134, 679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codd, V.; Nelson, C.P.; Albrecht, E.; Mangino, M.; Deelen, J.; Buxton, J.L.; Hottenga, J.J.; Fischer, K.; Esko, T.; Surakka, I.; et al. Identification of Seven Loci Affecting Mean Telomere Length and Their Association with Disease. Nat. Genet. 2013, 45, 422–427. [Google Scholar] [CrossRef] [Green Version]
- Mangino, M.; Hwang, S.-J.; Spector, T.D.; Hunt, S.C.; Kimura, M.; Fitzpatrick, A.L.; Christiansen, L.; Petersen, I.; Elbers, C.C.; Harris, T.; et al. Genome-Wide Meta-Analysis Points to CTC1 and ZNF676 as Genes Regulating Telomere Homeostasis in Humans. Hum. Mol. Genet. 2012, 21, 5385–5394. [Google Scholar] [CrossRef] [Green Version]
- Haycock, P.C.; Burgess, S.; Nounu, A.; Zheng, J.; Okoli, G.N.; Bowden, J.; Wade, K.H.; Timpson, N.J.; Evans, D.M.; Willeit, P.; et al. Association between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases a Mendelian Randomization Study. JAMA Oncol. 2017, 3, 636–651. [Google Scholar] [CrossRef]
- Iles, M.M.; Bishop, D.T.; Taylor, J.C.; Hayward, N.K.; Brossard, M.; Cust, A.E.; Dunning, A.M.; Lee, J.E.; Moses, E.K.; Akslen, L.A.; et al. The Effect on Melanoma Risk of Genes Previously Associated With Telomere Length. JNCI J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef]
- Zhang, C.; Doherty, J.A.; Burgess, S.; Hung, R.J.; Lindström, S.; Kraft, P.; Gong, J.; Amos, C.I.; Sellers, T.A.; Monteiro, A.N.A.; et al. Genetic Determinants of Telomere Length and Risk of Common Cancers: A Mendelian Randomization Study. Hum. Mol. Genet. 2015, 24, 5356–5366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luu, H.N.; Long, J.; Wen, W.; Zheng, Y.; Cai, Q.; Gao, Y.-T.; Zheng, W.; Shu, X.-O. Association between Genetic Risk Score for Telomere Length and Risk of Breast Cancer. Cancer Causes Control 2016, 27, 1219–1228. [Google Scholar] [CrossRef] [Green Version]
- Walsh, K.M.; Codd, V.; Rice, T.; Nelson, C.P.; Smirnov, I.V.; McCoy, L.S.; Hansen, H.M.; Elhauge, E.; Ojha, J.; Francis, S.S.; et al. Longer Genotypically-Estimated Leukocyte Telomere Length Is Associated with Increased Adult Glioma Risk. Oncotarget 2015, 6, 42468–42477. [Google Scholar] [CrossRef] [Green Version]
