Prolactin-Releasing Peptide Differentially Regulates Gene Transcriptomic Profiles in Mouse Bone Marrow-Derived Macrophages
Abstract
:1. Introduction
- (a)
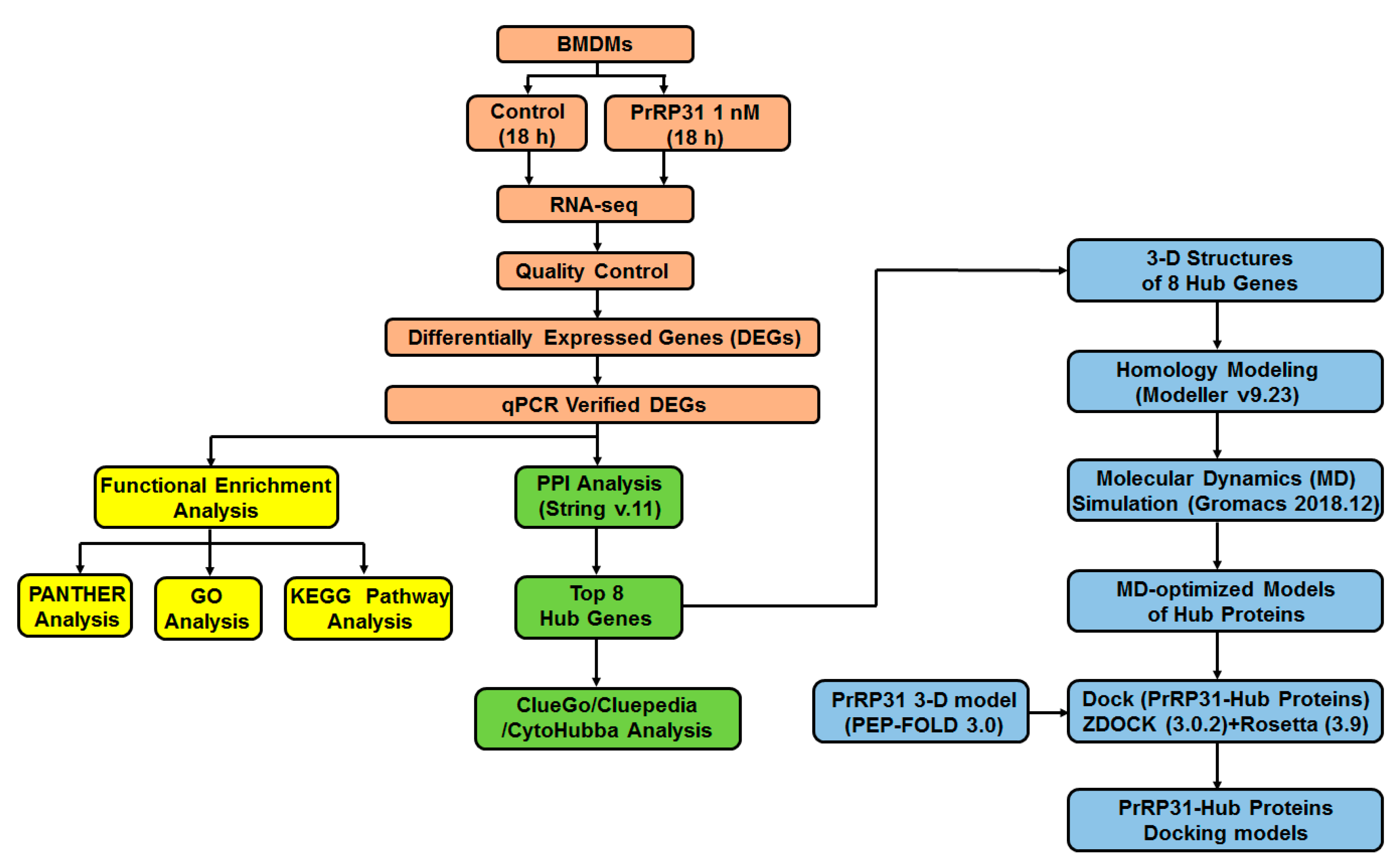
- PrRP31-triggered differentially expressed genes (DEGs) were acquired from murine bone marrow-derived macrophages (BMDMs). Cells treated with 1 nM PrRP31 (18 h) were detected by RNA-seq. A total of 410 DEGs were obtained. In addition, the influence of PrRP on the morphology of cells was observed with optical microscopy.
