Gene Coexpression Network Analysis Indicates that Hub Genes Related to Photosynthesis and Starch Synthesis Modulate Salt Stress Tolerance in Ulmus pumila
Abstract
1. Introduction
2. Results
2.1. Differences in Salt Tolerance among Cultivars
2.2. Physiology and Photosynthesis of Upu11 under Salt Stress
2.3. Functional Annotation and Classification of DEGs
2.4. Identification and Functional Annotation of Salt-Induced DEGs
2.5. Weighted Gene Correlation Network Analysis (WGCNA)
2.6. Transcript Regulation in the Photosynthesis and Starch-Synthesis Pathways
2.7. Hub Transcript Network Construction Based on WGCNA
3. Discussion
3.1. Increased Photosynthesis Maintains the Balance between Osmotic Regulation and Lipid Peroxidation
3.2. Mild Salt Stress Induces Expression of Genes Promoting Photosynthesis
3.3. Salt-Induced Genes Accelerate Synthesis and Accumulation of Starch
4. Materials and Methods
4.1. Plant Materials and Salt Tsreatments
4.2. Growth and Photosynthetic Traits
4.3. RNA Isolation, Library Preparation, and RNA-Seq
4.4. Functional Annotation of Unigenes
4.5. Gene Expression and Annotation
4.6. qRT-PCR
4.7. Weighted Gene Correlation Network Analysis
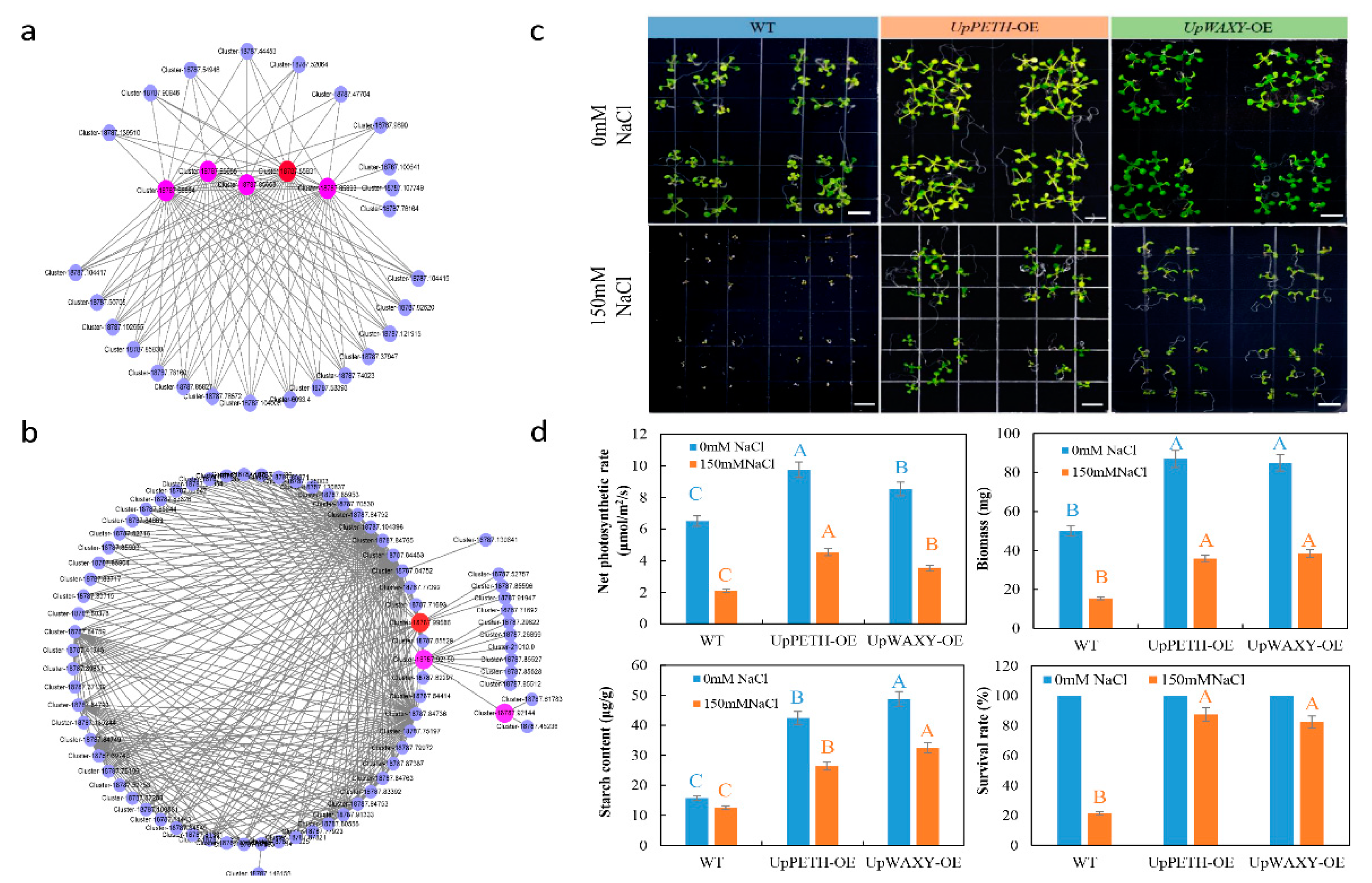
4.8. Overexpression of Salt-Responsive Hub Genes in Arabidopsis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shi, L.; Zhang, Z.J.; Zhang, C.Y.; Zhang, J.Z. Effects of sand burial on survival, growth, gas exchange and biomass allocation of Ulmus pumila seedlings in the hunshandak sandland, China. Ann. Bot. 2004, 94, 553–560. [Google Scholar] [CrossRef]
- Chen, X.; Xu, L. Phenological responses of Ulmus pumila (siberian elm) to climate change in the temperate zone of China. Int. J. Biometeorol. 2012, 56, 695–706. [Google Scholar] [CrossRef]
- Mn, C.G. A provenance test of white elm (Ulmus pumila L.) in China. Silvae Genet. 1989, 38, 37–44. [Google Scholar]
- You, Y.O.; Choi, N.Y.; Kim, K.J. Ethanol extract of Ulmus pumila root bark inhibits clinically isolated antibiotic-resistant bacteria. Evid. Based Complementary Altern. Med. 2013, 2013, 269874. [Google Scholar] [CrossRef] [PubMed]
- Solla, A.; Martín, J.A.; Corral, P.; Gil, L. Seasonal changes in wood formation of Ulmus pumila and U. minor and its relation with Dutch elm disease. New Phytol. 2005, 166, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Santini, A.; Fagnani, A.; Ferrini, F.; Mittempergher, L.; Macchioni, N. Elm breeding for DED resistance, the Italian clones and their wood properties. For. Syst. 2004, 13, 179–184. [Google Scholar]
- Chen, Z.L.; Tian, Y.; Zhang, C.Y.; Zhang, Z.Y. The principal component analysis on the physical and strength properties of Ulmus pumila clones wood. J. Sichuan Agric. Univ. 1998, 16, 140–144. [Google Scholar]
- Murdoch, C.W.; Campana, R.J.; Biermann, C.J. Physical and chemical-properties of wetwood in American elm (Ulmus americana). Can. J. Plant Pathol. 1987, 9, 20–23. [Google Scholar] [CrossRef]