- Campa, D.; Matarazzi, M.; Greenhalf, W.; Bijlsma, M.; Saum, K.-U.; Pasquali, C.; van Laarhoven, H.; Szentesi, A.; Federici, F.; Vodicka, P.; et al. Genetic Determinants of Telomere Length and Risk of Pancreatic Cancer: A PANDoRA Study. Int. J. Cancer 2018, 144, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Giaccherini, M.; Macauda, A.; Sgherza, N.; Sainz, J.; Gemignani, F.; Maldonado, J.M.S.; Jurado, M.; Tavano, F.; Mazur, G.; Jerez, A.; et al. Genetic Polymorphisms Associated with Telomere Length and Risk of Developing Myeloproliferative Neoplasms. Blood Cancer J. 2020, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef]
- Gentiluomo, M.; Katzke, V.A.; Kaaks, R.; Tjønneland, A.; Severi, G.; Perduca, V.; Boutron-Ruault, M.C.; Weiderpass, E.; Ferrari, P.; Johnson, T.; et al. Mitochondrial DNA Copy-Number Variation and Pancreatic Cancer Risk in the Prospective EPIC Cohort. Cancer Epidemiol. Biomark. Prev. 2020, 29, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Tiseo, B.C.; Cocuzza, M.; Bonfá, E.; Srougi, M.; Silva, C.A. Male Fertility Potential Alteration in Rheumatic Diseases: A Systematic Review. Int. Braz. J. Urol. 2016, 42, 11–21. [Google Scholar] [CrossRef] [Green Version]
- D’Cruz, O.J.; Haas, G.G.; Reichlin, M. Autoantibodies to Decondensed Sperm Nuclear Deoxyribonucleic Acid in Patients with Antisperm Antibodies and Systemic Lupus Erythematosus Detected by Immunofluorescence Flow Cytometry. Fertil. Steril. 1994, 62, 834–844. [Google Scholar] [CrossRef]
- Soares, P.M.F.; Borba, E.F.; Bonfa, E.; Hallak, J.; Corrêa, A.L.; Silva, C.A.A. Gonad Evaluation in Male Systemic Lupus Erythematosus. Arthritis Rheum. 2007, 56, 2352–2361. [Google Scholar] [CrossRef] [Green Version]
- Hickman, R.A.; Gordon, C. Causes and Management of Infertility in Systemic Lupus Erythematosus. Rheumatology 2011, 50, 1551–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krausz, C.; Giachini, C.; Forti, G. TSPY and Male Fertility. Genes 2010, 1, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Vodicka, R.; Vrtel, R.; Dusek, L.; Singh, A.R.; Krizova, K.; Svacinova, V.; Horinova, V.; Dostal, J.; Oborna, I.; Brezinova, J.; et al. TSPY Gene Copy Number as a Potential New Risk Factor for Male Infertility. Reproduct. BioMed. Online 2007, 14, 579–587. [Google Scholar] [CrossRef]