- (b)
- DEGs were analyzed by a series of bioinformatics approaches, including GO analysis, functional enrichment, and protein–protein interaction (PPI) studies. Next, eight hub genes were finally acquired for subsequent study.
- (c)
- The three-dimensional structures of hub proteins were studied. By using homology-modeling, the structure of proteins encoded by hub genes (hub proteins) was built. Subsequently, molecular dynamics simulations (at least 300 ns) were performed to capture the trajectory of hub proteins. Finally, molecular dynamics-optimized protein structures of hub proteins were obtained.
- (d)
- The docking models of PrRP and hub proteins were predicted with the peptide-protein docking module of the Rosetta program.
2. Results
2.1. PrRP Demonstrated No Significant Effect on the Shape of BMDMs
2.2. Identification of DEGs
2.3. Functional and Pathway Enrichment Analysis of DEGs
2.4. Protein–Protein Interaction (PPI) Network Analysis
2.5. Common Transcription Factors Tied to Genes Down-Regulated by PrRP
2.6. Verification of Hub Genes with qPCR and Western Blot
2.7. Protein Modeling of Hub Proteins
2.8. Molecular Dynamics Simulation of Hub Proteins
2.9. Peptide-Hub Protein Docking
2.10. Expression of GPR10 on BMDMs
3. Discussion
3.1. PrRP Modulated Different Functional Enrichment Pathways of BMDMs
3.2. Common Transcription Factors Tied to PrRP-Regulated DEGs in BMDMs
3.3. The Concentration of PrRP in the Experimental System for High-Throughput Sequencing
3.4. Expression of PrRP-GPR10 on BMDMs
3.5. Prolactin and Inflammatory Processes
3.6. Prolactin and Macrophages
3.7. PrRP and Microglia
3.8. Considerations of Double Positive Cells in the Control Group of Flow Cytometry Results
3.8.1. Non-Specific Fluorescent Signal Caused by Dead Cells
3.8.2. Non-Specific Fluorescent Signal from Cell Debris or Tiny Tissue Pieces
3.8.3. The Excitation Wavelength of Fluorescein Is the Same as the Emission Wavelength, Which Is a Wavelength Range, Rather Than a Specific Value
3.8.4. Non-Specific Fluorescence Signals Caused by BMDMs
3.9. The Limitations of Our Study
- (1)
- Hub genes responded to PrRP stimulation in protein levels might be late than in mRNA level. After PrRP (1 nM) treatment for 18 h, the mRNA levels of IFIT1 and USP18 were significantly increased. However, the influence of PrRP on the protein expression of hub genes might be longer than 18 h. In the process of gene expression (from mRNA to protein), it takes minutes to hours to translate mRNA into proteins [67].
- (2)
- PrRP may regulate the expression of the hub proteins of BMDMs in a variety of ways. Although PrRP (1 nM) treatment for 18 h significantly affected the expression of hub genes, this does not mean that PrRP will cause changes in hub proteins’ expression levels. In the step from mRNA to protein, a variety of post-transcriptional modifications can cause changes in protein levels, including mRNA degradation control, mRNA translocation control, protein degradation control, translation control, mRNA translocation control, and protein degradation control [67]. Therefore, these above links may be employed by PrRP to regulate the expression of the hub proteins of BMDMs.
- (3)
- PrRP may modulate the functions of the hub proteins of BMDMs in diverse manners. It is the function of proteins, rather than the protein expression, that plays a pivotal role in cellular activities [68]. Various biologically active molecules (including neuropeptides) may regulate each hub protein’s activity in a variety of ways, including phosphorylation, heterogeneous regulation, covalent modification, etc. [67]. However, this study’s focus is to investigate the effect of PrRP on the transcriptomic gene expression of BMDMs, which may provide clues for the subsequent exploration of the functional regulation of PrRP on BMDMs. Therefore, follow-up studies on the complex biological functions of PrRP on BMDMs are worth looking in to.
