Abstract
The short-chain dehydrogenases/reductases (SDR) superfamily is involved in multiple physiological processes. In this study, genome-wide identification and comprehensive analysis of SDR superfamily were carried out in 29 animal species based on the latest genome databases. Overall, the number of SDR genes in animals increased with whole genome duplication (WGD), suggesting the expansion of SDRs during evolution, especially in 3R-WGD and polyploidization of teleosts. Phylogenetic analysis indicated that vertebrates SDRs were clustered into five categories: classical, extended, undefined, atypical, and complex. Moreover, tandem duplication of hpgd-a, rdh8b and dhrs13 was observed in teleosts analyzed. Additionally, tandem duplications of dhrs11-a, dhrs7a, hsd11b1b, and cbr1-a were observed in all cichlids analyzed, and tandem duplication of rdh10-b was observed in tilapiines. Transcriptome analysis of adult fish revealed that 93 SDRs were expressed in more than one tissue and 5 in one tissue only. Transcriptome analysis of gonads from different developmental stages showed that expression of 17 SDRs were sexually dimorphic with 11 higher in ovary and 6 higher in testis. The sexually dimorphic expressions of these SDRs were confirmed by in situ hybridization (ISH) and qPCR, indicating their possible roles in steroidogenesis and gonadal differentiation. Taken together, the identification and the expression data obtained in this study contribute to a better understanding of SDR superfamily evolution and functions in teleosts.
1. Introduction
Short-chain dehydrogenases/reductases (SDR) superfamily comprises NAD(P)(H)-dependent oxidoreductases that possess the conserved Rossmann-fold motif. They are a large and ancient gene family found in organisms as diverse as viruses and vertebrates [1].With the rapid release of genome databases for numerous species, the size of the SDR superfamily has grown considerably since the initial family members were characterized in the late 1970′s [2,3,4,5]. Genome-wide identification of SDRs was performed in cyanobacteria [6], nematode and fruit fly [7], rat, mouse, and human [5,8,9], and plants [10]. Generally, SDRs are divided into seven different categories termed “classical”, “extended”, “intermediate”, “divergent”, “complex”, “atypical”, and “undefined” according to their different cofactor binding sites, substrate binding sites, functions, and sequence structures [1]. However, understanding the evolutionary relationships among the SDR superfamily genes is difficult because most family members share only 20–30% amino acid sequence similarity.
The diversification and the morphological innovations of vertebrates are attributed to large-scale gene or genome duplications at the origin of the group. These duplications are predicted to have occurred in two rounds of genome duplication, the “2R” hypothesis, or in one genome duplication plus many segmental duplications, although these hypotheses are still somewhat controversial [11]. In addition, teleost fish underwent three round of genome duplication (3R-WGD) [12]. Some teleosts such as rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar) underwent an additional round of genome duplication (4R) [13,14]. Therefore, teleost fish represent a unique model for studies on evolution. Although fish represent the largest and the most diverse group of vertebrates, comprehensive identification and genome-wide study on the SDR superfamily are still lacking. The available genome sequences of more and more species provided new resources to understand the evolution of SDR superfamily. Recently, genome sequences were released for elephant shark (Callorhinchus milii, a chondrichthyan) [15], coelacanth (Latimeria menadoensis, an early sarcopterygian) [16], and spotted gar (Lepisosteus productus, a non-teleost actinopterygian) [17], which represent the key nodes for studying WGD in vertebrates. Comparative genomic analysis and gene mapping of SDR superfamily in vertebrates provide insights into the evolution of this gene family after each round of WGDs.
Nowadays, identification and characterization of some subfamily of SDR superfamily are reported in several vertebrates. Genome wide identification and expression analysis of beta-hydroxysteroid dehydrogenase genes were reported in orange-spotted groupers (Epinephelus coioides) [18]. Upregulation and downregulation of beta-hydroxysteroid dehydrogenase genes are found in both brain and gonads during sex reversal, indicating their critical roles in neurogenesis of brain and sexual maintenance in orange spotted groupers [19]. In humans, at least 14 separate enzymes that all display catalytic activities towards 17β-hydroxy and keto-steroid substrates were identified [20,21]. In olive flounder (Paralichthys olivaceus), eight hsd17b family genes, including hsd17b3, -4, -7, -8, -10, -12a, -12b, and -14, were identified. Flounder hsd17b10, -12a, and -12b are highly expressed in the ovary, while hsd17b3 is dominantly expressed in the testis. In mammalian species, 11β-hydroxysteroid dehydrogenases (HSD11b1 and HSD11b2) catalyze the interconversion between active and inactive glucocorticoid [21]. In contrast, fish hsd11b1 proved to be a potent enzyme for the biosynthesis of 11-ketotestosterone (11-KT) from testosterone in testis [22]. Previous study revealed that dhrs11 can act as an NADPH-dependent hsd17b and play an important role in the metabolism of 11-oxygenated-C19-steroids as well as estrogens, androgens, and androgen precursors [23,24]. Rdh is another well-defined large subfamily of SDR which is involved in the first step of retinoic acid (RA) synthesis from vitamin A by catalyzing the oxidation of retinol to retinaldehyde [25]. However, these studies have thus far been restricted to expression and function data for a single gene or a subcluster of SDR superfamily in limited tissues or at limited stages of development [24,26]. The overall expression profiles of the SDRs in different tissues and at different stages of gonadal development are largely unknown in vertebrates, especially in teleosts.
The Nile tilapia (Oreochromis niloticus) is a commercially important farmed fish species in aquaculture worldwide. It is also a good model for study of sex determination and differentiation for the availability of genome sequences, mono-sex fry, and transcriptome data from different tissues [27] and gonads from different developmental stages [28,29]. Given the versatility and the fundamental importance of SDRs in lipid, amino acid, carbohydrate, hormone, and xenobiotic metabolism as well as in redox sensor mechanisms, we carried out an overall identification and comprehensive analysis of the chromosome location, phylogeny, synteny, and spatiotemporal expression profiles in Nile tilapia. Our results provide new insights into the evolution of SDRs and reveal their potential roles in vertebrates.
2. Results
2.1. Identification of SDRs from Nile tilapia and Representative Species
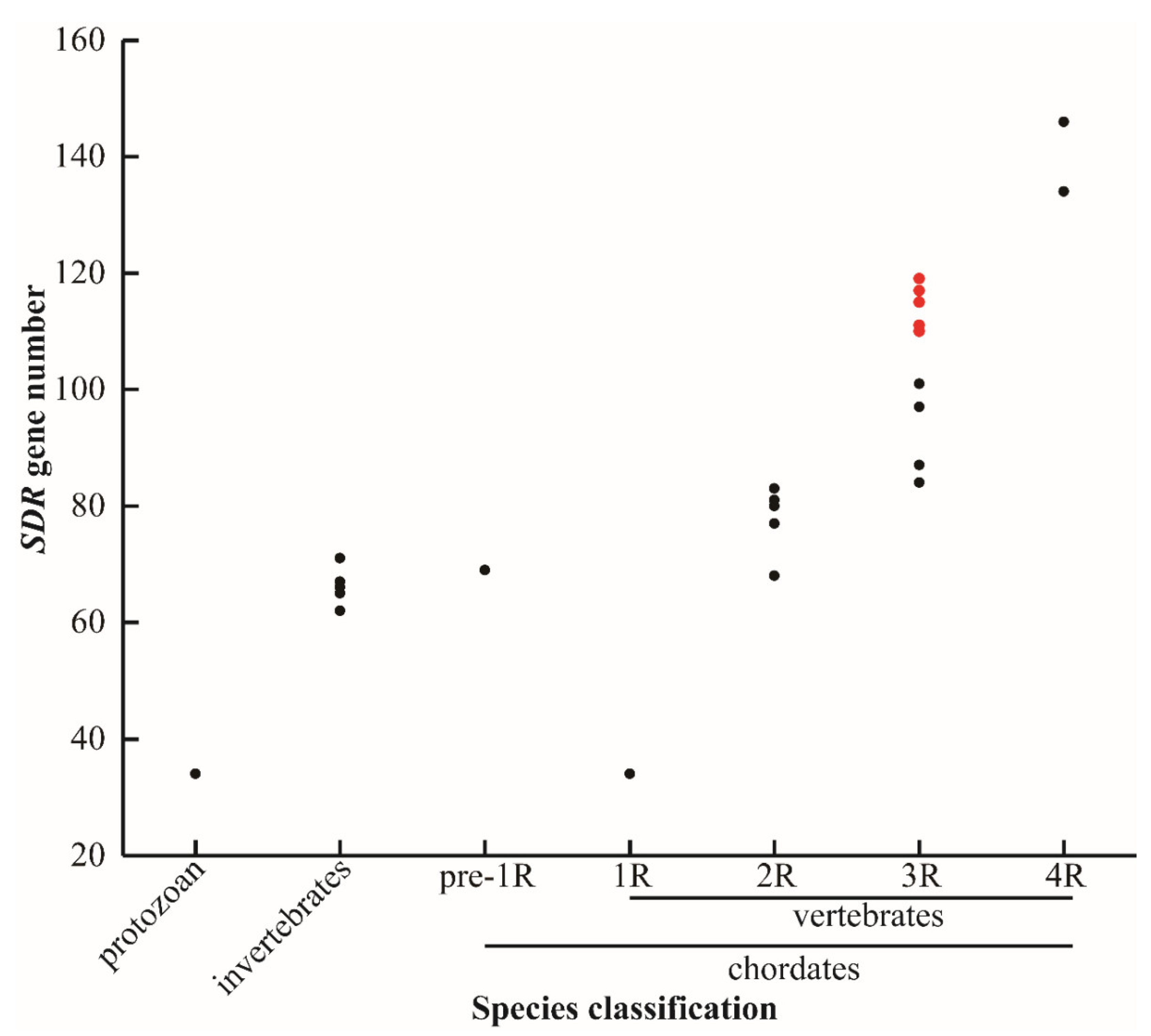
In total, 119 SDRs were identified in the Nile tilapia genome. Of them, nine SDRs including bdh1, c-factor, hsd17b7, rdh12, rdh13, blvrb, ak7, dhrsx, and hsd11b1 had two copies. In addition, some SDRs had multiple copies, for example, three replicates for hsd17b12, dhrs13, rdh14, cbr1, dhrs12, and rdh10, four replicates for rdh8, rdh12, and hpgd, and five replicates for dhrs7. Interestingly, 14 replicates of dhrs11 were isolated from Nile tilapia. Additionally, SDRs were isolated from other 28 animal species with different numbers of WGD (Figure 1). The sequences of SDRs in blue tilapia, Mozambique tilapia, and blackchin tilapia were identified from their genomes that were sequenced by our lab and are shown in Supplementary file 1. The SDR gene name, the accession number, and the chromosome location for all other species are listed in Tables S1–S26. The total number of SDRs changed remarkably in species that experienced 3R- and 4R-WGD, while it remained basically constant in protozoan, invertebrates, and species that experienced 1R- and 2R-WGD. It is noteworthy that there is a great difference in SDR numbers between cichlids and other teleost species.