- Iosub-Amir, A.; Bai, F.; Sohn, Y.S.; Song, L.; Tamir, S.; Marjault, H.B.; Mayer, G.; Karmi, O.; Jennings, P.A.; Mittler, R.; et al. The Anti-Apoptotic Proteins NAF-1 and IASPP Interact to Drive Apoptosis in Cancer Cells. Chem. Sci. 2019, 10, 665–673. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Wang, Y.; Zhu, M.; Wang, Y.; Huang, M.; Jin, G.; Guo, X.; Sha, J.; Dai, J.; Hu, Z. Transcriptome-Wide Association Study Revealed Two Novel Genes Associated with Nonobstructive Azoospermia in a Chinese Population. Fertil. Steril. 2017, 108, 1056–1062. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Li, Z.; Yu, J.; Tong, C.; Lin, Y.; Guo, X.; Lu, F.; Dong, J.; Xia, Y.; Wen, Y.; et al. Association Analysis Identifies New Risk Loci for Non-Obstructive Azoospermia in Chinese Men. Nat. Commun. 2014, 5, 3857. [Google Scholar] [CrossRef] [Green Version]
- Cawthon, R.M. Telomere Length Measurement by a Novel Monochrome Multiplex Quantitative PCR Method. Nucleic Acids Res. 2009, 37, 21. [Google Scholar] [CrossRef] [Green Version]
- Hosen, I.; Rachakonda, P.S.; Heidenreich, B.; de Verdier, P.J.; Ryk, C.; Steineck, G.; Hemminki, K.; Kumar, R. Mutations in TERTPromoter and FGFR3and Telomere Length in Bladder Cancer. Int. J. Cancer 2015, 137, 1621–1629. [Google Scholar] [CrossRef]
- Levy, D.; Neuhausen, S.L.; Hunt, S.C.; Kimura, M.; Hwang, S.J.; Chen, W.; Bis, J.C.; Fitzpatrick, A.L.; Smith, E.; Johnson, A.D.; et al. Genome-Wide Association Identifies OBFC1 as a Locus Involved in Human Leukocyte Telomere Biology. Proc. Natl. Acad. Sci. USA 2010, 107, 9293–9298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Total | ABCD Groups | ||||
|---|---|---|---|---|---|
| A (<5th) | B (5–50th) | C (50–95th) | D (>95th) | ||
| Age (years) | 34.8 (±7.5) (n = 599) | ||||
| Sperm concentration (106/mL) | 55.4 (±49.2) | 6.7 (±4.5) (n = 155) | 38.8 (±16.5) (n = 275) | 114.5 (±32.1) (n = 162) | 256 (±30.1) (n = 7) |
| Progressive motility (%) | 45.9 (±18.1) | 18.8 (±10.1) (n = 121) | 44.0 (±8.0) (n = 246)) | 61.1 (±8.3) (n = 214) | 74.4 (±1.9) (n = 18) |
| Total motility (%) | 48.3 (±16.8) | 26.3 (±11.4) (n = 152) | 50.0 (±12.3) (n = 279) | 64.9 (±7.5) (n = 159) | 78.2 (±7.9) (n = 8) |
| Sperm morphology (%) | 7.4 (±4.3) | 2.9 (±1.7) (n = 105) | 7.9 (±3.7) (n = 435) | 16.4 (±2.7) (n = 54) | 19.2 (±1.5) (n = 5) |
| Univariable Model | Multivariable Model * | |||
|---|---|---|---|---|