- Kim, S.I.; Sim, K.H.; Choi, H.Y. A comparative study of antioxidant activity in some korean medicinal plant used as food materials. Mol. Cell Toxicol. 2010, 6, 279–285. [Google Scholar] [CrossRef]
- Kim, K.B.; Jo, B.S.; Park, H.J.; Park, K.T.; Cho, Y.J. Healthy functional food properties of phenolic compounds isolated from Ulmus pumila. Korean J. Food Preserv. 2012, 19, 909–918. [Google Scholar] [CrossRef]
- Sun, Q.J. Effect of NaCl Stress on the Growth and Photosynthesis Characteristics of Ulmus pumila L. Seedlings; Shandong Normal University: Jinan, China, 2014. [Google Scholar]
- Lin, T.T. Effects of Drought Stress on Growth and Ecological Stoichiometry of Ulmus pumila Seedlings; Liaoning Technical University: Fuxing, China, 2019. [Google Scholar]
- Butkut, B.; Lemeien, N.; Ceseviien, J.; Liatukas, Z.; Dabkeviciene, G. Carbohydrate and lignin partitioning in switchgrass biomass (Panicum virgatum L.) as a bioenergy feedstock. Zemdirbyste 2013, 100, 251–260. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Zhou, X.P.; Tao, M.; Yuan, F.; Liu, L.L.; Wu, F.H.; Wu, X.M.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Z.; Zhang, H.; Song, C.P.; Zhu, J.K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Park, G.E.; Kim, K.W.; Lee, D.K.; Hyun, J.O. Adaptive phenotypic plasticity of Siberian elm in response to drought stress: Increased stomatal pore depth. Microsc. Microanal. 2013, 19, 178–181. [Google Scholar] [CrossRef]
- Mu, D.; Zwiazek, J.J.; Li, Z.; Zhang, W. Genotypic variation in salt tolerance of Ulmus pumila plants obtained by shoot micropropagation. Acta Physiol. Plant 2016, 38, 1–16. [Google Scholar] [CrossRef]
- Zalapa, J.E.; Brunet, J.; Guries, R.P. Genetic diversity and relationships among Dutch elm disease tolerant Ulmus pumila L. accessions from China. Genome 2008, 51, 492–500. [Google Scholar] [CrossRef]
- Mø, A.P.; Dongen, S.V. Ontogeny of asymmetry and compensational growth in elm Ulmus glabra leaves under different environmental conditions. Int. J. Plant Sci. 2003, 164, 519–526. [Google Scholar]
- Bosu, P.P.; Wagner, M.R. Effects of induced water stress on leaf trichome density and foliar nutrients of three elm (Ulmus) species: Implications for resistance to the elm leaf beetle. Environ. Entomol. 2007, 36, 595–601. [Google Scholar] [CrossRef]
- Feng, Z.T.; Deng, Y.Q.; Fan, H.; Sun, Q.J.; Sui, N.; Wang, B.S. Effects of NaCl stress on the growth and photosynthetic characteristics of Ulmus pumila L. seedlings in sand culture. Photosynthetica 2014, 52, 313–320. [Google Scholar] [CrossRef]
- Viswanathan, C.; Khanna–Chopra, R. Effect of heat stress on grain growth, starch synthesis and protein synthesis in grains of wheat (Triticum aestivum L.) varieties differing in grain weight stability. J. Agron. Crop Sci. 2010, 186, 1–7. [Google Scholar] [CrossRef]
- Zinselmeier, C.; Sun, Y.; Helentjaris, T.; Beatty, M.; Yang, S.; Smith, H.; Habben, J. The use of gene expression profiling to dissect the stress sensitivity of reproductive development in maize. Field Crop. Res. 2002, 75, 111–121. [Google Scholar] [CrossRef]