- (4)
- The regulation of PrRP on the transcriptome gene expression of BMDMs may be more complicated than we previously assumed. Since proteins and various protein–protein interaction (PPI) networks are prominent members that play an essential role in regulating various biological processes in cells, the influence of neuropeptide PrRP on BMDMs may be a complicated process. In the same vein, emerging data have indicated that PrRP may have a wide range of effects in regulating the neuroendocrine system [40,69].
4. Materials and Methods
4.1. Ethical Statement
4.2. Animals
4.3. Reagents
4.4. BMDM Preparation
4.5. RNA-seq Sample Preparation and Microscope Detection
4.6. Flow Cytometry Detection
4.7. RNA-seq Sample Collection and Preparation
4.7.1. RNA Quantification and Qualification
4.7.2. Library Construction for RNA Sequencing
4.7.3. Sequencing and Clustering
4.8. RNA-seq Data Interpretation
4.8.1. Quality Control, Mapping Reads to the Reference Genome and Quantification of Gene Expression
4.8.2. Differential Expression Interpretation
4.8.3. GO and KEGG Enrichment Interpretation
4.8.4. Protein-Protein Interaction (PPI) Network Analysis
4.9. qPCR Analysis
4.10. Western Blot Detection
4.11. Immunofluorescence Stain
4.12. Homology Modeling of Proteins
4.13. Molecular Dynamics (MD) Simulation
4.13.1. Molecular Dynamic Simulation: Protein in Water
4.13.2. Molecular Dynamic Simulation: Analysis
4.14. Dock
- (1)
- (2)
- The primary docking complexes were sent to the Flexible peptide docking module of the Rosetta program (3.9) for subsequent docking. (A) Pre-pack mode: 1 model was constructed. (B) Low-resolution ab-initio analysis: 100 models were obtained, and 1 model with the best docking score was chosen for subsequent analysis. (C) Refinement analysis: 100 docking models were acquired, and a docking model was finally selected based on the total_score.
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Welch, S.K.; O’Hara, B.F.; Kilduff, T.S.; Heller, H.C. Sequence and tissue distribution of a candidate G-coupled receptor cloned from rat hypothalamus. Biochem. Biophys. Res. Commun. 1995, 209, 606–613. [Google Scholar] [CrossRef]
- Hinuma, S.; Habata, Y.; Fujii, R.; Kawamata, Y.; Hosoya, M.; Fukusumi, S.; Kitada, C.; Masuo, Y.; Asano, T.; Matsumoto, H.; et al. A prolactin-releasing peptide in the brain. Nature 1998, 393, 272–276. [Google Scholar] [CrossRef]
- Fujimoto, M.; Takeshita, K.; Wang, X.; Takabatake, I.; Fujisawa, Y.; Teranishi, H.; Ohtani, M.; Muneoka, Y.; Ohta, S. Isolation and characterization of a novel bioactive peptide, Carassius RFamide (C-RFa), from the brain of the Japanese crucian carp. Biochem. Biophys. Res. Commun. 1998, 242, 436–440. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.Y.; Wu, Y.; Huang, G.; Li, J.; Leung, F.C. Identification of the receptors for prolactin-releasing peptide (PrRP) and Carassius RFamide peptide (C-RFa) in chickens. Endocrinology 2012, 153, 1861–1874. [Google Scholar] [CrossRef]
- Marchese, A.; Heiber, M.; Nguyen, T.; Heng, H.H.; Saldivia, V.R.; Cheng, R.; Murphy, P.M.; Tsui, L.C.; Shi, X.; Gregor, P.; et al. Cloning and chromosomal mapping of three novel genes, GPR9, GPR10, and GPR14, encoding receptors related to interleukin 8, neuropeptide Y, and somatostatin receptors. Genomics 1995, 29, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Roland, B.L.; Sutton, S.W.; Wilson, S.J.; Luo, L.; Pyati, J.; Huvar, R.; Erlander, M.G.; Lovenberg, T.W. Anatomical distribution of prolactin-releasing peptide and its receptor suggests additional functions in the central nervous system and periphery. Endocrinology 1999, 140, 5736–5745. [Google Scholar] [CrossRef]