| Sperm Parameters | Coefficient (95% CI) | p | Coefficient (95% CI) | p |
| Log STL | 0.006 (−0.004 to 0.015) | 0.25 | 0.005 (−0.005 to 0.016) | 0.31 |
| Concentration | 0.23 (−0.28 to 0.74) | 0.38 | 0.13 (−0.39 to 0.65) | 0.63 |
| Total number | 0.12 (−1.54 to 1.78) | 0.89 | −0.09 (−1.83 to 1.64) | 0.91 |
| Progressive motility | −0.31 (−0.50 to −0.13) | 1.00 × 10−3 | −0.31 (−0.50 to −0.11) | 2.00 × 10−3 |
| Non-progressive motility | 0.06 (0.00 to 0.12) | 0.07 | 0.04 (−0.01 to 0.09) | 0.09 |
| Total motility | −0.28 (−0.46 to −0.11) | 2.00 × 10−3 | −0.31 (−0.49 to −0.13) | 1.00 × 10−3 |
| Normal acrosome | −0.08 (−0.14 to −0.02) | 0.01 | −0.07 (−0.14 to −0.01) | 0.03 |
| Normal head | −0.13 (−0.23 to −0.03) | 0.01 | −0.12 (−0.22 to −0.01) | 0.03 |
| Normal flagella | −0.15 (−0.26 to −0.05) | 4.00 × 10−3 | −0.15 (−0.26 to −0.04) | 8.00 × 10−3 |
| ABCD score | −0.03 (−0.05 to 0.00) | 0.05 | −0.03 (−0.06 to 0.00) | 0.03 |
| ABCD_mot score | −0.03 (−0.04 to −0.01) | 2.00 × 10−3 | −0.03 (−0.05 to −0.01) | 1.00 × 10−3 |
| ABCD_mot score | −0.03 (−0.04 to −0.01) | 2.00 × 10−3 | −0.03 (−0.05 to −0.01) | 1.00 × 10−3 |
| Log STL Residuals (Linear) | 2nd vs. 1st Quintile | 3rd vs. 1st Quintile | 4th vs. 1st Quintile) | 5th vs. 1st Quintile | |
|---|---|---|---|---|---|
| Sperm Parameters | Coeff. (95% CI) | Coeff. (95% CI) | Coeff. (95% CI) | Coeff. (95% CI) | Coeff. (95% CI) |
| Concentration | −1.44 (−12.38–9.50) | 0.34 (−19.75–20.42) | 5.91 (−13.80–25.62) | −2.55 (−22.62–17.51) | 6.95 (−12.55–26.44) |
| Total number | −3.09 (−39.91–33.7) | −7.37 (−74.93–60.19) | 23.72 (−42.59–90.02) | 0.66 (−66.85–68.17) | 25.16 (−40.43–90.74) |
| Progressive motility | −0.42 (−4.5–3.65) | −1.19 (−8.67–6.30) | −2.47 (−9.82–4.87) | −2.60 (−10.08–4.88) | 0.94 (−6.33–8.2) |
| Non-progressive motility | 0.19 (−0.66–1.04) | −0.98 (−2.54–0.58) | −0.63 (−2.17–0.90) | −0.83 (−2.39–0.73) | −0.27 (−1.78–1.25) |
| Total motility | −1.80 (−5.64–2.03) | 0.00 (−7.05–7.06) | −2.13 (−9.06–4.79) | −2.48 (−9.53–4.57) | 0.26 (−6.59–7.11) |
| Normal morphology | −0.30 (−1.48–0.88) | −1.71 (−3.86–0.43) | −1.61 (−3.72–0.50) | −2.25 * (−4.40–−0.11) | −0.81 (−2.89–1.28) |
| Normal acrosome | 0.08 (−1.33–1.48) | −0.19 (−2.78–2.41) | 0.21 (−2.32–2.73) | −1.64 (−4.21–0.92) | 0.23 (−2.28–2.74) |