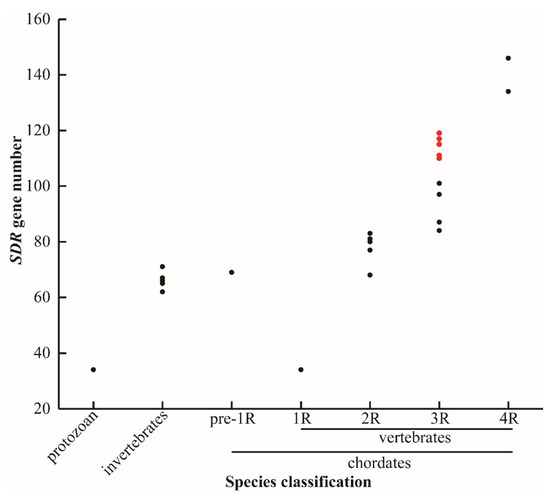
Figure 1.
SDR gene numbers in different species. Protozoan represents unicellular animals: paramecium (34). Invertebrates: fruit fly (62), domestic silkworm (65), nematode (66), sponge (67), and black tiger shrimp (71). Pre-1R species: vase tunicate (69). 1R species: lamprey (34). 2R species: elephant shark (68), human (77), spotted gar (80), coelacanth (80), tropical clawed frog (81), chicken (83), and python (83). 3R species: channel catfish (84), zebrafish (87), medaka (97), fugu (97), large yellow croaker (101), zebra mbuna (114), Flier cichlid (111), Eastern happy (117), blue tilapia (110), Mozambique tilapia (114), blackchin tilapia (115), and Nile tilapia (119). 4R species: common carp (134) and rainbow trout (146). Red dots represent seven cichlids. 1R, 2R, 3R, and 4R represent four rounds of whole genome duplication (WGD). The identified SDRs in lamprey is extraordinarily low, probably due to poor genome assembly.
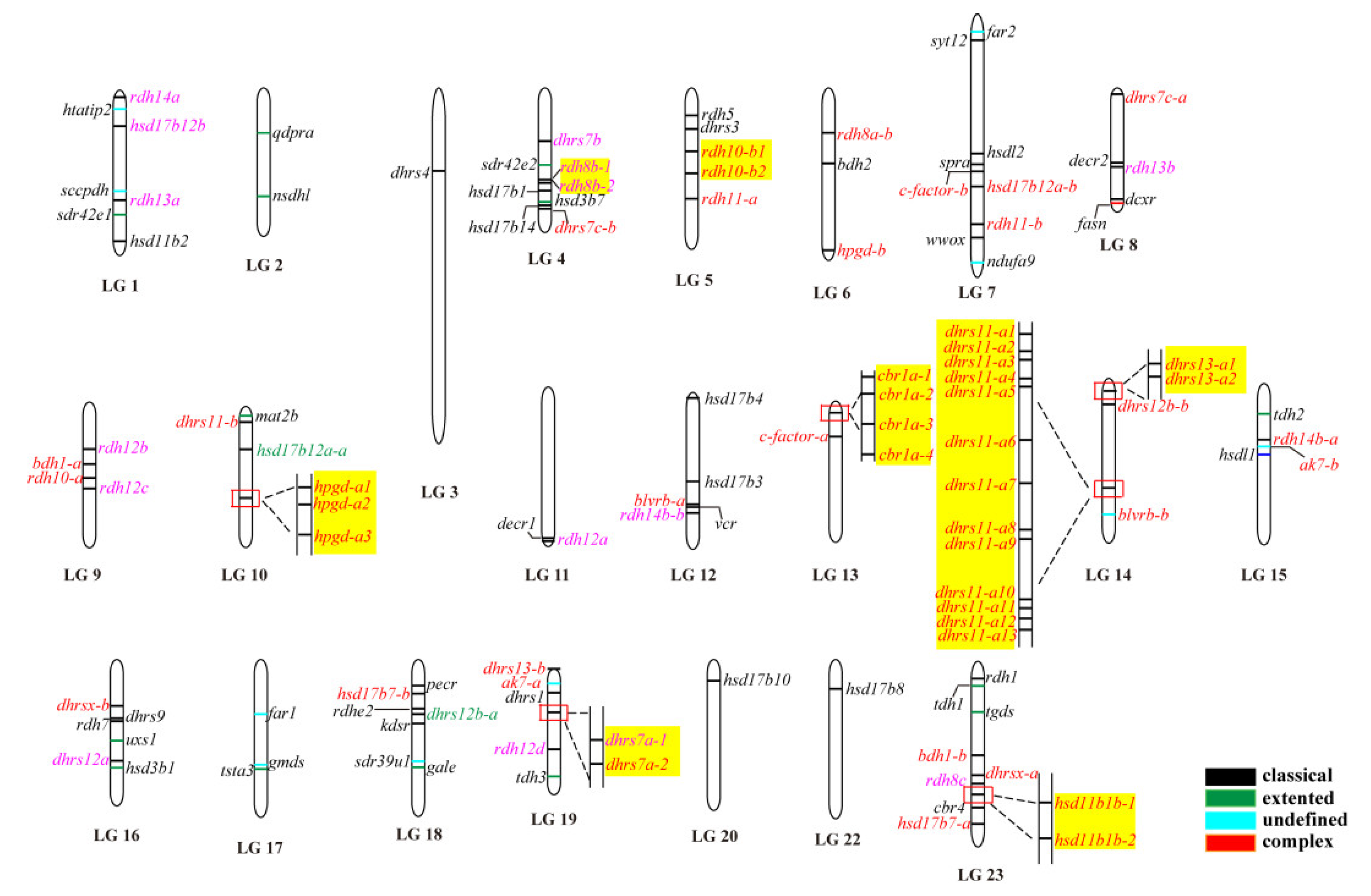
The 119 SDRs were unevenly distributed across all of the 22 linkage groups of Nile tilapia. For instance, there were 17 SDRs located on LG14, while there was only one on LG3, LG20, and LG22. Eight SDRs genes (rdh8, rdh10, hpgd, cbr1, dhrs7, dhrs11, dhrs13, and hsd11b1) had multiple copies located together in tandem arrays. These tandemly arrayed genes were on different chromosomes, except dhrs11 and dhrs13, which were located on LG14 (Figure 2). In total, eight SDRs (dhrs7a, dhrs11-a, dhrs13-a, rdh8b, rdh10-b, hpgd-b, cbr1a, and hsd11b1b) in Nile tilapia went through tandem duplication, which resulted in 2, 13, 2, 2, 2, 3, 4, and 2 copies, distributed on LG19, LG14, LG14, LG4, LG5, LG10, LG13, and LG23, respectively.
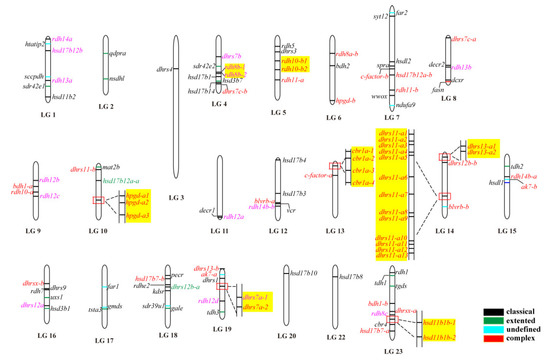
Figure 2.
Chromosomal maps of the SDRs in Nile tilapia. SDRs were depicted on linkage group (LG) of the genome. LG21 is absent because it was combined with LG16. Black, green, blue, and red horizontal lines indicate classical, extended, atypical, and complex SDRs, respectively. Gene names in pink and red indicate genes derived from 2R and 3R events, respectively, whereas gene names in green represent it undergoing both 2R and 3R events. Gene name in black is a single copy gene. Tandem duplicates are highlighted in a yellow shade.
2.2. Phylogenetic and Syntenic Analyses of SDRs
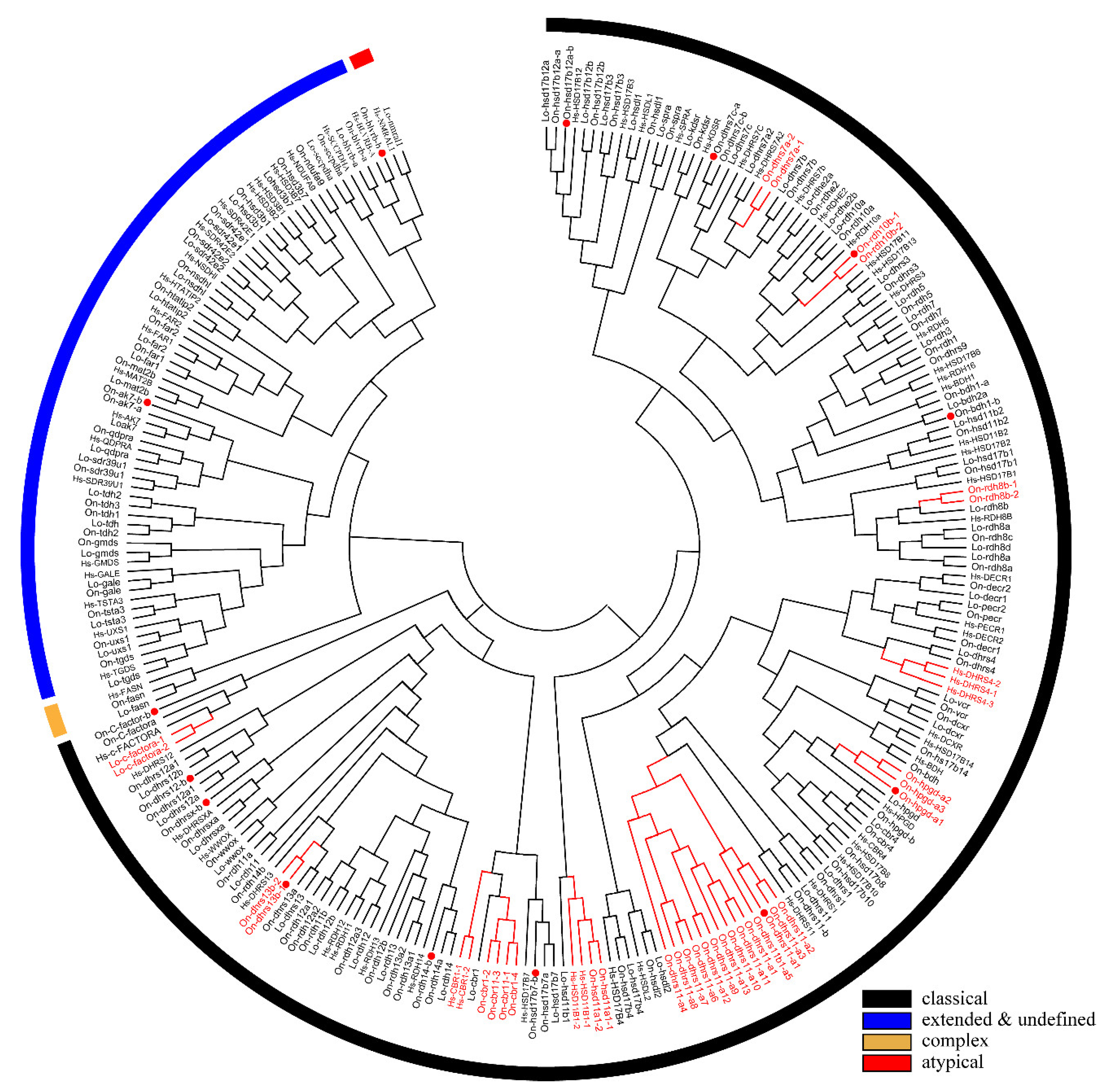
A phylogenetic tree was constructed using the conserved domain of amino acid sequences of all SDRs from Nile tilapia, spotted gar, and human. The SDRs were divided into five categories termed “classical”, “undefined”, “extended”, “complex”, and “atypical”. Most of the members belonged to the “classical” category. Only one member of the “complex” category was observed in human, spotted gar, and Nile tilapia, respectively. One member of “atypical” category was observed in human and spotted gar but none in Nile tilapia. “Classical”, “atypical”, and “complex” SDRs were clustered into different clades, while “extended” and “undefined” SDRs were clustered in one clade due to their high sequence similarity (Figure 3). “Intermediate” and “divergent” genes were not identified in the study. Among the 119 SDR members of Nile tilapia, 114 genes could be designated into 49 subfamilies according to previous publications [5]. Compared with human, Nile tilapia has two additional subfamilies, SDR112c and SDR348c, but lacks SDR48a. Subfamily designations of SDR genes in Nile tilapia, spotted gar and human are listed in Table S27.