- Yang, H.Y.; Tao, T.; Iadarola, M.J. Modulatory role of neuropeptide FF system in nociception and opiate analgesia. Neuropeptides 2008, 42, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, C.B.; Celsi, F.; Brennand, J.; Luckman, S.M. Alternative role for prolactin-releasing peptide in the regulation of food intake. Nat. Neurosci. 2000, 3, 645–646. [Google Scholar] [CrossRef]
- Ellacott, K.L.; Lawrence, C.B.; Pritchard, L.E.; Luckman, S.M. Repeated administration of the anorectic factor prolactin-releasing peptide leads to tolerance to its effects on energy homeostasis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 285, R1005–R1010. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, C.B.; Liu, Y.L.; Stock, M.J.; Luckman, S.M. Anorectic actions of prolactin-releasing peptide are mediated by corticotropin-releasing hormone receptors. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 286, R101–R107. [Google Scholar] [CrossRef]
- Maletínská, L.; Popelová, A.; Železná, B.; Bencze, M.; Kuneš, J. The impact of anorexigenic peptides in experimental models of Alzheimer’s disease pathology. J. Endocrinol. 2019, 240, R47–R72. [Google Scholar] [CrossRef] [Green Version]
- Maniscalco, J.W.; Rinaman, L. Interoceptive modulation of neuroendocrine, emotional, and hypophagic responses to stress. Physiol. Behav. 2017, 176, 195–206. [Google Scholar] [CrossRef]
- Romero, A.; Manríquez, R.; Alvarez, C.; Gajardo, C.; Vásquez, J.; Kausel, G.; Monrás, M.; Olavarría, V.H.; Yáñez, A.; Enríquez, R.; et al. Prolactin-releasing peptide is a potent mediator of the innate immune response in leukocytes from Salmo salar. Vet. Immunol. Immunopathol. 2012, 147, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kuang, Y.; Zuo, Z.; Zhang, J.; Ma, X.; Xing, X.; Liu, L.; Miao, Y.; Ren, T.; Li, H.; et al. Cellular processes involved in RAW 264.7 macrophages exposed to NPFF: A transcriptional study. Peptides 2021, 136, 170469. [Google Scholar] [CrossRef] [PubMed]
- Waqas, S.F.H.; Hoang, A.C.; Lin, Y.-T.; Ampem, G.; Azegrouz, H.; Balogh, L.; Thuróczy, J.; Chen, J.-C.; Gerling, I.C.; Nam, S.; et al. Neuropeptide FF increases M2 activation and self-renewal of adipose tissue macrophages. J. Clin. Investig. 2017, 127, 2842–2854. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Pidugu, V.K.; Pidugu, H.B.; Wu, M.M.; Liu, C.J.; Lee, T.C. Emerging Functions of Human IFIT Proteins in Cancer. Front. Mol. Biosci. 2019, 6, 148. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Ghosh, A.; Sarkar, S.N. OASL-a new player in controlling antiviral innate immunity. Curr. Opin. Virol. 2015, 12, 15–19. [Google Scholar] [CrossRef] [Green Version]
- Ciancanelli, M.J.; Abel, L.; Zhang, S.Y.; Casanova, J.L. Host genetics of severe influenza: From mouse Mx1 to human IRF7. Curr. Opin. Immunol. 2016, 38, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Fensterl, V.; Sen, G.C. Interferon-induced Ifit proteins: Their role in viral pathogenesis. J. Virol. 2015, 89, 2462–2468. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.A.; Jeon, Y.J. Emerging Roles of USP18: From Biology to Pathophysiology. Int. J. Mol. Sci. 2020, 21, 6825. [Google Scholar] [CrossRef]