| Normal head | −0.92 (−3.18–1.34) | 0.77 (−3.28–4.81) | 1.14 (−2.79–5.08) | −1.88 (−5.88–2.12) | 0.69 (−3.24–4.61) |
| Normal flagellum | −1.80 (−4.23–0.63) | −0.97 (−5.33–3.39) | −1.60 (−5.85–2.64) | −3.74 (−8.06–0.58) | −1.49 (−5.73–2.75) |
| ABCD | −0.24 (−0.74–0.26) | 0.14 (−0.77–1.06) | −0.18 (−1.07–0.72) | −0.19 (−1.11–0.72) | 0.09 (−0.80–0.97) |
| ABCD_mot | −0.15 (−0.50–0.20) | 0.10 (−0.55–0.74) | −0.16 (−0.79–0.47) | −0.25 (−0.89–0.39) | −0.08 (−0.71–0.54) |
| Coefficient (95% CI) | p | |
|---|---|---|
| U-GS (linear) | 0.01 (−0.03–0.04) | 0.60 |
| 2nd vs. 1st quintile | −0.05 (−0.27–0.18) | 0.69 |
| 3rd vs. 1st quintile | 0.06 (−0.17–0.30) | 0.60 |
| 4th vs. 1st quintile | 0.08 (−0.14–0.30) | 0.50 |
| 5th vs. 1st quintile | 0.07 (−0.19–0.33) | 0.60 |
| W-GS (linear) | 1 × 10−4 (−4 × 10−4–6 × 10−4) | 0.76 |
| 2nd vs. 1st quintile | −0.06 (−0.30–0.18) | 0.64 |
| 3rd vs. 1st quintile | −0.09 (−0.33–0.15) | 0.47 |
| 4th vs. 1st quintile | −0.08 (−0.33–0.16) | 0.50 |
| 5th vs. 1st quintile | 0.03 (−0.21–0.27) | 0.79 |
| rs11125529 | 0.18 (−0.02–0.37) | 0.07 |
| C/A vs. C/C | 0.19 (−0.04–0.43) | 0.10 |
| A/A vs. C/C | 0.29 (−0.38–0.96) | 0.39 |
| rs3027234 | 0.01 (−0.13–0.14) | 0.91 |
| C/T vs. C/C | −0.06 (−0.24–0.12) | 0.50 |
| T/T vs. C/C | 0.14 (−0.21–0.49) | 0.42 |
| rs6028466 | −0.13 (−0.34–0.09) | 0.25 |
| G/A vs. G/G | −0.15 (−0.38–0.09) | 0.22 |
| A/A vs. G/G | −0.02 (−1.17–1.13) | 0.97 |
| rs7675998 | −0.05 (−0.17–0.08) | 0.47 |
| G/A vs. G/G | −0.04 (−0.20–0.12) | 0.61 |
| A/A vs. G/G | −0.10 (−0.43–0.22) | 0.54 |
| rs9420907 | −0.09 (−0.23–0.06) | 0.25 |
| A/C vs. C/C | −0.16 (−0.33–0.01) | 0.06 |
| A/A vs. C/C | 0.12 (−0.33–0.56) | 0.60 |
| rs6772228 | 0.20 (−0.19–0.58) | 0.32 |
| T/A vs. T/T | 0.20 (−0.19–0.58) | 0.32 |
| rs10936599 | −0.10 (−0.23–0.03) | 0.14 |
| C/T vs. C/C | −0.17 (−0.33–−0.01) | 0.04 |
| T/T vs. C/C | 0.01 (−0.38–0.39) | 0.97 |
| rs2736100 | 0.11 (0.003–0.22) | 0.05 |
| C/A vs. C/C | 0.12 (−0.06–0.30) | 0.19 |
| A/A vs. C/C | 0.22 (0.001–0.44) | 0.05 |
| rs755017 | 0.10 (−0.10–0.30) | 0.31 |
| A/G vs. A/A | 0.10 (−0.10–0.30) | 0.31 |
| rs8105767 | 0.08 (−0.04–0.20) | 0.17 |
| A/G vs. A/A | 0.08 (−0.08–0.25) | 0.32 |
| G/G vs. A/A | 0.16 (−0.12–0.44) | 0.25 |
| rs412658 | 0.06 (−0.05–0.17) | 0.32 |
| C/T vs. C/C | −0.03 (−0.20–0.13) | 0.69 |