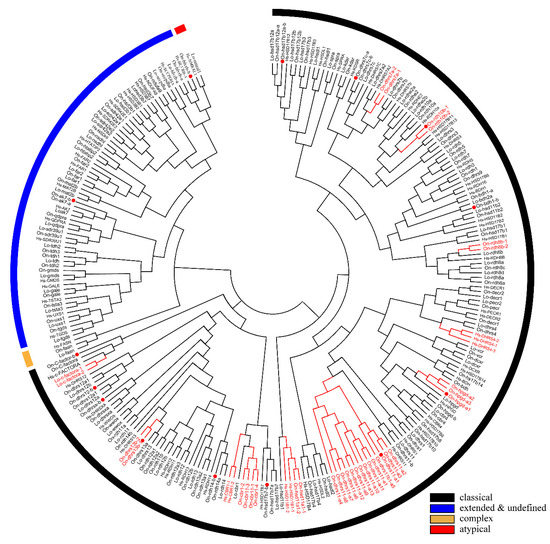
Figure 3.
Phylogenetic tree of Nile tilapia, human, and spotted gar SDR family. The NJ method was used to construct the tree by MEGA 6.0. The multiple alignment software Bioedit was used to align the amino acid sequences of the conserved domain. On, Oreochromis niloticus; Lo, Lepisosteus oculatus; Hs, Homo sapiens. Three different colored arcs on the outside represent different categories of SDRs. The red lines and the gene names represent genes with tandem duplications. The red dots represent genes stemmed from 3R. Two copies of cbr1 were observed in zebrafish and channel catfish and one copy in Nile tilapia, therefore, the 3R-event of this gene was presented in Figure S2. GenBank accession numbers of each sequence used are listed in Tables S4, S12, and S15.
Phylogenetic analyses revealed that duplicates of 15 SDRs (bdh1, c-factor, cbr1, dhrsx, hsd17b7, rdh12, blvrb, dhrs11, dhrs13, dhrs12b, dhrs7c, rdh14b, hsd17b12a, hpgd, and ak7) were retained after the 3R-WGD events (Figure 3). Further phylogenetic analyses revealed that duplicates of four SDRs (dhrs12, hsd11b1, rdh14, and hsd17b12) were retained after the 2R-WGD (Figure S1). cbr1 retained two copies in zebrafish and channel catfish, suggesting one of them was retained after 3R-WGD, and rdh8 had multiple copies in lower species they retained since early replication (Figure S2). In addition, phylogenetic trees of six genes (bdh1, c-factor, dhrsx, hsd17b7, blvrb, and ak7), which retained two copies and had no tandem duplicates, were constructed (Figure S3). Moreover, eight genes (dhrs11, rdh8, rdh10, hpgd, cbr1, dhrs7, hsd11b1, and dhrs13) were tandem duplicated in different species, which resulted in multiple copies on the same chromosome (Figure 3 and Figure 4, and Figure S4). Among these genes, hpgd-a, rdh8b, and dhrs13-b were tandem duplicated in teleosts, dhrs11-a, dhrs7a, hsd11b1-b, and cbr1-a were tandem duplicated in cichlids, and rdh10-b was tandem duplicated in tilapiines. A phylogenetic tree of dhrs11 in vertebrates demonstrated a duplication of dhrs11 corresponding to the teleost-specific 3R-WGD, and 8 to 14 tandem duplicates of dhrs11-a were observed in cichlids. Interestingly, two copies of dhrs13 were observed on adjacent positions of the same chromosome in all bony fishes, indicating that tandem duplication of dhrs13 occurred in their ancestor. In addition, species specific tandem duplication of some SDR genes was observed in some vertebrates, such as hsd11b1b in elephant shark and coelacanth (Figure S1, and cbr1-a in human (Figure S2).
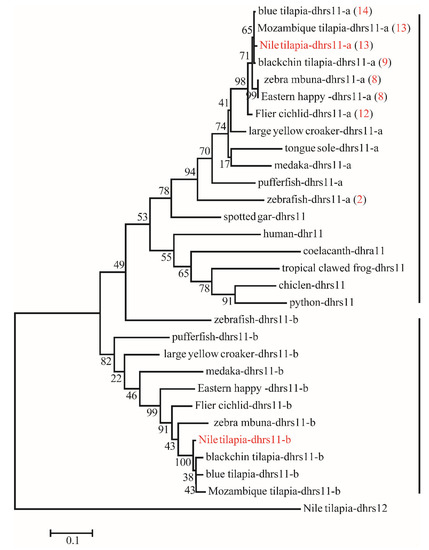
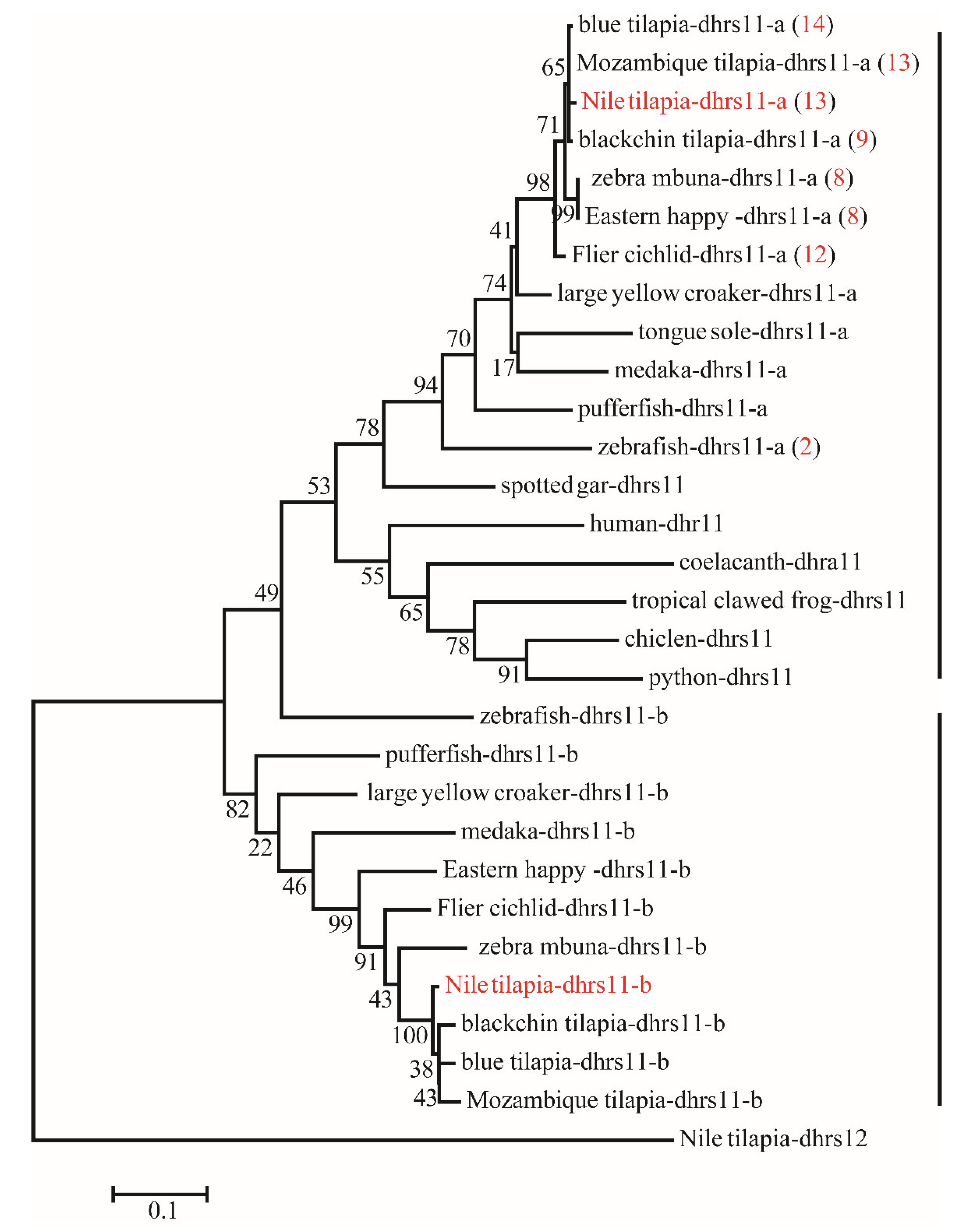
Figure 4.
Phylogenetic tree of dhrs11 in vertebrates. The ML tree was constructed using the amino acid sequences from different species using Nile tilapia dhrs12 as the outgroup. All sequences of dhrs11 were clustered into two branches termed dhrs11-a and dhrs11-b. The red numbers in brackets represent the number of tandem duplicates in corresponding species. Numbers at the branch of the phylogenetic tree stand for bootstrap.
