Integrative Analysis of Transcriptome and Metabolome Reveals Salt Stress Orchestrating the Accumulation of Specialized Metabolites in Lycium barbarum L. Fruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Measurement of Physiological Indexes
2.3. Transcriptome Sequencing Analysis
2.4. Untargeted Metabolomic Analysis
2.5. Carotenoids Quantitative Analysis
2.6. Quantitative Real-Time PCR Analysis
2.7. Statistical Analysis
3. Results and Discussion
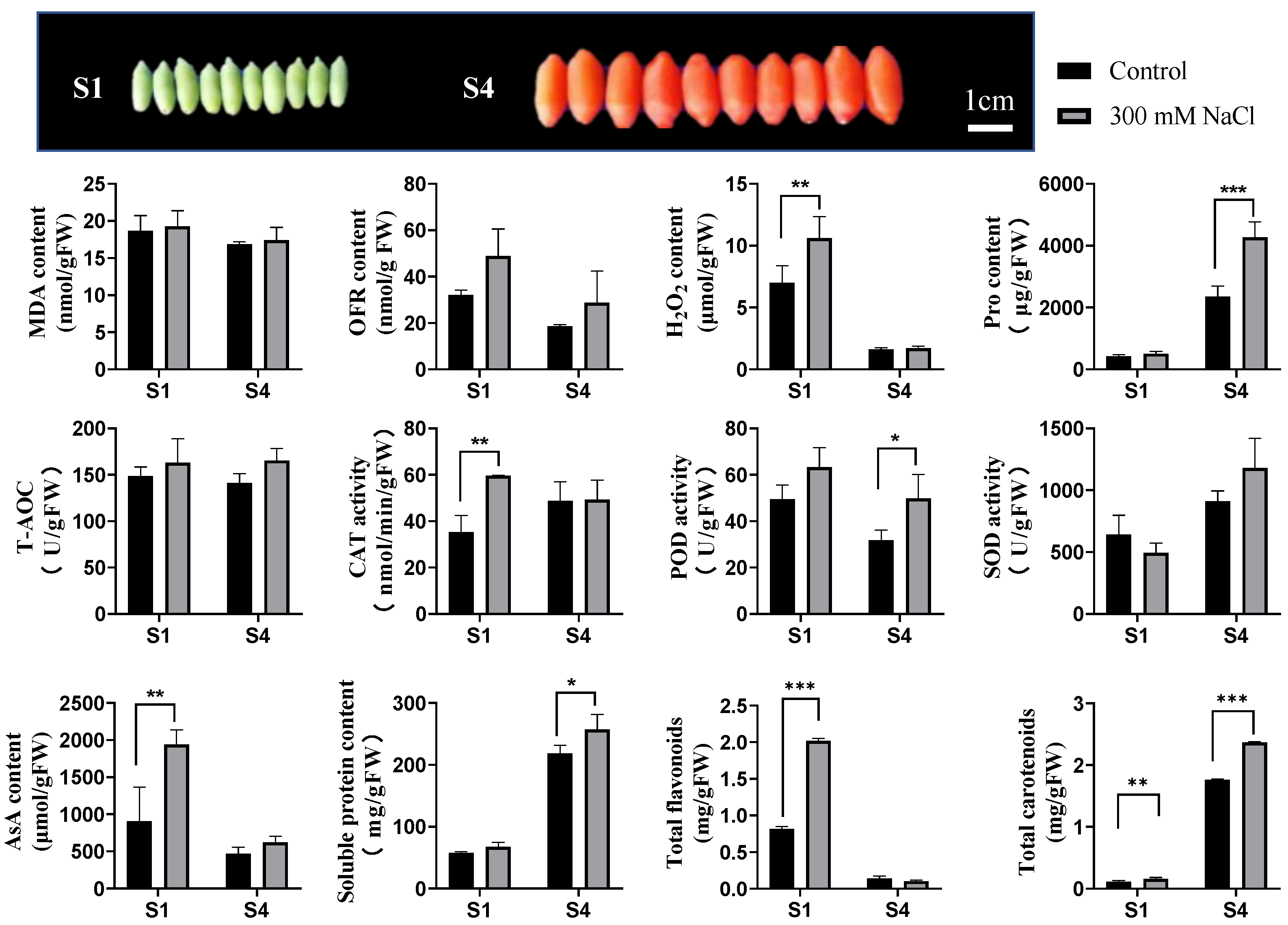
3.1. Physiological Response of LBF under Salt Stress
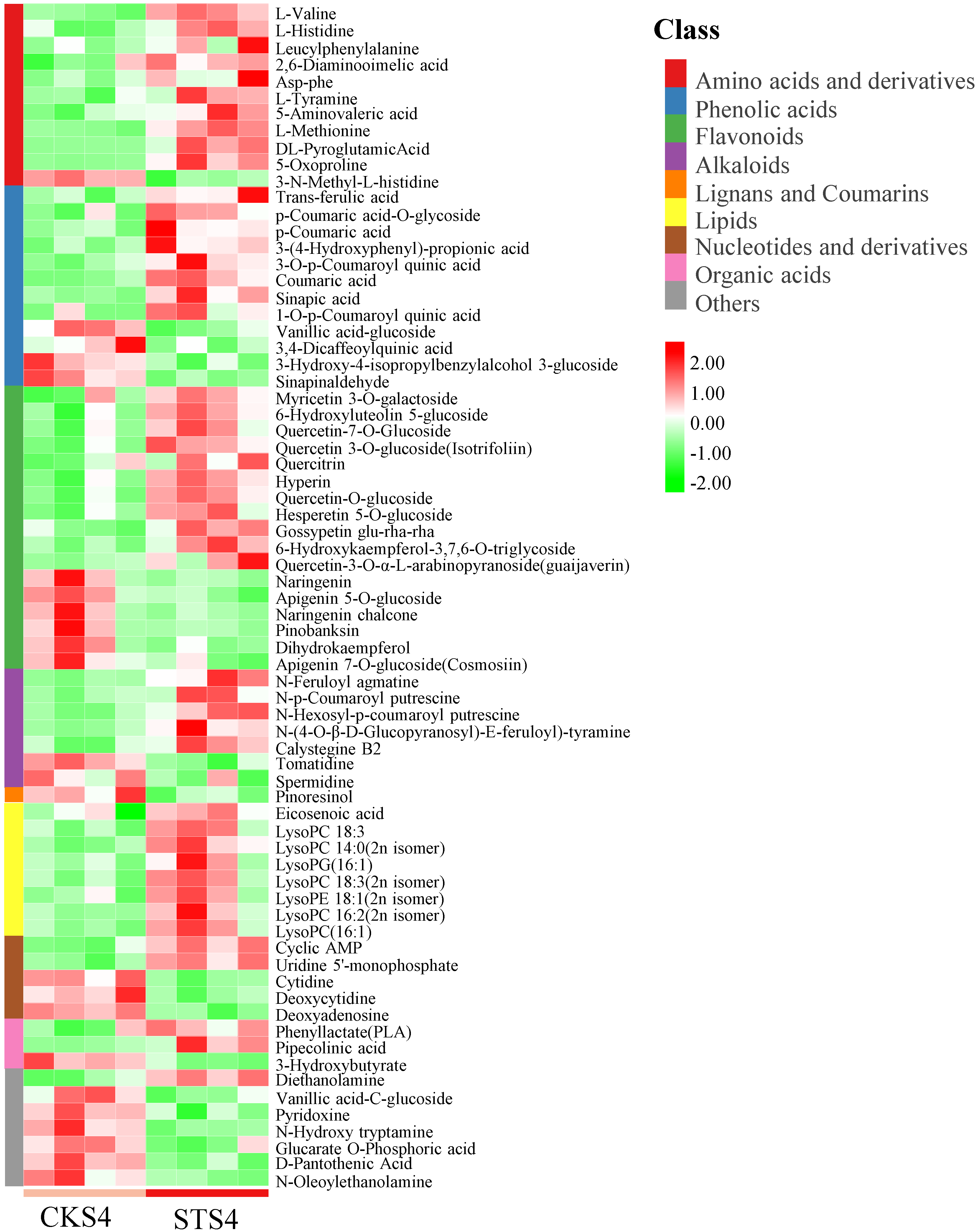
3.2. Non-Targeted and Targeted Metabolomic Analysis of CKS4 vs. STS4
3.3. Transcriptomic Analysis
3.4. Integrative Analysis of Transcriptome and Metabolome
3.5. Fatty Acid and Lipid Metabolisms
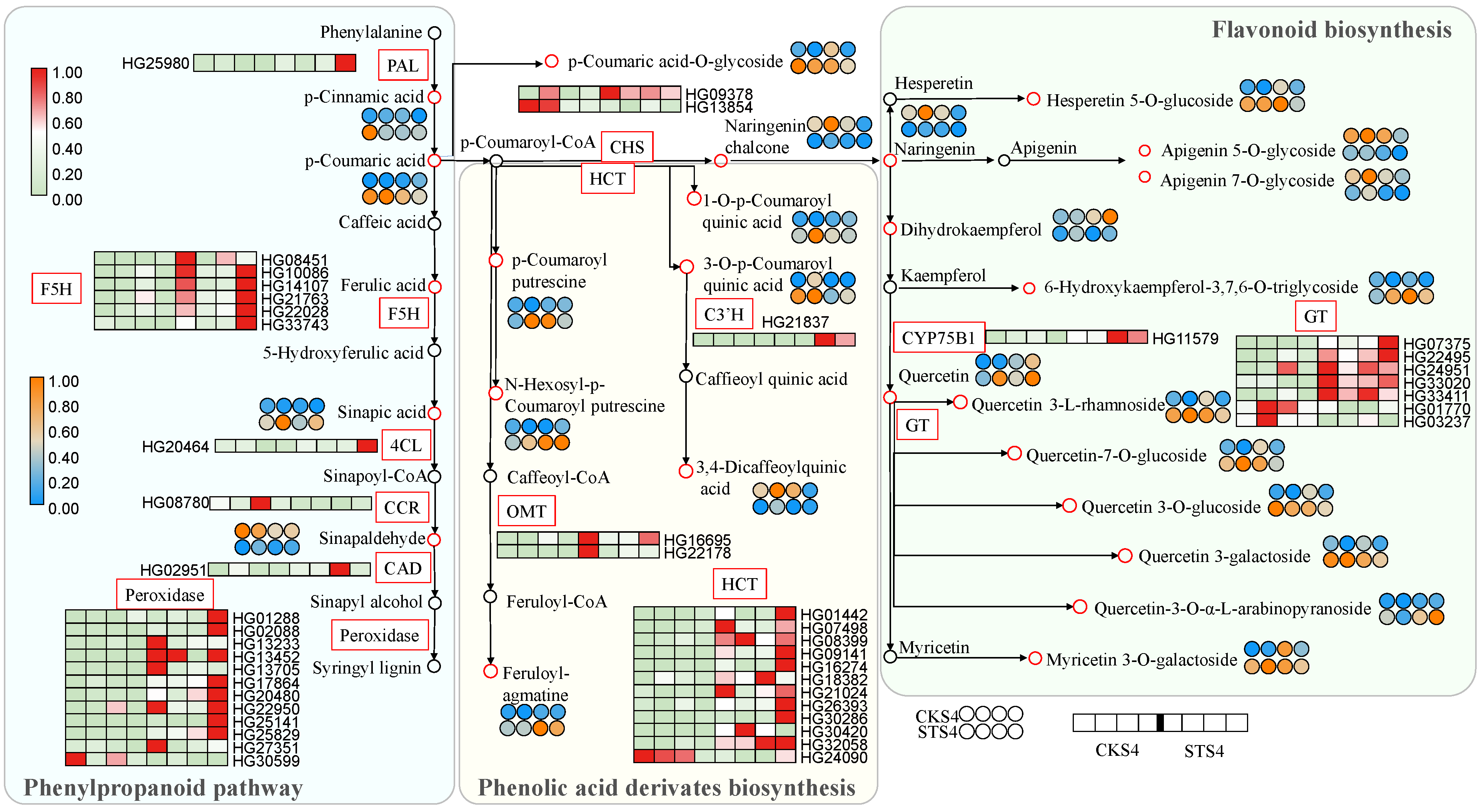
3.6. Flavonoid and Phenylpropanoid Pathways
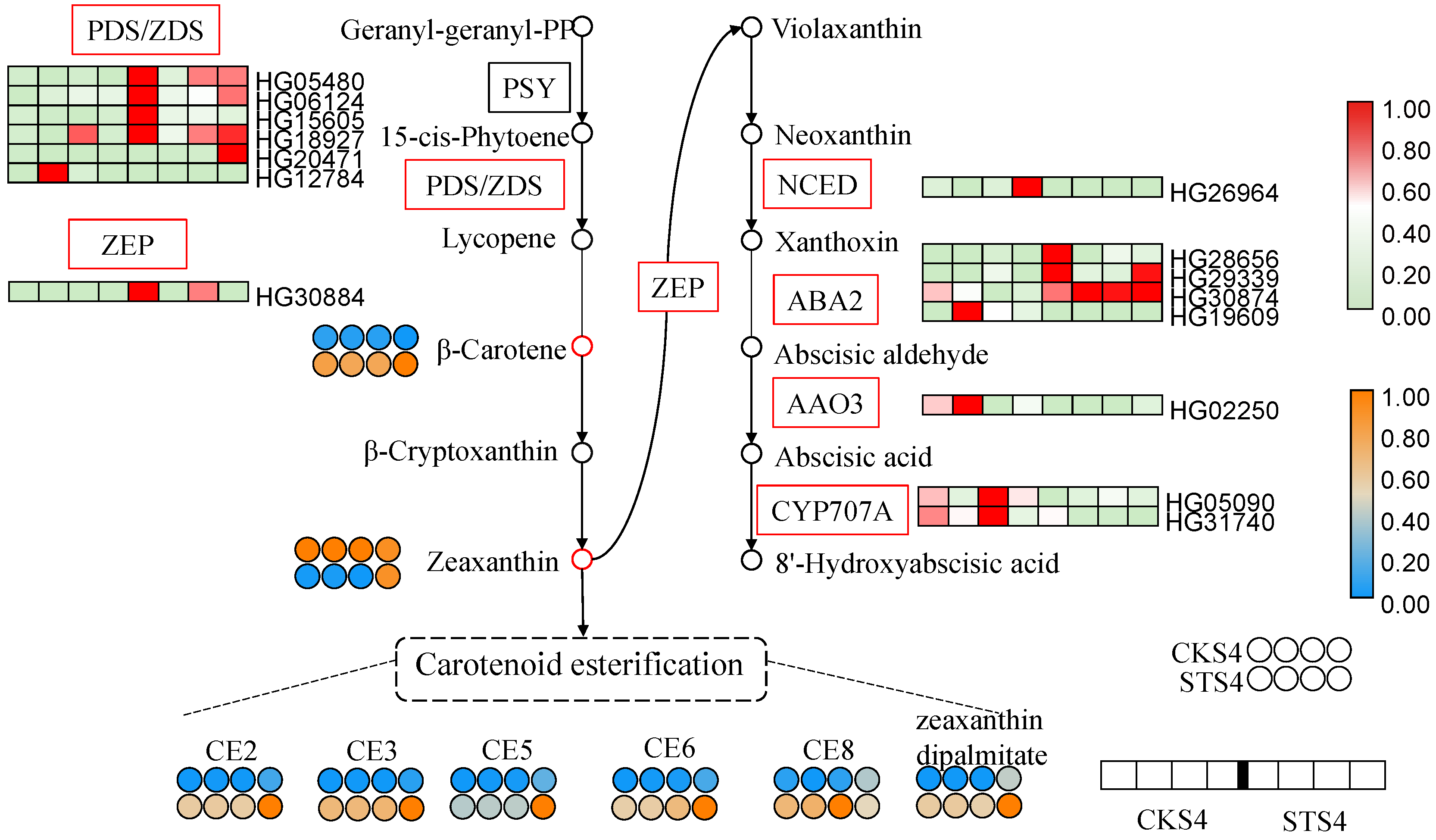
3.7. Carotenoid Biosynthetic Pathway
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaman, M.; Shahid, S.A.; Heng, L. Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Zorb, C.; Geilfus, C.M.; Dietz, K.J. Salinity and crop yield. Plant. Biol. 2019, 21, 31–38. [Google Scholar] [CrossRef]
- Ashraf, M. Stress-induced changes in wheat grain composition and quality. Crit. Rev. Food Sci. Nutr. 2014, 54, 1576–1583. [Google Scholar] [CrossRef]