| T/T vs. C/C | 0.17 (−0.06–0.41) | 0.15 |
| Coefficient (95% CI) | p | Coefficient (95% CI) | p | Coefficient (95% CI) | p | |
|---|---|---|---|---|---|---|
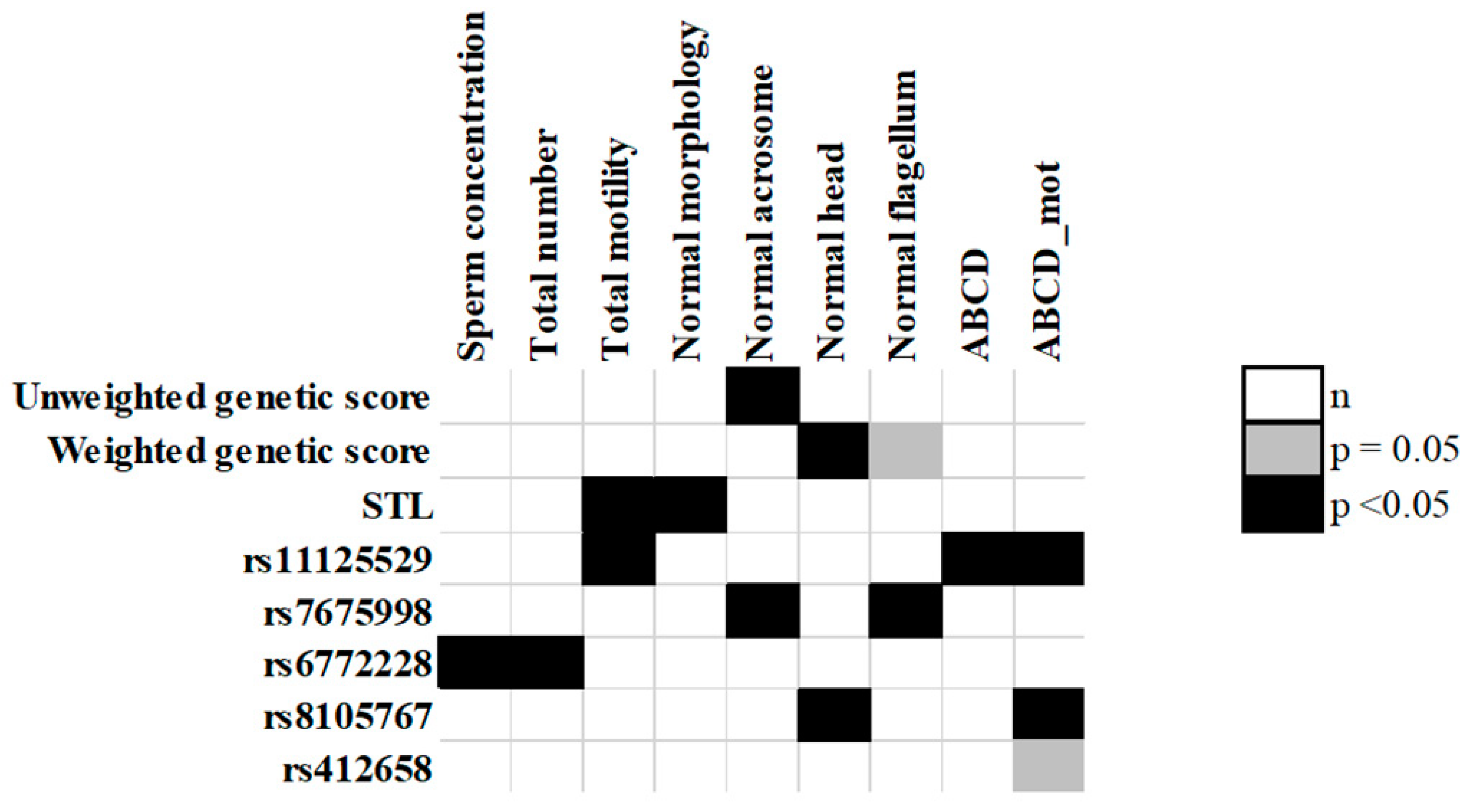
| Unweighted genetic score | normal acrosome | - | - | - | - | - |
| 2nd vs. 1st quintile | −1.26 (−2.88–0.36) | 0.13 | - | - | - | - |
| 3rd vs. 1st quintile | −0.64 (−2.21–0.94) | 0.43 | - | - | - | - |
| 4th vs. 1st quintile | −0.12 (−1.64–1.41) | 0.88 | - | - | - | - |
| 5th vs. 1st quintile | −1.90 (−3.74–−0.05) | 0.04 | - | - | - | - |
| Weighted genetic score | normal head | - | - | - | - | - |
| 2nd vs. 1st quintile | −2.79 (−5.40–−0.17) | 0.04 | - | - | - | - |
| 3rd vs. 1st quintile | −1.22 (−3.90–1.47) | 0.37 | - | - | - | - |
| 4th vs. 1st quintile | −1.28 (−3.88–1.31) | 0.33 | - | - | - | - |
| 5th vs. 1st quintile | −1.78 (−4.44–0.88) | 0.19 | - | - | - | - |
| STL (quintiles) | normal morphology | - | - | - | - | - |
| 2nd vs. 1st quintile | −1.71 (−3.86–0.43) | 0.12 | - | - | - | - |
| 3rd vs. 1st quintile | −1.61 (−3.72–0.50) | 0.14 | - | - | - | - |
| 4th vs. 1st quintile | −2.25 (−4.40–−0.11) | 0.04 | - | - | - | - |
| 5th vs. 1st quintile | −0.81 (−2.89–1.28) | 0.45 | - | - | - | - |
| rs11125529 | total motility | - | ABCD | - | ABCD_mot | - |
| additive model | 4.16 (0.52–7.80) | 0.03 | 0.56 (0.02–1.10) | 0.04 | 0.38 (0.04–0.72) | 0.03 |
| C/A vs. C/C | 4.08 (0.08–8.08) | 0.05 | 0.59 (0.00–1.18) | 0.05 | 0.37 (0.00–0.75) | 0.05 |
| A/A vs. C/C | 9.06 (−7.43–25.55) | 0.28 | 0.86 (−1.60–3.32) | 0.49 | 0.82 (−0.75–2.39) | 0.31 |
| rs7675998 | normal acrosome | - | normal flagellum | - | - | - |
| additive model | 0.86 (−0.02–1.74) | 0.06 | 1.04 (−0.40–2.48) | 0.16 | - | - |
| G/A vs. G/G | 0.46 (−0.66–1.58) | 0.42 | 0.12 (−1.71–1.95) | 0.90 | - | - |
| A/A vs. G/G | 2.54 (0.26–4.81) | 0.03 | 4.00 (0.29–7.72) | 0.04 | - | - |
| rs6772228 | sperm concentration | - | total number | - | - | - |
| additive model | 26.31 (4.52–48.09) | 0.02 | 76.24 (3.25–149.23) | 0.04 | - | - |
| T/A vs. T/T | 26.31 (4.52–48.09) | 0.02 | 76.24 (3.25–149.23) | 0.04 | - | - |
| rs8105767 | normal head | - | ABCD_mot | - | - | - |
| additive model | −0.98 (−2.29–0.34) | 0.14 | −0.08 (−0.30–0.14) | 0.49 | - | - |
| A/G vs. A/A | −2.39 (−4.16–−0.61) | 0.01 | −0.37 (−0.66–−0.07) | 0.01 | - | - |