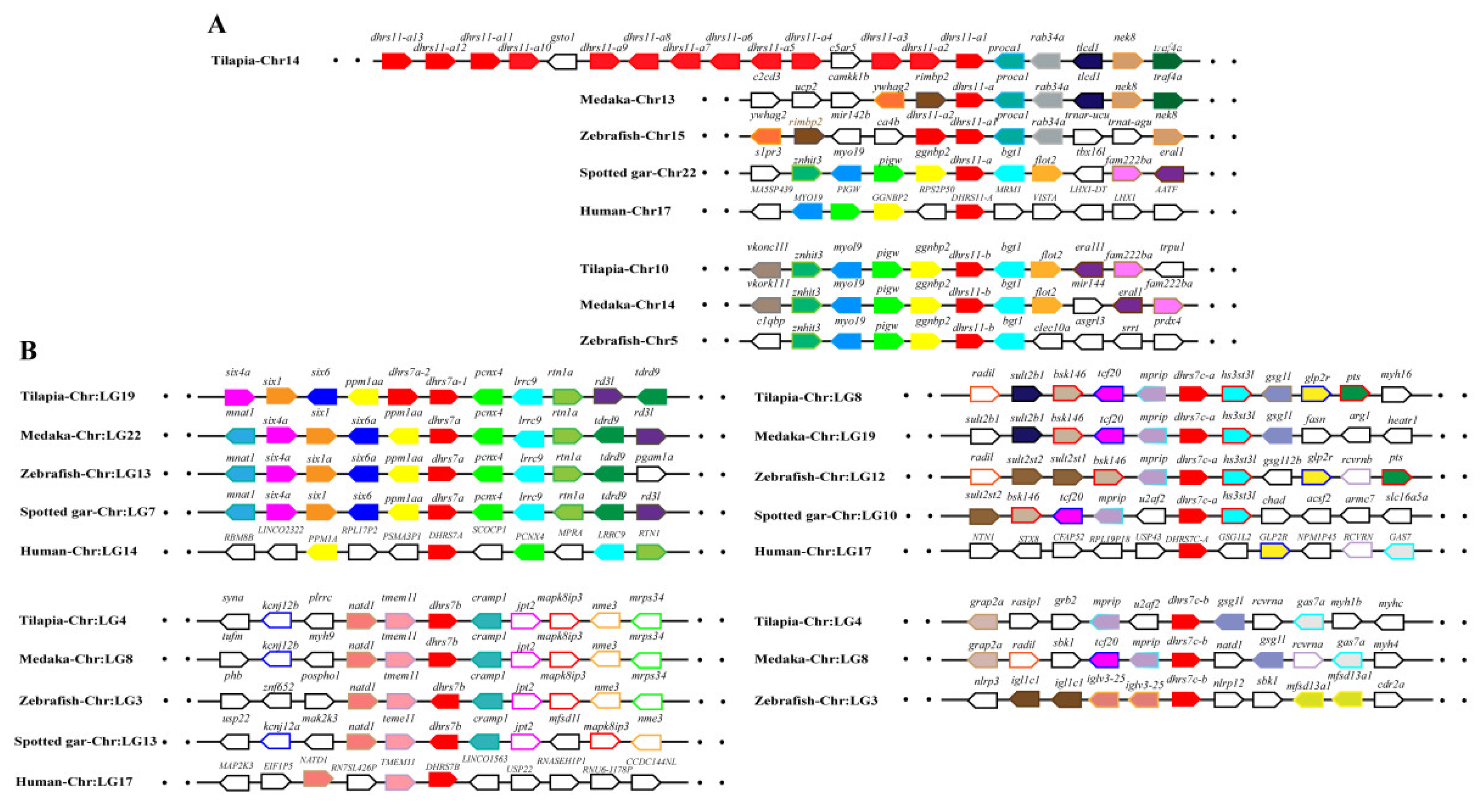
Synteny analysis was performed for duplicated SDR genes to further conform their origin. Duplicates of dhrs12, rdh14, hsd11b1, and hsd17b12 were derived from 2R-WGD, as the synteny was shared in vertebrates (Figure S5), while duplicates of bdh1, c-factor, cbr1, dhrsx, hsd17b7, rdh12, blvrb, dhrs11, dhrs13, dhrs12b, dhrs7c, rdh14b, hsd17b12a, hpgd, and ak7 were derived from 3R-WGD, as the synteny was conserved only in teleosts (Figure 5, Figures S5 and S6). In addition, tandem duplication was observed for dhrs7, dhrs11, dhrs13, hpgd, hsd11b1, and rdh10. Synteny analysis also revealed tandem duplication for cbr1 and rdh8 (Figure S7). Conserved synteny of dhrs11-b and its upstream genes (myo19, pigw, and ggnbp2) and downstream genes (big2, fam22ba, and cra11) were observed in Nile tilapia, zebrafish, and medaka. Similarly, conserved synteny of dhrs11-a and its upstream genes (tlcd1 and traf4a) and its downstream genes (ywhag2, nek8, rimbp2, procal) were observed in Nile tilapia, zebrafish, and medaka but not in non-teleost vertebrates. Interestingly, 14 copies of dhrs11 genes were found in Nile tilapia, while there were only two copies in most teleosts and three copies in zebrafish (Figure 5A). Conserved synteny of three dhrs7, named as dhrs7a, dhrs7b, and dhrs7c, was observed throughout vertebrates, and dhrs7c was further duplicated in teleosts, as reflected by conserved synteny of dhrs7c-a and dhrs7c–b with their adjacent genes. In contrast, tandem duplication of dhrs7a (dhrs7a-1 and dhrs7a-2) was found in Nile tilapia (Figure 5B). These synteny analyses provide strong evidence for determining the origin of different genes.
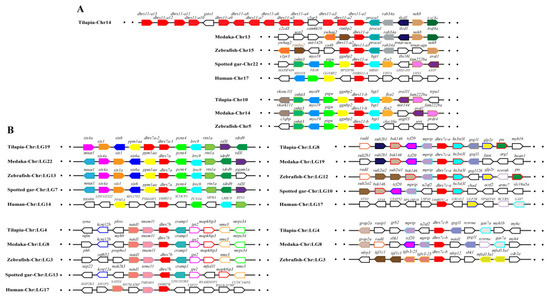
Figure 5.
Syntenic analysis of dhrs11 (A) and dhrs7 (B) and their adjacent genes in Nile tilapia and other vertebrates. Rectangles represent genes in chromosome/scaffold. Dots represent omitted genes of the chromosome/scaffold. The direction of the arrows indicates the gene orientation. The SDRs are shown in red, while the other genes are shown in different colors.
2.3. Tissue Distribution and Ontogenic Expression of SDRs in Nile Tilapia Gonads
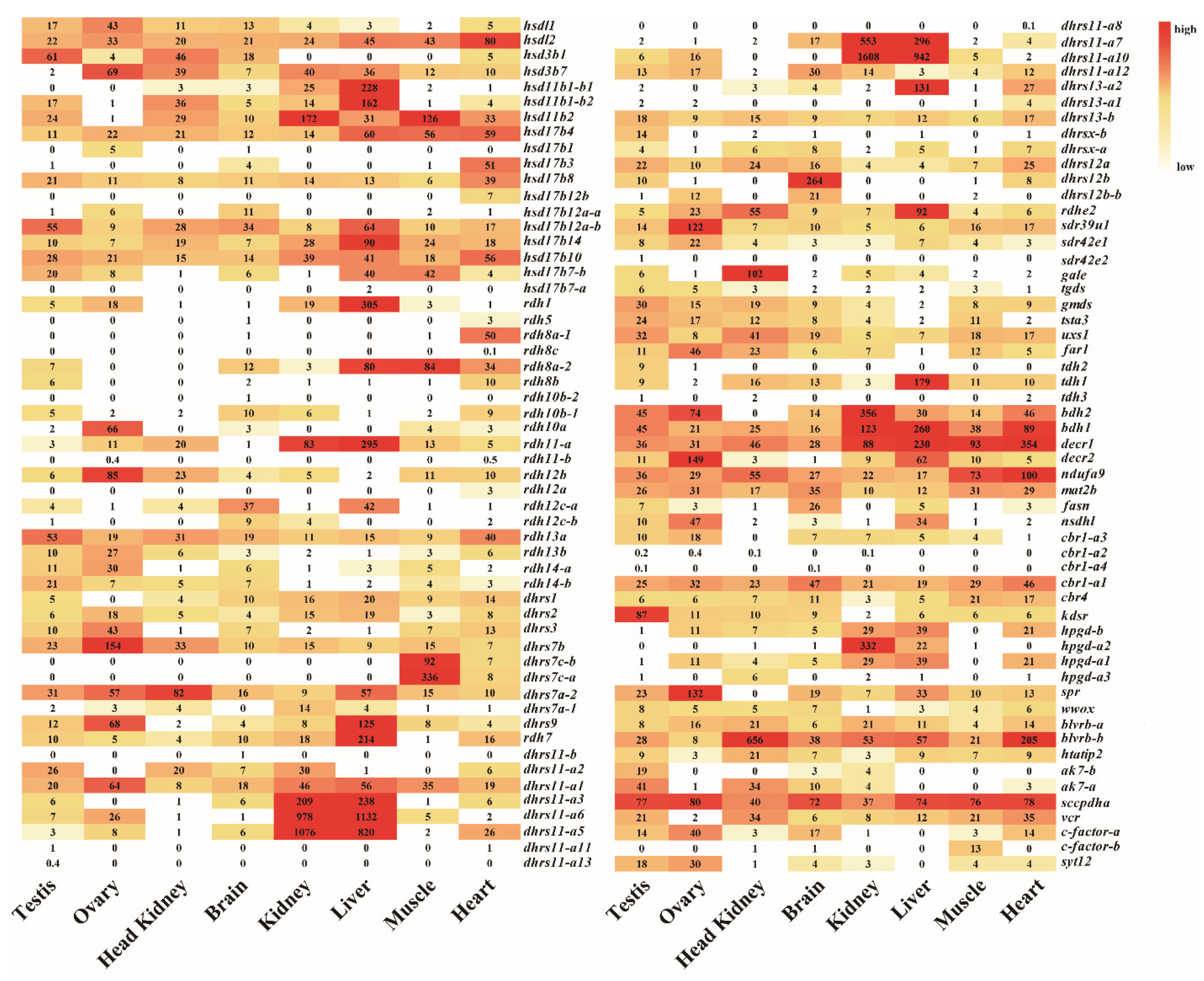
Transcriptome data from eight adult tissues and gonads from four developmental stages of Nile tilapia allowed us to analyze the spatial and the temporal expression profiles of SDRs (Figure 6). There were 92 SDRs expressed in at least one tissue with 85 genes in the brain, 88 genes in the heart, 74 genes in the kidney, 73 genes in the liver, 85 genes in the testis, 73 genes in the ovary, 78 genes in the muscle, and 68 genes in the head kidney. In total, 35 genes were ubiquitously expressed in all 8 tissues, while 18 genes were expressed at background levels. It is noteworthy that dhrs12a, decr1, dhrs11-a10, dhrs11-a6, kdsr, decr2, dhrs7c-a, and blverb-b showed the highest expression in brain, heart, kidney, liver, testicle, ovary, muscle, and head kidney, respectively. The two dhrs7c (dhrs7c-a and dhrs7c-b) were specifically expressed in the muscle, while gale was expressed only in the head kidney. Moreover, ten SDRs (rdhe2, hsd11b1a-1, hsd11b1b-2, hsd17b14, rdh1, dhrs9, rdh11a, rdh7-a, dhrs13-b2, and tdh1) were predominantly expressed in the liver. Importantly, hsd3b1 and kdsr were exclusively expressed in the testis, while hsd3b7, rdh12b, decr2, dhrs7b, sdr39u1, and spra were dominantly expressed in the ovary. Seven copies of the 13 dhrs11-a were highly expressed in both liver and kidney.