- Wang, J.; Shi, K.; Lu, W.P.; Lu, D.L. Effects of post-silking shading stress on enzymatic activities and phytohormone contents during grain development in spring maize. J. Plant Growth Regul. 2020. [Google Scholar] [CrossRef]
- Wang, S.J.; Liu, L.F.; Chen, C.K.; Chen, L.W. Regulations of granule-bound starch synthase I gene expression in rice leaves by temperature and drought stress. Biol. Plantarum. 2006, 50, 537–541. [Google Scholar] [CrossRef]
- Zhu, Y.; Cai, X.L.; Wang, Z.Y.; Hong, M.M. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J. Biol. Chem. 2003, 278, 47803–47811. [Google Scholar] [CrossRef]
- Fu, F.F.; Xue, H.W. Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef]
- Li, L.; Pan, X.; Li, H. Responses of Photosystem II (PSII) Function in leaves and samaras of Ulmus pumila to chilling and freezing temperatures and subsequent recovery. Int. J. Agric. Biol. 2012, 14, 739–744. [Google Scholar]
- Long, B.M.; Hee, W.Y.; Sharwood, R.E.; Rae, B.D.; Kaines, S.; Lim, Y.L.; Nguyen, N.D.; Massey, B.; Bala, S.; Caemmerer, S.V.; et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 2018, 9, 3570. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Sousa, B.; Lopes, J.; Soares, C.; Machado, J.; Carvalho, S.; Fidalgo, F.; Teixeira, J. Diclofenac shifts the role of root glutamine synthetase and glutamate dehydrogenase for maintaining nitrogen assimilation and proline production at the expense of shoot carbon reserves in Solanum lycopersicum L. Environ. Sci. Pollut Res. 2020, 27, 29130–29142. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.R.; Wang, L.M.; Lin, X.L.; Yao, Z.; Xu, H.W.; Zhu, C.H.; Teng, H.Y.; Cui, L.L.; Liu, E.E.; Zhang, J.J.; et al. Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice. Mol. Plant. 2019, 12, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Zheng, T.C.; Shao, L.T.; Xiao, Z.H.; Wang, F.W.; Li, S.C.; Zang, L.; Zheng, M.; Li, Y.; Qu, G.Z. Variation analysis of physiological traits in Betula platyphylla overexpressing TaLEA-ThbZIP gene under salt stress. PLoS ONE 2016, 11, 0164820. [Google Scholar] [CrossRef]
- Ranney, T.G.; Whitlow, T.H.; Bassuk, N.L. Response of five temperate deciduous tree species to water stress. Tree Physiol. 1990, 6, 439. [Google Scholar] [CrossRef]
- Ma, H.Z.; Liu, C.; Li, Z.X.; Ran, Q.J.; Xie, G.N.; Wang, B.M.; Fang, S.; Chu, J.F.; Zhang, J.R. ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.F.; Zuo, L.H.; Yu, X.Y.; Dong, Y.; Zhang, S.; Yang, M.S. Response mechanism in populus × euramericana cv. ‘74/76’ revealed by rna-seq under salt stress. Acta Physiol. Plant. 2018, 40, 96. [Google Scholar] [CrossRef]
- Woolley, J.T. Sodium and silicon as nutrients for the tomato plant. Plant Physiol. 1957, 32, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Z.; Zhao, K.F. Effect of low NaCl concentration on the growth of Zea mays L. Plant Physiol. Commun. 2006, 42, 628–632. [Google Scholar]