| G/G vs. A/A | 0.00 (−3.10–3.11) | 1.00 | 0.27 (−0.25–0.79) | 0.31 | - | - |
| ABCD | ABCD_mot | |||
|---|---|---|---|---|
| Coefficient (95% CI) | p | Coefficient (95% CI) | p | |
| rs11125529 | 0.56 (0.02–1.10) | 0.04 | 0.38 (0.04–0.72) | 0.03 |
| C/A vs. C/C | 0.59 (0.00–1.18) | 0.05 | 0.37 (0.00–0.75) | 0.05 |
| A/A vs. C/C | 0.86 (−1.60–3.32) | 0.49 | 0.82 (−0.75–2.39) | 0.31 |
| rs3027234 | 0.03 (−0.34–0.41) | 0.86 | −0.01 (−0.25–0.23) | 0.94 |
| C/T vs. C/C | 0.25 (−0.22–0.73) | 0.30 | 0.17 (−0.13–0.47) | 0.28 |
| T/T vs. C/C | −0.43 (−1.42–0.57) | 0.40 | −0.41 (−1.04–0.22) | 0.20 |
| rs6028466 | −0.2 (−0.86–0.46) | 0.56 | −0.03 (−0.45–0.38) | 0.88 |
| G/A vs. G/G | −0.2 (−0.91–0.50) | 0.58 | −0.03 (−0.48–0.41) | 0.88 |
| A/A vs. G/G | −0.31 (−3.81–3.20) | 0.86 | −0.04 (−2.27–2.19) | 0.97 |
| rs7675998 | 0.04 (−0.32–0.39) | 0.83 | 0.01 (−0.21–0.23) | 0.93 |
| G/A vs. G/G | 0.07 (−0.39–0.53) | 0.77 | 0.04 (−0.25–0.34) | 0.76 |
| A/A vs. G/G | 0.02 (−0.87–0.91) | 0.96 | −0.04 (−0.61–0.52) | 0.88 |
| rs9420907 | −0.25 (−0.65–0.15) | 0.23 | −0.18 (−0.43–0.08) | 0.18 |
| A/C vs. C/C | −0.16 (−0.66–0.33) | 0.52 | −0.07 (−0.39–0.25) | 0.66 |
| A/A vs. C/C | −0.75 (−1.92–0.43) | 0.21 | −0.66 (−1.40–0.09) | 0.09 |
| rs6772228 | 0.83 (−0.33–1.98) | 0.16 | 0.62 (−0.12–1.35) | 0.10 |
| T/A vs. T/T | 0.83 (−0.33–1.98) | 0.16 | 0.62 (−0.12–1.35) | 0.10 |
| rs10936599 | −0.10 (−0.49–0.28) | 0.60 | −0.06 (−0.31–0.18) | 0.60 |
| C/T vs. C/C | −0.27 (−0.74–0.20) | 0.26 | −0.18 (−0.47–0.12) | 0.24 |
| T/T vs. C/C | 0.29 (−0.81–1.39) | 0.60 | 0.21 (−0.48–0.91) | 0.55 |
| rs2736100 | −0.06 (−0.38–0.27) | 0.72 | −0.08 (−0.29–0.12) | 0.44 |
| C/A vs. C/C | −0.08 (−0.60–0.44) | 0.77 | −0.14 (−0.47–0.19) | 0.41 |
| A/A vs. C/C | −0.11 (−0.77–0.54) | 0.74 | −0.15 (−0.56–0.27) | 0.49 |
| rs755017 | 0.43 (−0.15–1.01) | 0.15 | 0.26 (−0.11–0.62) | 0.17 |
| A/G vs. A/A | 0.36 (−0.23–0.95) | 0.23 | 0.19 (−0.19–0.57) | 0.32 |
| G/G vs. A/A | 3.34 (−1.61–8.28) | 0.19 | 3.04 (−0.10–6.18) | 0.06 |
| rs8105767 | 0.00 (−0.35–0.35) | 1.00 | −0.08 (−0.30–0.14) | 0.49 |
| A/G vs. A/A | −0.38 (−0.84–0.08) | 0.11 | −0.37 (−0.66–−0.07) | 0.01 |
| G/G vs. A/A | 0.57 (−0.26–1.40) | 0.18 | 0.27 (−0.25–0.79) | 0.31 |
| rs412658 | −0.10 (−0.43–0.22) | 0.54 | −0.09 (−0.3–0.11) | 0.37 |