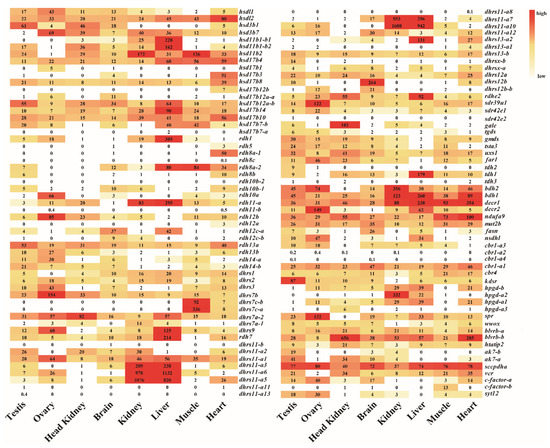
Figure 6.
The expression profiles (RPKM) of SDRs in eight tissues based on transcriptome data in Nile tilapia. Color gradients indicate differences in expression levels. Each row represents a different gene, and each column represents an independent tissue sample. The widespread complex expression patterns in all tissues were readily discernable.
Analysis of the gonadal transcriptomic data from four critical development stages revealed that 95 SDRs were expressed in Nile tilapia gonads according to the threshold we set, while 56 SDRs were highly expressed (total RPKM > 100). The expression of SDRs (RPKM) in Nile tilapia gonads at four developmental stages was listed Table S28. Based on the transcriptome data, 17 genes displayed high and sexually dimorphic expression in adult gonads. The expression levels of 11 ovary-enriched genes (hsd3b7, rdh10a, rdh12b, dhrs3, dhrs7b, dhrs9 sdr39u1, hpgd-b2, far1, spra, decr2) were found to peak at 90 dah, followed by 180, 30, and 5 dah. Expression levels of six testis-enriched genes (hsd3b1, hsd11b1-b2, hsd11b2, rdh14b, kdsr, and ak7-a) displayed similar expression profiles as the ovary-enriched genes but with higher expression in XY than in XX gonads (Figure S8).
2.4. Validation of the Transcriptome Expression Profile by ISH and qPCR
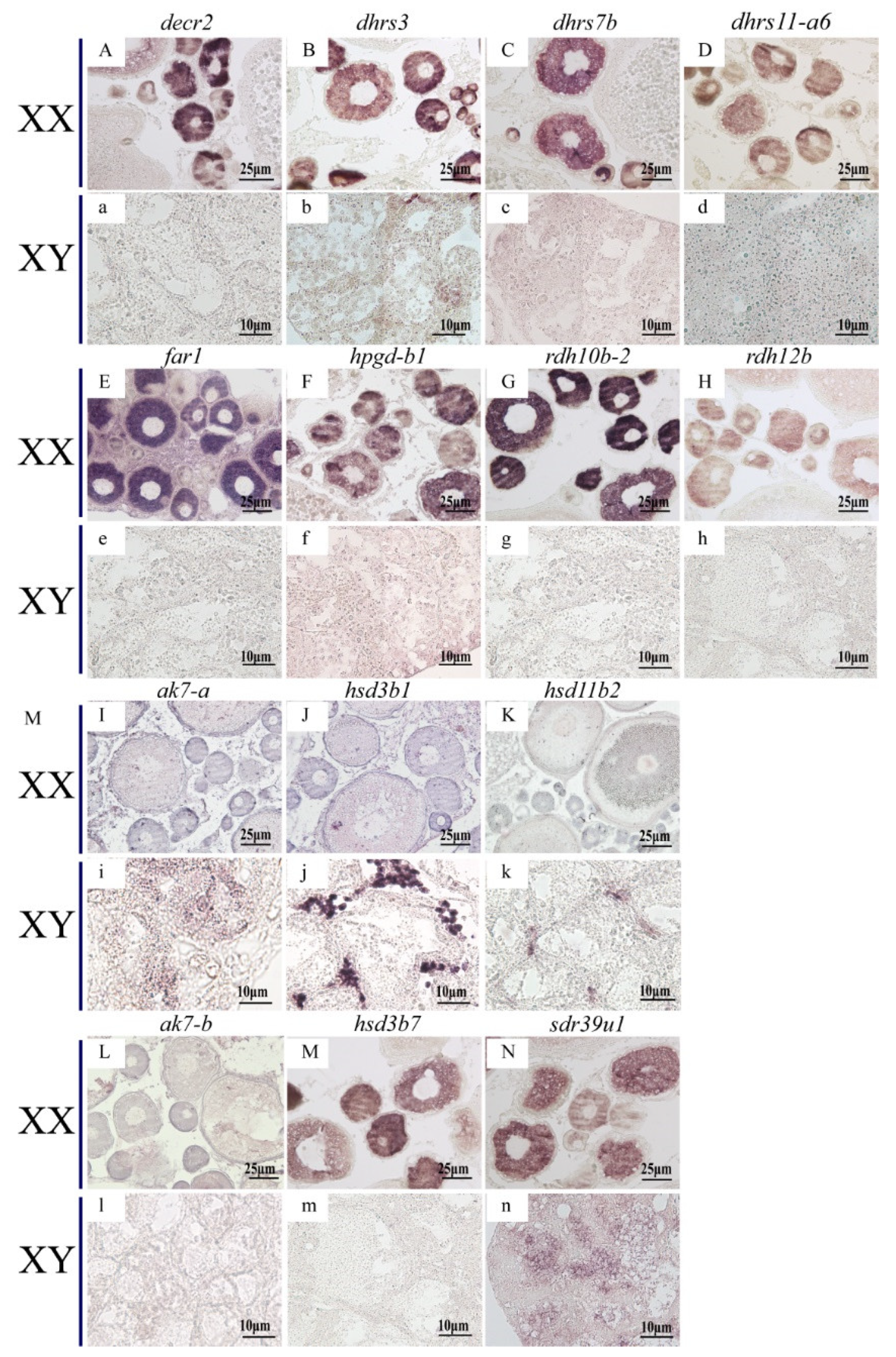
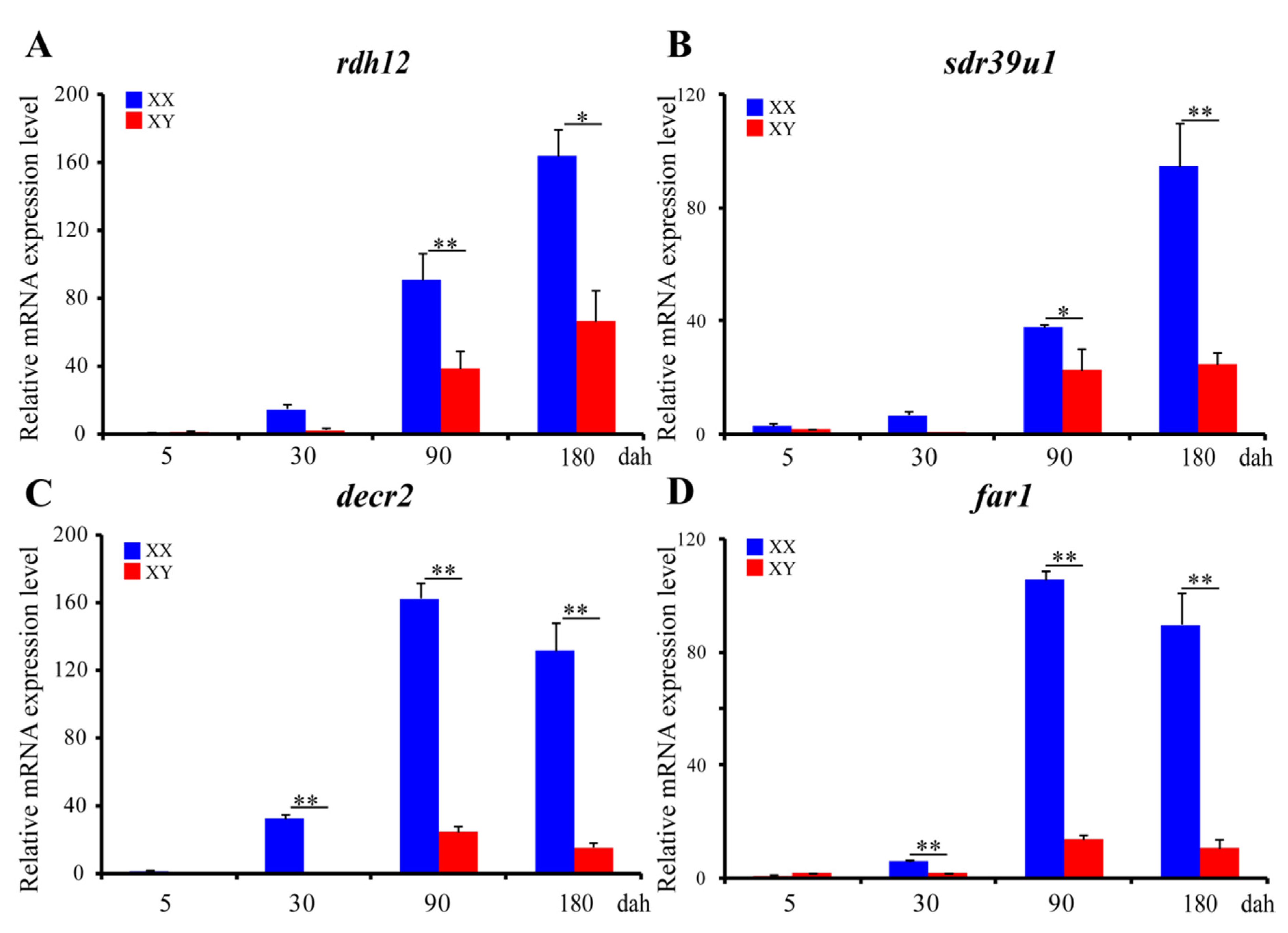
ISH was performed to detect the cellular localization of the selected 14 SDRs (decr2, dhrs3, dhrs7b, dhrs11-a6, far1, hpgd-b1, hsd3b1, ak7-a, ak7-b, hsd3b7, hsd11b2, rdh10b-2, rdh12b, sdr39u1) in gonads of Nile tilapia at 180 dah. Nine (decr2, dhrs3, dhrs7b, dhrs11-a6, far1, hpgd-b1, rdh10b-2, rdh12b, and hsd3b7) of them were expressed dominantly in the cytoplasm of phase I and II oocytes in the ovary. On the contrary, hsd3b1 and hsd11b2 were expressed in the Leydig cells, and ak7-a was expressed in somatic cells of the testis. Expression of sdr39u1 was detected in both the spermatocytes of the testis and the cytoplasm of phase II oocytes in the ovary (Figure 7). Additionally, qPCR was performed to validate the transcriptome data of rdh12b, sdr39u1, far1, and decr2. Consistently, very low or background expression level was detected for the four genes at 5 dah. The expressions of rdh12b and sdr39u1 displayed no significant differences between XX and XY gonad at 30 dah, but expressions of both genes were significantly higher in XX ovary than in XY testis at 90 and 180 dah, while the expressions of decr2 and far1 were significantly higher in XX ovary than in XY testis at 30, 90, and 180 dah (Figure 8). Primers used for ISH and qPCR in this study are listed in Table S29.