- Li, B.; Tester, M.; Gilliham, M. Chloride on the move. Trends Plant Sci. 2017, 22, 236–248. [Google Scholar] [CrossRef]
- Munns, R. Physiological processes limiting plant growth in saline soil: Some dogmas and hypotheses. Plant Cell Enciron. 1993, 16, 15–24. [Google Scholar] [CrossRef]
- Chen, S.L. Proteomics reveal both photochemical and biochemical limitations involved in salt-induced suppression of photosynthesis in trees. Tree Physiol. 2018, 38, 1599–1604. [Google Scholar] [CrossRef]
- Smirnoff, N. The role of active oxygen in the response of plant to water deficit and desiccation. New Phytol. 1993, 125, 27–58. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Schrader, S.M.; Wise, R.R.; Wacholtz, W.F.; Ort, D.R.; Sharkey, T.D. Thylakoid membrane responses to moderately high leaf temperature in Pima cotton. Plant Cell Environ. 2004, 27, 725–735. [Google Scholar] [CrossRef]
- Ascencio–Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.J.; Parkin, I.A. Differential SAGE analysis in Arabidopsis uncovers increased transcriptome complexity in response to low temperature. BMC Genom. 2008, 9, 434. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.E.; Haldrup, A.; Rosgaard, L.; Scheller, V. Molecular dissection of photosystem I in higher plants: Topology, structure and function. Physiol. Plantarum. 2003, 119, 313–321. [Google Scholar] [CrossRef]
- Seok, M.S.; You, Y.N.; Park, H.J.; Lee, S.S.; Aigen, F.; Luan, S.; Ahn, J.C.; Cho, H.S. AtFKBP16-1, a chloroplast lumenal immunophilin, mediates response to photosynthetic stress by regulating PsaL stability. Physiol. Plantarum. 2014, 150, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Livingston, A.K.; Cruz, J.A.; Kohzuma, K.; Dhingr, A.; Kramera, D.M. An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 2010, 22, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; McAusland, L.; Lawson, T.; Raines, C.A. Overexpression of the RieskeFeS protein increases electron transport rates and biomass yield. Plant Physiol. 2017, 175, 134–145. [Google Scholar] [CrossRef]
- Lintala, M.; Lehtimäki, N.; Benz, J.P.; Jungfer, A.; Soll, J.; Aro, E.M.; Bölter, B.; Mulo, P. Depletion of leaf-type ferredoxin-NADP(+) oxidoreductase results in the permanent induction of photoprotective mechanisms in Arabidopsis chloroplasts. Plant J. 2012, 70, 809–817. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Murata, N. Revised scheme for the mechanism of photoinhibition and its application to enhance the abiotic stress tolerance of the photosynthetic machinery. Appl. Microbiol. Biot. 2014, 98, 8777–8796. [Google Scholar] [CrossRef]
- Watkins, K.P.; Williams-Carrier, R.; Chotewutmontri, P.; Friso, G.; Teubner, M.; Belcher, S.; Ruwe, H.; Schmitz-Linneweber, C.; van Wijk, K.J.; Barkan, A. Exploring the proteome associated with the mRNA encoding the D1 reaction center protein of Photosystem II in plant chloroplasts. Plant J. 2020, 102, 369–382. [Google Scholar] [CrossRef]