- Cheluvappa, R. Identification of New Potential Therapies for Colitis Amelioration Using an Appendicitis-Appendectomy Model. Inflamm. Bowel Dis. 2019, 25, 436–444. [Google Scholar] [CrossRef]
- Fujita, W. The Possible Role of MOPr-DOPr Heteromers and Its Regulatory Protein RTP4 at Sensory Neurons in Relation to Pain Perception. Front. Cell. Neurosci. 2020, 14, 609362. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Hayamizu, T.F.; Finger, J.H.; Bello, S.M.; McCright, I.J.; Xu, J.; Baldarelli, R.M.; Beal, J.S.; Campbell, J.; Corbani, L.E.; et al. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 2019, 47, D774–D779. [Google Scholar] [CrossRef] [Green Version]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wall, K.D.; Olivos, D.R.; Rinaman, L. High Fat Diet Attenuates Cholecystokinin-Induced cFos Activation of Prolactin-Releasing Peptide-Expressing A2 Noradrenergic Neurons in the Caudal Nucleus of the Solitary Tract. Neuroscience 2020, 447, 113–121. [Google Scholar] [CrossRef]
- Davis, X.S.; Grill, H.J. The hindbrain is a site of energy balance action for prolactin-releasing peptide: Feeding and thermic effects from GPR10 stimulation of the nucleus tractus solitarius/area postrema. Psychopharmacology 2018, 235, 2287–2301. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, Y.; Matsumoto, H.; Nakata, M.; Mera, T.; Fukusumi, S.; Hinuma, S.; Ueta, Y.; Yada, T.; Leng, G.; Onaka, T. Endogenous prolactin-releasing peptide regulates food intake in rodents. J. Clin. Investig. 2008, 118, 4014–4024. [Google Scholar] [CrossRef] [Green Version]
- Bjursell, M.; Lennerås, M.; Göransson, M.; Elmgren, A.; Bohlooly, Y.M. GPR10 deficiency in mice results in altered energy expenditure and obesity. Biochem. Biophys. Res. Commun. 2007, 363, 633–638. [Google Scholar] [CrossRef]
- Lawrence, C.B.; Ellacott, K.L.; Luckman, S.M. PRL-releasing peptide reduces food intake and may mediate satiety signaling. Endocrinology 2002, 143, 360–367. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Yu, H.; Dilbaz, S.; Coßmann, J.; Hoang, A.C.; Diedrich, V.; Herwig, A.; Harauma, A.; Hoshi, Y.; Moriguchi, T.; Landgraf, K.; et al. Breast milk alkylglycerols sustain beige adipocytes through adipose tissue macrophages. J. Clin. Investig. 2019, 129, 2485–2499. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, H.; Murakami, Y.; Horikoshi, Y.; Noguchi, J.; Habata, Y.; Kitada, C.; Hinuma, S.; Onda, H.; Fujino, M. Distribution and characterization of immunoreactive prolactin-releasing peptide (PrRP) in rat tissue and plasma. Biochem. Biophys. Res. Commun. 1999, 257, 264–268. [Google Scholar] [CrossRef]
- Fujii, R.; Fukusumi, S.; Hosoya, M.; Kawamata, Y.; Habata, Y.; Hinuma, S.; Sekiguchi, M.; Kitada, C.; Kurokawa, T.; Nishimura, O.; et al. Tissue distribution of prolactin-releasing peptide (PrRP) and its receptor. Regul. Pept. 1999, 83, 1–10. [Google Scholar] [CrossRef]
- Nieminen, M.L.; Brandt, A.; Pietilä, P.; Panula, P. Expression of mammalian RF-amide peptides neuropeptide FF (NPFF), prolactin-releasing peptide (PrRP) and the PrRP receptor in the peripheral tissues of the rat. Peptides 2000, 21, 1695–1701. [Google Scholar] [CrossRef]
- Matafome, P.; Seiça, R. The Role of Brain in Energy Balance. Adv. Neurobiol. 2017, 19, 33–48. [Google Scholar]