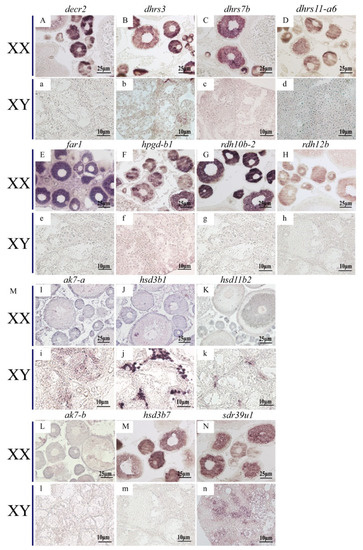
Figure 7.
Cellular localization of 14 SDRs in the Nile tilapia gonads at 180 dah by in situ hybridization. Nine of them were expressed exclusively in the cytoplasm of phase I and II oocytes (A–H,M), but not expressed in testis (a–h,m). ak7-b was not expressed in neither ovary nor testis (L,l). sdr39u1 was expressed in phase II oocytes of ovary (N) and spermatocytes of testis (n). The other three SDRs were expressed in the Leydig cells of testis exclusively (i–k), but not expressed in ovary (I–K).
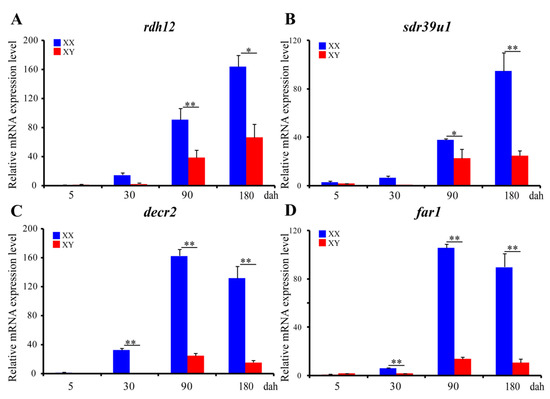
Figure 8.
Ontogeny expression of rdh12 (A), sdr39u1 (B), decr2 (C), and far1 (D) in gonads at 5, 30, 90, and 180 dah analyzed by qPCR (n = 3). eef1a1a was used as internal control. Data were expressed as the mean ± SD. “*” and “**” above the error bar indicate statistically significant differences between XX and XY gonad at p < 0.05 and p< 0.01 by Student’s t-test, respectively. All examined genes displayed similar expression profiles to those from the transcriptome data.
3. Discussion
SDR enzymes play critical roles in various physiological and metabolic processes from archaea and bacteria to eukaryotes. However, the genome-wide evolution, expression, and function of the SDR superfamily in the animal kingdom, especially in teleosts, has yet to be elucidated. In the present study, we identified and performed phylogenetic and syntenic analyses of SDRs from the genomes of representative vertebrates and invertebrates. We then quantified the expression of Nile tilapia SDRs in different adult tissues and gonads from four critical development stages.
3.1. Identification, Phylogenetic, and Syntenic Analyses of SDRs
Studies on SDR superfamily mainly focused on the molecular structure, the substrates, and the catalytic property of a single gene or different subclusters. Thus far, genome wide identification of SDR superfamily has been done in a few species, including cyanobacteria [6], nematode and fruit fly [7], plants [10], and humans [30]. Overall identification and expression analyses of SDRs members are of great importance to define their diverse biological functions. Fish, being the largest group of vertebrates with more than 30,000 species, diverse habitats, reproductive patterns, and strong environmental adaptability, occupy an important position in evolution [31]. In this study, we isolated SDRs from the genomes of 29 animal species, including Nile tilapia. Most SDRs in Nile tilapia belong to the “classical” category, followed by the “extended” category, which is consistent with the results reported for human [5]. In contrast, both “complex” and “atypical” categories have one member only. Among the 119 SDRs of Nile tilapia, 114 members were assigned into 49 subfamilies, while the classifications of the remaining 5 genes were uncertain. SDR48A, which also exists in human genome, was not detected in Nile tilapia. In contrast, two subfamilies, SDR112C and SDR348C, which are absent in human genome, were identified in Nile tilapia genome, with one SDR for each subfamily. Gene duplication is assumed to have played a crucial role in the evolution of vertebrates. Four rounds of large-scale genome duplications (referred to as 1R, 2R, 3R, and 4R) shaped genome evolution in fish. In combination with phylogenetic and syntenic analyses, we concluded that SDR superfamily members increased evidently due to the 3R- and the 4R-WGD in fish. Although teleost fish experienced the 3R-WGD, the extended SDRs has not expanded much, indicating that a large number of the extended SDR subfamilies in teleost fishes were lost during evolution. Variation in the number of SDR superfamily in different representative species is consistent with multiple WGD events. Tandem duplications of dhrs11-a, hsd11b1b, cbr1-a, and dhrs7a were observed in seven cichlids analyzed, and tandem duplication of rdh10-b was only observed in four tilapiine species, indicating that they are lineage specific. In addition, tandem duplications of SDRs were also observed in other species, such as far, hsd11b1, and dhrs4 in humans and c-factor in spotted gar. These results demonstrated that tandem duplication is common in the SDR superfamily in vertebrates. The different number of SDRs observed in teleosts might be mainly attributed to different tandem duplication of different SDR genes among them. It is worth noting that some SDR genes have multiple copies located on different chromosomes in some invertebrates. These duplicates probably resulted from transposon-mediated replication. The origin of these copies remains to be elucidated.
3.2. Spatial and Temporal Expression of SDRs in Nile Tilapia
SDR superfamily encoded a large number of enzymes and displayed a broad spectrum of metabolic functions [1]. Transcriptome data revealed that the majority of SDRs are expressed in multiple tissues, indicating that SDRs may play roles in various physiological and metabolic processes in Nile tilapia. In humans, DHRS7 is expressed in prostate, adrenal glands, liver, and intestine and participates in the reductive metabolism of both steroids and retinoids [32]. Over-expression of SRP-35 (DHRS7C) in mouse skeletal muscles induces enhanced glucose metabolism [33]. In the present study, five dhrs7, dhrs7a-1, dhrs7a-2, dhrs7b, dhrs7c-a, and dhrs7c-b, were identified in Nile tilapia. Dhrs7b was expressed in ovary, indicating its possible role in steroid biosynthesis. Dhrs7c-a and dhrs7c-b, which originated from fish specific 3R-WGD, were highly expressed in muscle, suggesting their possible role in glucose metabolism. Previous reports demonstrated that human BLVRB has several biochemical functions, including biliverdin and riboflavin reductase (NADPH) activity. It catalyzes the reduction of biliverdin tetrapyrrole as an intermediary redox substrate in bilirubin generation in the heme degradation pathway [34]. In Nile tilapia, high levels of expression of blvrb were detected in both heart and head kidney, suggesting their possible role in heme metabolism. Human DHRS11 exhibits enzymatic activities of both 17β-hydroxysteroid dehydrogenase and 3β-hydroxysteroid dehydrogenase, which efficiently catalyze the reduction of 11KA4, 11K-Adione, and 11KAST [23,35]. The tandem duplication of dhrs11 in cichlids and the high expression of different copies in different tissues suggested their importance in multiple functions. High expression of dhrs11-a5, dhrs11-a6, and dhrs11-a10 in both kidney and liver indicated their essential roles in metabolism and homeostasis in Nile tilapia. In zebrafish, dhrs12 (named as 36K) was identified in myelin of the central nervous system to regulate membrane lipid composition and influence oligodendrocyte precursor cell differentiation and further myelination by altering the amount of transmembrane Notch ligands [36]. Consistently, dhrs12a gene was found to be expressed exclusively in Nile tilapia brain, indicating its critical role in brain functions.
It is well documented that sex steroids play essential roles in fish sex differentiation, gametogenesis, and reproduction [37,38]. Recent studies showed that RA is critical for initiation of meiosis in both mammals and teleosts [39,40]. It is well known that SDRs play important roles in steroids and retinoids metabolism [25]. Expression, substrate specificity, and functions of hsd17b, hsd3b, and hsd11b in sex steroids biosynthesis were extensively investigated in several teleosts [41,42,43,44]. In this study, hsd3b7, rdh10b-2, rdh12b, dhrs3, dhrs7b, dhrs9, sdr39u1, hpgd-b1, far1, spra, and decr2 were identified as ovary-enriched genes, while hsd3b1, hsd11b1-b2, hsd11b2, rdh14b, kdsr, and ak7-a were identified as testis-enriched genes. Sexually dimorphic expression of SDRs in Nile tilapia gonads indicated their essential roles in steroids and RA metabolism. However, the exact role of the newly identified ovary- and testis-enriched SDR genes remains to be elucidated. Further investigations of the functions of these SDRs in fish gonads will greatly promote the understanding of their roles in sex steroids and RA metabolism.
Transcriptome analyses revealed that all of the ovary-enriched genes and the testis-enriched genes were highly expressed at 90 and 180 dah, indicating that they might be important for oogenesis and spermatogenesis, respectively. rdh12b, sdr39u1, decr2, and far1 were significantly up-regulated from 90 dah in females, indicating their indispensable roles in oogenesis. Previous studies showed that rdh12b recognizes both retinoids and lipid peroxidation products (C9 aldehydes) as substrates and contributes to the reduction of all-trans-retinaldehyde [45]. Decr2 gene encodes the peroxisomal 2,4-dienoyl-CoA reductase, which primarily converts 2,4-dienoyl-CoA to trans-3-enoyl-CoA. Biochemical studies and structural analysis suggest that human DECRs can catalyze the shortening of six-carbon-long substrates and shorten very long chain fatty acids [46,47]. Far1 catalyzes the reduction of saturated and unsaturated C16 or C18 fatty acyl-CoA to fatty alcohols and plays an essential role in the production of ether lipids/plasmalogens in mammals [48]. These three genes might be responsible for the formation of oil droplets in oocytes in fish, which contain mainly neutral lipids rich in monounsaturated fatty acids, which serve as metabolic energy reserves [48]. In contrast, the role of sdr39u1 in vertebrates is still largely unknown. Taken together, the abundant expression of SDRs in ovary indicated that anabolism of RA and fatty acid is critical for oogenesis in fish.