- Méteignier, L.V.; Ghandour, R.; Meierhoff, K.; Zimmerman, A.; Chicher, J.; Baumberger, N.; Alioua, A.; Meurer, J.; Zoschke, R.; Hammani, K. The Arabidopsis mTERF-repeat MDA1 protein plays a dual function in transcription and stabilization of specific chloroplast transcripts within the psbE and ndhH operons. New Phytol. 2020, 227, 1376–1391. [Google Scholar] [CrossRef]
- Takahashi, M.; Shigeto, J.; Sakamoto, A.; Morikawa, H. Selective nitration of PsbO1, PsbO2, and PsbP1 decreases PSII oxygen evolution and photochemical efficiency in intact leaves of Arabidopsis. Plant Signal. Behav. 2017, 12, 1376157. [Google Scholar] [CrossRef] [PubMed]
- Suorsa, M.; Sirpiö, S.; Paakkarinen, V.; Kumari, N.; Holmström, M.; Aro, E.M. Two proteins homologous to PsbQ are novel subunits of the chloroplast NAD(P)H dehydrogenase. Plant Cell Physiol. 2010, 51, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Tsunoda, H.; Suzuki, Y.; Makino, A.; Ishida, H. RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J. Exp. Bot. 2012, 63, 2159–2170. [Google Scholar] [CrossRef] [PubMed]
- Maurino, V.G.; Peterhansel, C. Photorespiration: Current status and approaches for metabolic engineering. Curr. Opin. Plant Biol. 2010, 13, 249–256. [Google Scholar] [CrossRef]
- Eisenhut, M.; Roell, M.S.; Weber, A.P.M. Mechanistic understanding of photorespiration paves the way to a new green revolution. New Phytol. 2019, 223, 1762–1769. [Google Scholar] [CrossRef]
- Yang, T.; Wang, L.; Li, C.; Liu, Y.; Zhu, S.; Qi, Y.; Liu, X.; Lin, Q.; Luan, S.; Yu, F. Receptor protein kinase FERONIA controls leaf starch accumulation by interacting with glyceraldehyde-3-phosphate dehydrogenase. Biochem. Bioph. Res. Commun. 2015, 465, 77–82. [Google Scholar] [CrossRef]
- Lu, W.; Tang, X.; Huo, Y.; Xu, R.; Qi, S.; Huang, J.; Zheng, C.; Wu, C.A. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 2012, 503, 65–74. [Google Scholar] [CrossRef]
- Henkes, S.; Sonnewald, U.; Badur, R.; Flachmann, R.; Stitt, M. A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 2001, 13, 535–551. [Google Scholar] [CrossRef]
- Khozaei, M.; Fisk, S.; Lawson, T.; Gibon, Y.; Sulpice, R.; Stitt, M.; Lefebvre, S.C.; Raines, C.A. Overexpression of plastid transketolase in tobacco results in a thiamine auxotrophic phenotype. Plant Cell 2015, 27, 432–447. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Seung, D.; Soyk, S.; Coiro, M.; Maier, B.A.; Eicke, S.; Zeeman, S.C. Protein targeting to starch is required for localising GRANULE–BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 2015, 13, 1002080. [Google Scholar] [CrossRef] [PubMed]