- Prazienkova, V.; Funda, J.; Pirnik, Z.; Karnosova, A.; Hruba, L.; Korinkova, L.; Neprasova, B.; Janovska, P.; Benzce, M.; Kadlecova, M.; et al. GPR10 gene deletion in mice increases basal neuronal activity, disturbs insulin sensitivity and alters lipid homeostasis. Gene 2021, 774, 145427. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S. Peptides and Peptidomimetics as Potential Antiobesity Agents: Overview of Current Status. Front. Nutr. 2019, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Ibata, Y.; Iijima, N.; Kataoka, Y.; Kakihara, K.; Tanaka, M.; Hosoya, M.; Hinuma, S. Morphological survey of prolactin-releasing peptide and its receptor with special reference to their functional roles in the brain. Neurosci. Res. 2000, 38, 223–230. [Google Scholar] [CrossRef]
- Pražienková, V.; Popelová, A.; Kuneš, J.; Maletínská, L. Prolactin-Releasing Peptide: Physiological and Pharmacological Properties. Int. J. Mol. Sci. 2019, 20, 5297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borba, V.V.; Zandman-Goddard, G.; Shoenfeld, Y. Prolactin and autoimmunity: The hormone as an inflammatory cytokine. Best Pr. Res. Clin. Endocrinol. Metab. 2019, 33, 101324. [Google Scholar] [CrossRef] [PubMed]
- Brand, J.M.; Frohn, C.; Cziupka, K.; Brockmann, C.; Kirchner, H.; Luhm, J. Prolactin triggers pro-inflammatory immune responses in peripheral immune cells. Eur. Cytokine Netw. 2004, 15, 99–104. [Google Scholar] [PubMed]
- Shelly, S.; Boaz, M.; Orbach, H. Prolactin and autoimmunity. Autoimmun. Rev. 2012, 11, A465–A470. [Google Scholar] [CrossRef]
- Borba, V.V.; Zandman-Goddard, G.; Shoenfeld, Y. Prolactin and Autoimmunity. Front. Immunol 2018, 9, 73. [Google Scholar] [CrossRef]
- Costanza, M.; Binart, N.; Steinman, L.; Pedotti, R. Prolactin: A versatile regulator of inflammation and autoimmune pathology. Autoimmun. Rev. 2015, 14, 223–230. [Google Scholar] [CrossRef]
- Carvalho-Freitas, M.I.; Anselmo-Franci, J.A.; Palermo-Neto, J.; Felicio, L.F. Prior reproductive experience alters prolactin-induced macrophage responses in pregnant rats. J. Reprod. Immunol. 2013, 99, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.W.; Garcia, S.; Malvar Fernandez, B.; Gerlag, D.M.; Tak, P.P.; Reedquist, K.A. Rheumatoid arthritis and psoriatic arthritis synovial fluids stimulate prolactin production by macrophages. J. Leukoc. Biol. 2017, 102, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Sodhi, A. Prolactin-induced production of cytokines in macrophages in vitro involves JAK/STAT and JNK MAPK pathways. Int. Immunol. 2008, 20, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Tang, M.W.; Garcia, S.; Gerlag, D.M.; Reedquist, K.A.; Tak, P.P. Prolactin Is Locally Produced in the Synovium of Patients with Inflammatory Arthritic Diseases and Promotes Macrophage Activation. Arthritis Rheumatol. 2014, 66, S526. [Google Scholar] [CrossRef]
- Tang, M.W.; Garcia, S.; Gerlag, D.M.; Tak, P.P.; Reedquist, K.A. Insight into the Endocrine System and the Immune System: A Review of the Inflammatory Role of Prolactin in Rheumatoid Arthritis and Psoriatic Arthritis. Front. Immunol 2017, 8, 720. [Google Scholar] [CrossRef] [Green Version]