In fish, both RA and 11-KT proved to play essential roles in testicular differentiation and spermatogenesis. Previous reports showed that both HSD17b3 and HSD11b2 might be involved in the biosynthesis of 11-KT in human [41]. RDH14 [49] is a ubiquitously expressed microsomal enzyme, which might function as a reductase and contribute to the reduction of retinaldehyde to retinol in most human tissues. The abundant expression of rdh14 in testis of Nile tilapia further emphasizes the essentiality of retinol for testicular differentiation and functions. In humans, 11HSD1, which is ubiquitously expressed in most tissues, catalyzes cortisone to cortisol, while 11HSD2, expressed in the kidney and other classical target tissues for aldosterone, is responsible for the inactivation of cortisol to cortisone [50]. In fish, the recombinant Hsd11b, with 11β-dehydrogenase activity metabolizing from cortisol to cortisone and 11β-hydroxytestosterone to 11-KT, is involved in mitosis of spermatogonia and spermatogenesis [41]. In this study, dominant expression of both 11hsdb1-a1, -a2 and 11hsdb2 was found in testis, suggesting their possible roles in 11-KT production.
4. Materials and Methods
4.1. Animal Rearing
Nile tilapia fish used in this study were reared in recirculating freshwater tanks at 26 °C and under natural photoperiod. All female (XX) progenies were obtained by crossing the normal female (XX) with the sex-reversed XX pseudomales. All male (XY) progenies were obtained by crossing the normal female (XX) with YY supermales. Animal experiments were performed following the regulations of the Guide for Care and approved by the Institutional Animal Care and Use Committee of Southwest University.
4.2. Identification and Nomenclature of the SDRs
SDR sequences of human (Homo sapiens) and zebrafish (Danio rerio) were used as the query sequences to blast the genome sequences by tblastn (E = 2 × 10−5)) to identify the SDR genes in each genome analyzed. The identified SDRs were used to search against the NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 5 May 2020) by blastx to reduce redundant matches. In addition, SDR sequences of the rainbow trout (Oncorhynchus mykiss), common carp (Cyprinus carpio), Nile tilapia (Oreochromis niloticus), zebra mbuna (Maylandia zebra), Flier cichlid (Archocentrus centrarchus), Eastern happy (Astatotilapia calliptera), large yellow croaker (Larimichthys crocea), medaka (Oryzias latipes), fugu (Takifugu rubripes), channel catfish (Ictalurus punctatus), spotted gar (Lepisosteus oculatus), coelacanth (Latimeria chalumnae), elephant shark (Callorhinchus milii), tropical clawed frog (Xenopus tropicalis), chicken (Gallus gallus), python (Python bivittatus), lamprey (Lampetra japonicavase), tunicate (Ciona intestinalis), black tiger shrimp (Penaeus monodon), domestic silkworm (Bombyx mori), fruit fly (Drosophila melanogaster), nematode (Caenorhabditis elegans), sponge (Amphimedon queenslandica), and paramecium (Paramecium tetraurelia) were collected from NCBI and Ensembl (http://asia.ensembl.org/index.html, accessed on 18 May 2020) databases. SDR sequences of blue tilapia (Oreochromis aureus), Mozambique tilapia (Oreochromis mossambicus), and blackchin tilapia (Sarotherodon melanotheron) were collected from unpublished genome database generated by our lab. Relatively high-quality genome sequences in these species allowed us to isolate more SDR members to reflect the true evolutionary history of the superfamily. These 29 species provided a broad evolutionary coverage of different animal groups to test the SDR superfamily variation. In order to distinguish genes derived from different rounds of WGD or tandem duplication, we developed a new system of nomenclature by adding numbers or letters as suffixes of the gene name. The gene derived before 3R-WGD was represented by the letters a, b after the gene name. The gene derived from 3R-WGD was represented by a hyphen followed by a, b, etc. The gene produced by tandem duplication was represented by a number after the hyphen or a letter after the hyphen.
4.3. Phylogenetic Analysis and Genomic Distribution of SDRs
The amino acid sequences of the conserved domain of SDRs from Nile tilapia, humans, and spotted gar were aligned by Clustal W with default parameters using the multiple alignment software BioEdit (Carlsbad, USA). The neighbor-joining (NJ) method was used for phylogenetic analyses of large numbers of SDRs in Nile tilapia, spotted gar, and humans, and the maximum likelihood (ML) method was used for phylogenetic analyses of specific SDRs in many different species by MEGA 6.0 software (Tempe, USA) [51]. A bootstrap of 1000 replicates was used to assess the confidence in all phylogenies. SDRs from representative species in vertebrates were used in these trees, including those from several species of Cichlidae (SDRs from Nile tilapia, zebra mbuna, flier cichlid and eastern happy were downloaded from NCBI, while those from blue tilapia, Mozambique tilapia, and blackchin tilapia were predicted according to their unpublished genome database in our lab. For syntenic analysis, position and orientation of SDRs and their adjacent genes on the chromosome were determined using NCBI database.
4.4. Expression Analysis of Nile Tilapia SDRs in Adult Tissues and Gonads at Four Critical Developmental Stages
The transcriptomes of eight tissues from adult Nile tilapia, including brain, heart, liver, ovary, testis, kidney, muscle, and head kidney, were downloaded from NCBI database (Accession codes: PRJNA78915 and SRR1916191). RPKM was used to normalize the expression profile of SDR genes. Bidirectional hierarchical clustering analyses were performed using the heatmap package [52]. The transcriptomes (accession codes: SRA055700) of four pairs of XX and XY gonads from Nile tilapia at 5, 30, 90, and 180 days after hatching (dah) were downloaded from the NCBI database. The members with total RPKM < 10 in all tissues or RPKM < 1.25 in a single tissue were regarded as the background expression [53].
4.5. Validation of Expression Profile of SDRs by qPCR and ISH
The 14 SDRs were selected for validation of their gonadal expression at 180 dah by in situ hybridization (ISH). Among them, 4 SDRs were selected for further validation of expression profile by qPCR. To perform qPCR, gonads dissected from XX and XY Nile tilapia at 5, 30, 90, and 180 dah (about 6–200 fish for each sample depending on the fish size) were collected. The total RNA was isolated from each sample and reverse-transcribed using MMLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Subsequently, qPCR examination was carried out according to the manufacturer’s protocol of SYBR Green I Master Mix (TaKaRa, Dalian, China). Nile tilapia eef1a1a was used as an internal control to normalize the expression of these 4 genes. The relative abundance of mRNA transcripts was evaluated using the formula R = 2−ΔΔCt. Data were expressed as mean ± SD for triplicates. Statistical analyses were carried out using Student’s t-test of the SPSS package, version 18.0.
To ascertain cellular localization of the SDR family members in the developing gonads, ISH was performed using ovaries and testes from Nile tilapia at 180 dah. Fixation, embedding, and sectioning of dissected gonads and ISH were performed as described previously. Probes of sense and antisense digoxigenin (DIG)-labeled RNA strands were transcribed in vitro from linearized pGEM-Teasy-decr2/dhrs3/dhrs7b/dhrs11-a6/far1/hpgd-b1/ak7-a/ak7-b/hsd3b1/hsd3b7/hsd11b2/rdh10b-2/rdh12b/sdr39u1 DNA using an RNA labeling kit (Roche, Mannheim, Germany).
5. Conclusions
In this study, 119 SDR members assigned into 49 subfamilies and four categories were identified by comprehensive analyses of genome and transcriptome data in Nile tilapia. Phylogenetic and syntenic analyses demonstrated that tandem duplications in combination with multiple WGDs contributed to the expansion of SDR superfamily in vertebrates. In general, the variation of SDR numbers reveals the evolution process of SDR superfamily. Most SDRs were expressed in different tissues, including developing gonads. Some SDRs displayed tissue specificity or gender dimorphism. This study provided a new perspective for the evolution of the SDR superfamily and laid a solid foundation for revealing the role of the SDR genes in bony fishes and even vertebrates, especially in oogenesis and spermatogenesis.
Supplementary Materials
All supplementary materials are available online at https://www.mdpi.com/article/10.3390/ijms22084201/s1.
Author Contributions
Conceptualization, D.W., L.X. and S.Z. (Shuai Zhang); investigation, S.Z. (Shuai Zhang), W.T., B.L. and X.W.; writing—original draft preparation, S.Z. (Shuai Zhang); writing—review and editing, S.Z. (Shuqing Zheng), T.D.K., L.Z. and D.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants 31861123001, 31630082, 31872556 and 31972778 from the National Natural Science Foundation of China; grant 2018IB019 from Yunnan Science and Technology project; grant cstc2019jscx-msxmX0320 from the Chongqing Science and Technology Bureau.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Acknowledgments
The authors thank Deyong Pu for providing technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| SDR | short-chain dehydrogenases/reductases |
| WGD | whole genome duplication |
| 1R | first round of genome duplication |
| 2R | second round of genome duplication |
| 3R | third round of genome duplication |
| 4R | fourth round of genome duplication |
| qRT-PCR | quantitative real time polymerase chain reaction |
| IHC | Immunohistochemistry |
| NJ | neighbor-joining |
| dah | day after hatching |
| RPKM | reads per kb per million reads |
References
- Kavanagh, K.L.; Jornvall, H.; Persson, B.; Oppermann, U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef] [PubMed]
- Mayoral, J.G.; Leonard, K.T.; Nouzova, M.; Noriega, F.G.; Defelipe, L.A.; Turjanski, A.G. Functional analysis of a mosquito short-chain dehydrogenase cluster. Arch. Insect Biochem. Physiol. 2013, 82, 96–115. [Google Scholar] [CrossRef]
- Jornvall, H.; Persson, M.; Jeffery, J. Alcohol and polyol dehydrogenases are both divided into two protein types, and structural properties cross-relate the different enzyme activities within each type. Proc. Natl. Acad. Sci. USA 1981, 78, 4226–4230. [Google Scholar] [CrossRef]
- Persson, B.; Krook, M.; Jornvall, H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 1991, 200, 537–543. [Google Scholar] [CrossRef]