- Rao, P.S.; Mishra, B.; Gupta, S.R. Effects of soil salinity and alkalinity on grain quality of tolerant, semi-tolerant and sensitive rice genotypes. Rice Sci. 2013, 20, 284–291. [Google Scholar] [CrossRef]
- Zheng, C.; Jiang, D.; Dai, T.; Jing, Q.; Cao, W. Effects of salt and waterlogging stress at post-anthesis stage on wheat grain yield and quality. Chin. J. Appl. Ecol. 2009, 20, 2391–12398. [Google Scholar]
- Borghesi, E.; Gonzalez-Miret, M.L.; Escudero-Gilete, M.L.; Malorgio, F.; Heredia, F.J.; Melendez-Martinez, A.J. Effects of salinity stress on carotenoids, anthocyanins, and color of diverse tomato genotypes. J. Agric. Food Chem. 2011, 59, 11676–11682. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Matsukura, C. Effect of Salt Stress on the Growth and Fruit Quality of Tomato Plants. In Abiotic Stress Biology in Horticultural Plants; Springer: Tokyo, Japan, 2015; pp. 3–16. [Google Scholar]
- Tang, H.; Zhang, X.; Gong, B.; Yan, Y.; Shi, Q. Proteomics and metabolomics analysis of tomato fruit at different maturity stages and under salt treatment. Food Chem. 2020, 311, 126009. [Google Scholar] [CrossRef]
- Kpinkoun, J.K.; Amoussa, A.M.; Mensah, A.C.G.; Komlan, F.A.; Kinsou, E.; Lagnika, L.; Gandonou, C.B. Effect of salt stress on flowering, fructification and fruit nutrients concentration in a local cultivar of chili pepper (Capsicum frutescens L.). Int. J. Plant. Physiol. Biochem. 2019, 11, 1–7. [Google Scholar]