- Orbach, H.; Zandman-Goddard, G.; Amital, H.; Barak, V.; Szekanecz, Z.; Szucs, G.; Danko, K.; Nagy, E.; Csepany, T.; Carvalho, J.F.; et al. Novel biomarkers in autoimmune diseases: Prolactin, ferritin, vitamin D, and TPA levels in autoimmune diseases. Ann. N. Y. Acad. Sci. 2007, 1109, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Holubová, M.; Hrubá, L.; Popelová, A.; Bencze, M.; Pražienková, V.; Gengler, S.; Kratochvílová, H.; Haluzík, M.; Železná, B.; Kuneš, J.; et al. Liraglutide and a lipidized analog of prolactin-releasing peptide show neuroprotective effects in a mouse model of β-amyloid pathology. Neuropharmacology 2019, 144, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Lim, S.; Hoeffel, G.; Low, D.; Huber, T. Origin and differentiation of microglia. Front. Cell. Neurosci. 2013, 7, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filiano, A.J.; Gadani, S.P.; Kipnis, J. Interactions of innate and adaptive immunity in brain development and function. Brain Res. 2015, 1617, 18–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushong, E.A.; Martone, M.E.; Jones, Y.Z.; Ellisman, M.H. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J. Neurosci. 2002, 22, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Kreutzberg, G.W. Microglia, the first line of defence in brain pathologies. Arzneimittelforschung 1995, 45, 357–360. [Google Scholar]
- Noubade, R.; Wong, K.; Ota, N.; Rutz, S.; Eidenschenk, C.; Valdez, P.A.; Ding, J.; Peng, I.; Sebrell, A.; Caplazi, P.; et al. NRROS negatively regulates reactive oxygen species during host defence and autoimmunity. Nature 2014, 509, 235–239. [Google Scholar] [CrossRef]
- Rao, P.; Hayden, M.S.; Long, M.; Scott, M.L.; West, A.P.; Zhang, D.; Oeckinghaus, A.; Lynch, C.; Hoffmann, A.; Baltimore, D.; et al. IkappaBbeta acts to inhibit and activate gene expression during the inflammatory response. Nature 2010, 466, 1115–1119. [Google Scholar] [CrossRef] [Green Version]
- Weischenfeldt, J.; Porse, B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH Protoc. 2008, 2008, pdb.prot5080. [Google Scholar] [CrossRef] [Green Version]
- Perfetto, S.P.; Chattopadhyay, P.K.; Lamoreaux, L.; Nguyen, R.; Ambrozak, D.; Koup, R.A.; Roederer, M. Amine reactive dyes: An effective tool to discriminate live and dead cells in polychromatic flow cytometry. J. Immunol. Methods 2006, 313, 199–208. [Google Scholar] [CrossRef]
- Hulspas, R.; O’Gorman, M.R.; Wood, B.L.; Gratama, J.W.; Sutherland, D.R. Considerations for the control of background fluorescence in clinical flow cytometry. Cytom. B Clin. Cytom. 2009, 76, 355–364. [Google Scholar] [CrossRef]
- Maecker, H.T.; Trotter, J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytom. A 2006, 69, 1037–1042. [Google Scholar] [CrossRef]
- Tung, J.W.; Heydari, K.; Tirouvanziam, R.; Sahaf, B.; Parks, D.R.; Herzenberg, L.A.; Herzenberg, L.A. Modern flow cytometry: A practical approach. Clin. Lab. Med. 2007, 27, 453–468. [Google Scholar] [CrossRef] [Green Version]
- Tung, J.W.; Parks, D.R.; Moore, W.A.; Herzenberg, L.A.; Herzenberg, L.A. New approaches to fluorescence compensation and visualization of FACS data. Clin. Immunol. 2004, 110, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Roederer, M. Spectral compensation for flow cytometry: Visualization artifacts, limitations, and caveats. Cytometry 2001, 45, 194–205. [Google Scholar] [CrossRef]