- Hebelstrup, K.H.; Nielsen, M.M.; Carciofi, M.; Andrzejczak, O.; Shaik, S.S.; Blennow, A.; Palcic, M.M. Waxy and non-waxy barley cultivars exhibit differences in the targeting and catalytic activity of GBSS1a. J. Exp. Bot. 2017, 68, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Seung, D.; Echevarría-Poza, A.; Steuernagel, B.; Smith, A.M. Natural polymorphisms in Arabidopsis result in wide variation or loss of the amylose component of starch. Plant Physiol. 2020, 182, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Grisolia, M.J.; Peralta, D.A.; Valdez, H.A.; Barchiesi, J.; Gomez-Casati, D.F.; Busi, M.V. The targeting of starch binding domains from starch synthase III to the cell wall alters cell wall composition and properties. Plant Mol. Biol. 2017, 93, 121–135. [Google Scholar] [CrossRef]
- Pfister, B.; Lu, K.J.; Eicke, S.; Feil, R.; Lunn, J.E.; Streb, S.; Zeeman, S.C. Genetic evidence that chain length and branch point distributions are linked determinants of starch granule formation in Arabidopsis. Plant Physiol. 2014, 165, 1457–1474. [Google Scholar] [CrossRef]
- Kleczkowski, L.A. A phosphoglycerate to inorganic phosphate ratio is the major factor in controlling starch levels in chloroplasts via ADP-glucose pyrophosphorylase regulation. FEBS Lett. 1999, 448, 153–156. [Google Scholar] [CrossRef]
- Caspar, T. Genetic dissection of the biosynthesis, degradation and biological functions of starch. Arabidopsis 1994, 27, 913–936. [Google Scholar]
- Sokolov, L.N.; Déjardin, A.; Kleczkowski, L.A. Sugars and light/dark exposure trigger differential regulation of ADP-glucose pyrophosphorylase genes in Arabidopsis thaliana (thale cress). Biochem. J. 1998, 336, 681–687. [Google Scholar] [CrossRef]
- Rojas-González, J.A.; Soto-Súarez, M.; García-Díaz, Á.; Romero-Puertas, M.C.; Sandalio, L.M.; Mérida, Á.; Thormählen, I.; Geigenberger, P.; Serrato, A.J.; Sahrawy, M. Disruption of both chloroplastic and cytosolic FBPase genes results in a dwarf phenotype and important starch and metabolite changes in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 2673–2689. [Google Scholar] [CrossRef]
- Simkin, A.J.; Lopez-Calcagno, P.E.; Davey, P.A.; Headland, L.R.; Lawson, T.; Timm, S.; Bauwe, H.; Raines, C.A. Simultaneous stimulation of sedoheptulose 1,7-bisphosphatase, fructose 1,6-bisphophate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnol. J. 2017, 15, 805–816. [Google Scholar] [CrossRef]
- Sánchez-López, Á.M.; Bahaji, A.; De Diego, N.; Baslam, M.; Li, J.; Muñoz, F.J.; Almagro, G.; García-Gómez, P.; Ameztoy, K.; Ricarte-Bermejo, A.; et al. Arabidopsis responds to alternaria alternata Volatiles by triggering plastid phosphoglucose isomerase-independent mechanisms. Plant Physiol. 2016, 172, 1989–2001. [Google Scholar] [CrossRef]
- Kunz, H.H.; Zamani-Nour, S.; Häusler, R.E.; Ludewig, K.; Schroeder, J.I.; Malinova, I.; Fettke, J.; Flügge, U.I.; Gierth, M. Loss of cytosolic phosphoglucose isomerase affects carbohydrate metabolism in leaves and is essential for fertility of Arabidopsis. Plant Physiol. 2014, 166, 753–765. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- De Bianchi, S.; Betterle, N.; Kouril, R.; Cazzaniga, S.; Boekema, E.; Bassi, R.; Dall’Osto, L. Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 2011, 23, 2659–2679. [Google Scholar] [CrossRef]