- Abdallah, S.B.; Aung, B.; Lachaˆal, M.; Amyot, L.; Karray-Bouraoui, N.; Lalin, I.; Hannoufa, A. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016, 38, 1–13. [Google Scholar] [CrossRef]
- Fini, A.; Brunetti, C.; Ferdinando, M.D.; Ferrini, F.; Tattini, M. Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant. Signal. Behav. 2011, 6, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Gruszecki, W.I. Carotenoids in membranes. In The Photochemistry of Carotenoids; Kluwer Academic: Amsterdam, The Netherlands, 1999; pp. 363–379. [Google Scholar]
- Potterat, O. Goji (Lycium barbarum and L. chinense): Phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med. 2010, 76, 7–19. [Google Scholar] [CrossRef] [Green Version]
- Chang, R.C.; So, K.F. Use of anti-aging herbal medicine, Lycium barbarum, against aging-associated diseases. What do we know so far? Cell Mol. Neurobiol. 2008, 28, 643–652. [Google Scholar] [CrossRef]
- Zhang, Z.; He, K.; Zhang, T.; Tang, D.; Li, R.; Jia, S. Physiological responses of Goji berry (Lycium barbarum L.) to saline-alkaline soil from Qinghai region, China. Sci. Rep. 2019, 9, 12057. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, S.; Sun, W.; Wu, M.; Hu, W.; Shen, X.; Wang, Y. Comparative analysis of carotenoid accumulation in two goji (Lycium barbarum L. and L. ruthenicum Murr.) fruits. BMC Plant. Biol. 2014, 14, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartley, S.W.; Mullikin, J.C. QoRTs: A comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinform. 2015, 16, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant. Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid-A potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant. Sci. 2017, 8, 613. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; He, S.; Zhai, H.; Ren, Z.; Liu, N.; Song, X.; Zhang, D.; Liu, Q. IbSIMT1, a novel salt-induced methyltransferase gene from Ipomoea batatas, is involved in salt tolerance. Plant. Cell Tissue Organ. Cult. 2014, 120, 701–715. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L.C. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant. Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Zhang, Q.; Peng, Z.; Miao, X. Lipid droplets mediate salt stress tolerance in Parachlorella kessleri. Plant. Physiol. 2019, 181, 510–526. [Google Scholar] [CrossRef] [PubMed]
- Altintas, A.; Kosar, M.; Kirimer, N.; Baser, K.H.C.; Demirci, B. Composition of the essential oils of Lycium barbarum and L. ruthenicum fruits. Chem. Nat. Compd. 2006, 42, 24–25. [Google Scholar] [CrossRef]