| C/T vs. C/C | −0.37 (−0.85–0.10) | 0.12 | −0.30 (−0.60–0.00) | 0.05 |
| T/T vs. C/C | 0.00 (−0.70–0.70) | 1.00 | −0.04 (−0.48–0.40) | 0.86 |
| Sperm Parameter | SNP | M/m | Pos.38 | Gene | Reg.DB | GTEx | R/A | Gene Symbol | p-Value | NES | Tissue |
|---|---|---|---|---|---|---|---|---|---|---|---|
| nH;ABCD_mot | rs8105767 | A/G | 19:22032639 | LOC112268248 | 1f | s/eQTL | A/G | ZNF676 | 1.20 × 10−11 | −0.36 | Testis |
| eQTL | RP11-157B13.7 | 1.70 × 10−8 | −0.2 | Testis | |||||||
| eQTL | ZNF729 | 1.00 × 10−4 | −0.21 | Testis | |||||||
| nA;nF | rs7675998 | G/A | 4:163086668 | None | 6 | eQTL | A/G | RP11-563E2.2 | - | −0.25 | no-testis |
| eQTL | NAF1 | - | [+/-]* | no-testis | |||||||
| sQTL | NAF1 | 1.50 × 10−6 | 0.31 | no-testis | |||||||
| TM; ABCD; ABCD_mot | rs11125529 | C/A | 2:54248729 | ACYP2: Intron Variant | 5 | eQTL | C/A | TSPYL6 | 1.80 × 10−69 | −0.95 | Testis |
| SC, STn | rs6772228 | T/A | 3:58390292 | PXK: Intron Variant | 7 | sQTL | T/A | PXK | 1.70 × 10−16 | 1.40 | Testis |
| eQTL | FAM107A | no-testis | |||||||||
| eQTL | PDHB | no-testis | |||||||||
| eQTL | RP11-359I18.5 | no-testis | |||||||||
| eQTL | RPP14 | no-testis | |||||||||
| STL | rs10936599 | C/T | 3:169774313 | MYNN: Missense Variant | 3a | eQTL | C/T | MYNN | 2.10 × 10−11 | 0.35 | Testis |
| LRRIQ4 | 1.90 × 10−6 | 0.19 | Testis | ||||||||
| STL | rs2736100 | C/A | 5:1286401 | TERT: Intron Variant | 5 | eQTL | C/A | TERT | 4.80 × 10−5 | 0.23 | no-testis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gentiluomo, M.; Luddi, A.; Cingolani, A.; Fornili, M.; Governini, L.; Lucenteforte, E.; Baglietto, L.; Piomboni, P.; Campa, D. Telomere Length and Male Fertility. Int. J. Mol. Sci. 2021, 22, 3959. https://doi.org/10.3390/ijms22083959
Gentiluomo M, Luddi A, Cingolani A, Fornili M, Governini L, Lucenteforte E, Baglietto L, Piomboni P, Campa D. Telomere Length and Male Fertility. International Journal of Molecular Sciences. 2021; 22(8):3959. https://doi.org/10.3390/ijms22083959
Chicago/Turabian StyleGentiluomo, Manuel, Alice Luddi, Annapaola Cingolani, Marco Fornili, Laura Governini, Ersilia Lucenteforte, Laura Baglietto, Paola Piomboni, and Daniele Campa. 2021. "Telomere Length and Male Fertility" International Journal of Molecular Sciences 22, no. 8: 3959. https://doi.org/10.3390/ijms22083959