- Marin, A.; Passarini, F.; van Stokkum, I.H.; van Grondelle, R.; Croce, R. Minor complexes at work: Light-harvesting by carotenoids in the photosystem II antenna complexes CP24 and CP26. Biophys. J. 2011, 100, 2829–2838. [Google Scholar] [CrossRef]
- Xia, Y.; Ning, Z.; Bai, G.; Li, R.; Yan, G.; Siddique, K.H.; Baum, M.; Guo, P. Allelic variations of a light harvesting chlorophyll a/b-binding protein gene (Lhcb1) associated with agronomic traits in barley. PLoS ONE 2012, 7, 37573. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Prioul, J.L.; Chartier, P. Partitioning of transfer and carboxylation components of intracellular resistance to photosynthetic CO2 fixation: A critical analysis of the methods used. Ann. Bot. 1997, 41, 789–800. [Google Scholar] [CrossRef]
- Frank, A.B.; Barker, R.E.; Berdahl, J.D. Water-use efficiency of grasses grown under controlled and field conditions. Agron. J. 1987, 79, 541–544. [Google Scholar] [CrossRef]
- Liu, G.; Yang, C.; Xu, K.; Zhang, Z.; Li, D.; Wu, Z.; Chen, Z. Development of yield and some photosynthetic characteristics during 82 years of genetic improvement of soybean genotypes in northeast China. Aust. J. Crop Sci. 2012, 6, 1416–1422. [Google Scholar]
- Olsson, T.; Leverenz, J.W. Non-uniform stomatal closure and the apparent convexity of the photosynthetic photon flux density response curve. Plant Cell Environ. 1994, 17, 701–710. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.; Liu, P.; Zhang, Q.; Bu, C.; Lu, C.; Srivastava, S.; Zhang, D.; Song, Y. Gene Coexpression Network Analysis Indicates that Hub Genes Related to Photosynthesis and Starch Synthesis Modulate Salt Stress Tolerance in Ulmus pumila. Int. J. Mol. Sci. 2021, 22, 4410. https://doi.org/10.3390/ijms22094410
Chen P, Liu P, Zhang Q, Bu C, Lu C, Srivastava S, Zhang D, Song Y. Gene Coexpression Network Analysis Indicates that Hub Genes Related to Photosynthesis and Starch Synthesis Modulate Salt Stress Tolerance in Ulmus pumila. International Journal of Molecular Sciences. 2021; 22(9):4410. https://doi.org/10.3390/ijms22094410
Chicago/Turabian StyleChen, Panfei, Peng Liu, Quanfeng Zhang, Chenhao Bu, Chunhao Lu, Sudhakar Srivastava, Deqiang Zhang, and Yuepeng Song. 2021. "Gene Coexpression Network Analysis Indicates that Hub Genes Related to Photosynthesis and Starch Synthesis Modulate Salt Stress Tolerance in Ulmus pumila" International Journal of Molecular Sciences 22, no. 9: 4410. https://doi.org/10.3390/ijms22094410
APA StyleChen, P., Liu, P., Zhang, Q., Bu, C., Lu, C., Srivastava, S., Zhang, D., & Song, Y. (2021). Gene Coexpression Network Analysis Indicates that Hub Genes Related to Photosynthesis and Starch Synthesis Modulate Salt Stress Tolerance in Ulmus pumila. International Journal of Molecular Sciences, 22(9), 4410. https://doi.org/10.3390/ijms22094410