- Persson, B.; Kallberg, Y. Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs). Chem. Biol. Interact. 2013, 202, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Kramm, A.; Kisiela, M.; Schulz, R.; Maser, E. Short-chain dehydrogenases/reductases in cyanobacteria. FEBS J. 2012, 279, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, Y.; Oppermann, U.; Jornvall, H.; Persson, B. Short-chain dehydrogenase/reductase (SDR) relationships: A large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 2002, 11, 636–641. [Google Scholar] [CrossRef]
- Kallberg, Y.; Oppermann, U.; Persson, B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010, 277, 2375–2386. [Google Scholar] [CrossRef]
- Bray, J.E.; Marsden, B.D.; Oppermann, U. The human short-chain dehydrogenase/reductase (SDR) superfamily: A bioinformatics summary. Chem. Biol. Interact. 2009, 178, 99–109. [Google Scholar] [CrossRef]
- Moummou, H.; Kallberg, Y.; Tonfack, L.B.; Persson, B. The plant short-chain dehydrogenase (SDR) superfamily: Genome-wide inventory and diversification patterns. BMC Plant Biol. 2012, 12, 219. [Google Scholar] [CrossRef]
- Simakov, O.; Marletaz, F.; Yue, J.X.; O’Connell, B.; Jenkins, J.; Brandt, A. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 2020, 4, 820–830. [Google Scholar] [CrossRef]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef]
- Moghsadam, H.K.; Ferguson, M.M.; Danzmann, R.G. Evolution of Hox clusters in Salmonidae: A comparative analysis between Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss). J. Mol. Evol. 2005, 61, 636–649. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, C.T.; Alfoldi, J.; Lee, A.P.; Fan, S.; Philippe, H.; Maccallum, I.; Braasch, I.; Manousaki, T.; Schneider, I.; Rohner, N.; et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013, 496, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Catchen, J.; Ferrara, A.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef]
- Xiao, L.; Guo, Y.; Wang, D.D.; Zhao, M.; Hou, X.; Li, S.S.; Lin, H.R.; Zhang, Y. Beta-hydroxysteroid dehydrogenase genes in orange-spotted grouper (Epinephelus coioides): Genome-wide identification and expression analysis during sex reversal. Front. Genet. 2020, 11, 161. [Google Scholar] [CrossRef]
- Zou, C.C.; Wang, L.J.; Zou, Y.X.; Wu, Z.H.; Wang, W.X.; Liang, S.S.; You, F. Characteristics and sex dimorphism of 17β-hydroxysteroid dehydrogenase family genes in the olive flounder Paralichthys olivaceus. J. Steroid. Biochem. Mol. Biol. 2020, 199, 17. [Google Scholar] [CrossRef]
- Hiltunen, J.K.; Kastaniotis, A.J.; Autio, K.J.; Jiang, G.; Chen, Z.; Glumoff, T. 17B-hydroxysteroid dehydrogenases as acyl thioester metabolizing enzymes. Mol. Cell. Endocrinol. 2019, 489, 107–118. [Google Scholar] [CrossRef]
- Saloniemi, T.; Jokela, H.; Strauss, L.; Pakarinen, P.; Poutanen, M. The diversity of sex steroid action: Novel functions of hydroxysteroid (17β) dehydrogenases as revealed by genetically modified mouse models. J. Endocrinol. 2012, 212, 27–40. [Google Scholar] [CrossRef]
- Yazawa, T.; Uesaka, M.; Inaoka, Y.; Mizutani, T.; Sekiguchi, T.; Kajitani, T.; Kitano, T.; Umezawa, A.; Miyamoto, K. Cyp11b1 is induced in the murine gonad by luteinizing hormone/human chorionic gonadotropin and involved in the production of 11-ketotestosterone, a major fish androgen: Conservation and evolution of the androgen metabolic pathway. Endocrinology 2008, 149, 1786–1792. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Miyagi, N.; Matsunaga, T.; Hara, A.; Ikari, A. Human dehydrogenase/reductase (SDR family) member 11 is a novel type of 17beta-hydroxysteroid dehydrogenase. Biochem. Biophys. Res. Commun. 2016, 472, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Morikawa, Y.; Kudo, Y.; Suenami, K.; Matsunaga, T.; Ikari, A.; Hara, A. Human dehydrogenase/reductase SDR family member 11 (DHRS11) and aldo-keto reductase 1C isoforms in comparison: Substrate and reaction specificity in the reduction of 11-keto-C19-steroids. J. Steroid. Biochem. Mol. Biol. 2020, 199, 105586. [Google Scholar] [CrossRef] [PubMed]
- Albalat, R.; Brunet, F.; Laudet, V.; Schubert, M. Evolution of retinoid and steroid signaling: Vertebrate diversification from an amphioxus perspective. Genome Biol. Evol. 2011, 3, 985–1005. [Google Scholar] [CrossRef] [PubMed]
- Kisiela, M.; Faust, A.; Ebert, B.; Maser, E.; Scheidig, A.J. Crystal structure and catalytic characterization of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans. FEBS J. 2018, 285, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.A.; Gammerdinger, W.J.; Bartie, K.L.; Penman, D.J.; Kocher, T.D. A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genom. 2017, 18, 3017–3723. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.N.; Jiang, X.L.; Xie, Q.P.; Yuan, J.; Huang, B.F.; Tao, W.J.; Zhou, L.Y.; Nagahama, Y.; Wang, D.S. Transdifferentiation of differentiated ovary into functional testis by long-term treatment of aromatase inhibitor in Nile tilapia. Endocrinology 2014, 155, 1476–1488. [Google Scholar] [CrossRef]
- Tao, W.J.; Yuan, J.; Zhou, L.Y.; Sun, L.N.; Sun, Y.L.; Yang, S.J.; Li, M.H.; Zeng, S.; Huang, B.F.; Wang, D.S. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS ONE 2013, 8, e63604. [Google Scholar] [CrossRef]
- Tang, N.T.; Le, L. Comparative study on sequence-structure-function relationship of the human short-chain dehydrogenases/reductases protein family. Evol. Bioinform. Online 2014, 10, 165–176. [Google Scholar] [CrossRef]
- Brawand, D.; Wagner, C.E.; Li, Y.I.; Malinsky, M.; Keller, I.; Fan, S.; Simakov, O.; Ng, A.Y.; Lim, Z.W.; Bezault, E.; et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 2014, 513, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Stambergova, H.; Zemanova, L.; Lundova, T.; Malcekova, B.; Skarka, A.; Safr, M.; Wsol, V. Human DHRS7, promising enzyme in metabolism of steroids and retinoids? J. Steroid. Biochem. Mol. Biol. 2016, 155, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Dror, E.; Handschin, C.; Furrer, R.; Perez-Schindler, J.; Bachmann, C.; Treves, S.; Zorzato, F. Over-expression of a retinol dehydrogenase (SRP35/DHRS7C) in skeletal muscle activates mTORC2, enhances glucose metabolism and muscle performance. Sci. Rep. 2018, 8, 636. [Google Scholar] [CrossRef]
- Wu, S.; Li, Z.D.; Gnatenko, D.V.; Zhang, B.B.; Zhao, L.; Malone, L.E.; Markova, N.; Mantle, T.J.; Nesbitt, N.M.; Bahou, W.F. BLVRB redox mutation defines heme degradation in a metabolic pathway of enhanced thrombopoiesis in humans. Blood 2016, 128, 699–709. [Google Scholar] [CrossRef]
- Nagarajan, B.; Harder, A.; Japp, A.; Häberlein, F.; Mingardo, E.; Kleinert, H.; Yilmaz, Ö.; Zoons, A.; Rau, B.; Christ, A.; et al. CNS myelin protein 36K regulates oligodendrocyte differentiation through Notch. Glia 2020, 68, 509–527. [Google Scholar] [CrossRef]
- Morris, J.K.; Willard, B.B.; Yin, X.; Jeserich, G.; Kinter, M.; Trapp, B.D. The 36K protein of zebrafish CNS myelin is a short-chain dehydrogenase. Glia 2004, 45, 378–391. [Google Scholar] [CrossRef]
- Nagahama, Y.; Yoshikuni, M.; Yamashita, M.; Tokumoto, T.; Katsu, Y. Regulation of oocyte growth and maturation in fish. Curr. Top. Dev. Biol. 1995, 30, 103–145. [Google Scholar] [PubMed]
- Li, M.H.; Sun, L.N.; Wang, D.S. Roles of estrogens in fish sexual plasticity and sex differentiation. Gen. Comp. Endocrinol. 2019, 277, 9–16. [Google Scholar] [CrossRef]
- Feng, R.J.; Fang, L.L.; Cheng, Y.Y.; He, X.; Jiang, W.T.; Dong, R.R.; Shi, H.J.; Jiang, D.N.; Sun, L.N.; Wang, D.S. Retinoic acid homeostasis through aldh1a2 and cyp26a1 mediates meiotic entry in Nile tilapia (Oreochromis niloticus). Sci. Rep. 2015, 5, 3657. [Google Scholar] [CrossRef]
- Koubova, J.; Menke, D.B.; Zhou, Q.; Capel, B.; Griswold, M.D.; Page, D.C. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. USA 2006, 103, 2474–2479. [Google Scholar] [CrossRef] [PubMed]
- Ijiri, S.; Shibata, Y.; Takezawa, N.; Kazeto, Y.; Takatsuka, N.; Kato, E.; Hagihara, S.; Ozaki, Y.; Adachi, S.; Yamauchi, K.; et al. 17beta-HSD type 12-like is responsible for maturation-inducing hormone synthesis during oocyte maturation in masu salmon. Endocrinology 2017, 158, 627–639. [Google Scholar] [PubMed]
- Mindnich, R.; Haller, F.; Halbach, F.; Moeller, G.; De Angelis, M.H.; Adamski, J. Androgen metabolism via 17beta-hydroxysteroid dehydrogenase type 3 in mammalian and non-mammalian vertebrates: Comparison of the human and the zebrafish enzyme. J. Mol. Endocrinol. 2005, 35, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.Y.; Wang, D.S.; Senthilkumaran, B.; Yoshikuni, M.; Shibata, Y.; Kobayashi, T.; Sudhakumari, C.C.; Nagahama, Y. Cloning, expression and characterization of three types of 17beta-hydroxysteroid dehydrogenases from the Nile tilapia, Oreochromis niloticus. J. Mol. Endocrinol. 2005, 35, 103–116. [Google Scholar] [CrossRef]
- Ozaki, Y.; Higuchi, M.; Miura, C.; Yamaguchi, S.; Tozawa, Y.; Miura, T. Roles of 11beta-hydroxysteroid dehydrogenase in fish spermatogenesis. Endocrinology 2006, 147, 5139–5146. [Google Scholar] [CrossRef]
- Belyaeva, O.V.; Korkina, O.V.; Stetsenko, A.V.; Kim, T.; Nelson, P.S.; Kedishvili, N.Y. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): Catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry 2005, 44, 7035–7047. [Google Scholar]
- Hua, T.; Wu, D.; Ding, W.; Wang, J.; Shaw, N.; Liu, Z.J. Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal β-oxidation of unsaturated fatty acids. J. Biol. Chem. 2012, 287, 28956–28965. [Google Scholar] [CrossRef] [PubMed]
- Alphey, M.S.; Yu, W.; Byres, E.; Li, D.; Hunter, W.N. Structure and reactivity of human mitochondrial 2,4-dienoyl-CoA reductase: Enzyme-ligand interactions in a distinctive short-chain reductase active site. J. Biol. Chem. 2005, 280, 3068–3077. [Google Scholar] [CrossRef]
- Cheng, J.B.; Russell, D.W. Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem. 2004, 279, 37789–37797. [Google Scholar] [CrossRef]
- Haeseleer, F.; Jang, G.F.; Imanishi, Y.; Driessen, C.; Matsumura, M.; Nelson, P.S.; Palczewski, K. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J. Biol. Chem. 2002, 277, 45537–45546. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.; Holmes, M.; Seckl, J. 11β-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 2013, 93, 1139–1206. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.Q.; Long, J.; Liu, Z.L.; Tao, W.J.; Wang, D.S. Identification and evolution of TGF-beta signaling pathway members in twenty-four animal species and expression in tilapia. Int. J. Mol. Sci. 2018, 19, 1154. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Tao, W.J.; Cheng, Y.Y.; Huang, B.F.; Wang, D.S. Genome-wide identification, phylogeny, and gonadal expression of fox genes in Nile tilapia, Oreochromis niloticus. Fish Physiol. Biochem. 2014, 40, 1239–1252. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).