- Mercadante, A.Z.; Rodrigues, D.B.; Petry, F.C.; Mariutti, L.R.B. Carotenoid esters in foods—A review and practical directions on analysis and occurrence. Food Res. Int. 2016, 99, 830–850. [Google Scholar] [CrossRef]
- Lippold, F.; Dorp, K.V.; Abraham, M.; Hölzl, G.; Wewer, V.; Yilmaz, J.L.; Lager, I.; Montandon, C.; Besagni, C.; Kessler, F.; et al. Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant. Cell 2012, 24, 2001–2014. [Google Scholar] [CrossRef] [Green Version]
- Ariizumi1, T.; Kishimoto, S.; Kakami, R.; Maoka, T.; Hirakawa, H.; Suzuki, Y.; Ozeki, Y.; Shirasawa, K.; Bernillon, S.; Okabe, Y.; et al. Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant. J. 2014, 79, 453–465. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Salama, K.H.A.; Allam, H.Y.H. Role of the plasma membrane in saline conditions: Lipids and proteins. Bot. Rev. 2015, 81, 416–451. [Google Scholar] [CrossRef]
- Dong, H.; Lu, C.; Deng, Y.; Zuo, J.; Mu, J.; Lu, Q.; Wang, Y.; Xu, Y.; Chu, C. The Arabidopsis Spontaneous Cell Death1 gene, encoding a zeta-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res. 2007, 17, 458–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellado-Ortega, E.; Hornero-Méndez, D. Isolation and identification of lutein esters, including their regioisomers, in tritordeum (xTritordeum Ascherson et Graebner) grains: Evidence for a preferential xanthophyll acyltransferase activity. Food Chem. 2012, 135, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Watkins, J.L.; Li, M.; McQuinn, R.P.; Chan, K.X.; McFarlane, H.E.; Ermakova, M.; Furbank, R.T.; Mares, D.; Dong, C.; Chalmers, K.J.; et al. A GDSL esterase/lipase catalyzes the esterification of lutein in bread wheat. Plant. Cell 2019, 31, 3092–3112. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Zeng, S.; A, B.; Yang, X.; Yang, T.; Zheng, G.; Mao, G.; Wang, Y. Integrative Analysis of Transcriptome and Metabolome Reveals Salt Stress Orchestrating the Accumulation of Specialized Metabolites in Lycium barbarum L. Fruit. Int. J. Mol. Sci. 2021, 22, 4414. https://doi.org/10.3390/ijms22094414
Lin S, Zeng S, A B, Yang X, Yang T, Zheng G, Mao G, Wang Y. Integrative Analysis of Transcriptome and Metabolome Reveals Salt Stress Orchestrating the Accumulation of Specialized Metabolites in Lycium barbarum L. Fruit. International Journal of Molecular Sciences. 2021; 22(9):4414. https://doi.org/10.3390/ijms22094414
Chicago/Turabian StyleLin, Shuang, Shaohua Zeng, Biao A, Xiaoman Yang, Tianshun Yang, Guoqi Zheng, Guilian Mao, and Ying Wang. 2021. "Integrative Analysis of Transcriptome and Metabolome Reveals Salt Stress Orchestrating the Accumulation of Specialized Metabolites in Lycium barbarum L. Fruit" International Journal of Molecular Sciences 22, no. 9: 4414. https://doi.org/10.3390/ijms22094414