- Hristov, M.; Schmitz, S.; Schuhmann, C.; Leyendecker, T.; von Hundelshausen, P.; Krötz, F.; Sohn, H.Y.; Nauwelaers, F.A.; Weber, C. An optimized flow cytometry protocol for analysis of angiogenic monocytes and endothelial progenitor cells in peripheral blood. Cytom. A 2009, 75, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Bray, D.; Hopkin, K.; Johnson, A.; Lewis, J.; Raff, M.C.; Roberts, K.; Walter, P. Essential Cell Biology; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Wu, L.; Candille, S.I.; Choi, Y.; Xie, D.; Jiang, L.; Li-Pook-Than, J.; Tang, H.; Snyder, M. Variation and genetic control of protein abundance in humans. Nature 2013, 499, 79–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zmeskalova, A.; Popelova, A.; Exnerova, A.; Zelezna, B.; Kunes, J.; Maletinska, L. Cellular Signaling and Anti-Apoptotic Effects of Prolactin-Releasing Peptide and Its Analog on SH-SY5Y Cells. Int. J. Mol. Sci. 2020, 21, 6343. [Google Scholar] [CrossRef]
- Davidson, E.H.; Erwin, D.H. Gene regulatory networks and the evolution of animal body plans. Science 2006, 311, 796–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Matteucci, C.; Argaw-Denboba, A.; Balestrieri, E.; Giovinazzo, A.; Miele, M.; D’Agostini, C.; Pica, F.; Grelli, S.; Paci, M.; Mastino, A.; et al. Deciphering cellular biological processes to clinical application: A new perspective for Talpha1 treatment targeting multiple diseases. Expert Opin. Biol. Ther. 2018, 18 (Suppl. 1), 23–31. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Hwang, S.H.; Cho, H.S.; Lee, M. Differential Gene Expression Common to Acquired and Intrinsic Resistance to BRAF Inhibitor Revealed by RNA-Seq Analysis. Biomol. Ther. 2019, 27, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wu, M.; Lin, Q.; Wang, S.; Chen, T.; Jiang, H. Key genes and integrated modules in hematopoietic differentiation of human embryonic stem cells: A comprehensive bioinformatic analysis. Stem Cell Res. Ther. 2018, 9, 301. [Google Scholar] [CrossRef]
- Sun, Y.L.; Chen, Z.H.; Chen, X.H.; Yin, C.; Li, D.J.; Ma, X.L.; Zhao, F.; Zhang, G.; Shang, P.; Qian, A.R. Diamagnetic Levitation Promotes Osteoclast Differentiation from RAW264.7 Cells. IEEE Trans. Biomed. Eng. 2015, 62, 900–908. [Google Scholar] [CrossRef]
- Sun, Y.L.; Sun, T.; Zhang, X.Y.; He, N.; Zhuang, Y.; Li, J.Y.; Fang, Q.; Wang, K.R.; Wang, R. NPFF2 Receptor is Involved in the Modulatory Effects of Neuropeptide FF for Macrophage Cell Line. Protein Pept. Lett. 2014, 21, 490–502. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Sun, Y.L.; Zhang, X.Y.; Sun, T.; He, N.; Li, J.Y.; Zhuang, Y.; Zeng, Q.; Yu, J.; Fang, Q.; Wang, R. The anti-inflammatory potential of neuropeptide FF in vitro and in vivo. Peptides 2013, 47, 124–132. [Google Scholar] [CrossRef]
- Raman, S.; Vernon, R.; Thompson, J.; Tyka, M.; Sadreyev, R.; Pei, J.; Kim, D.; Kellogg, E.; DiMaio, F.; Lange, O.; et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 2009, 77 (Suppl. 9), 89–99. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; DiMaio, F.; Wang, R.Y.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-resolution comparative modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., 3rd; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E.J.S. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Hourai, Y.; Weng, Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE 2011, 6, e24657. [Google Scholar] [CrossRef] [PubMed]
- Mintseris, J.; Pierce, B.; Wiehe, K.; Anderson, R.; Chen, R.; Weng, Z. Integrating statistical pair potentials into protein complex prediction. Proteins 2007, 69, 511–520. [Google Scholar] [CrossRef] [PubMed]




























| Gene_Symbol | log2FoldChange | Padj | Gene_Chrosome | Change |
|---|---|---|---|---|
| Entpd4b | −3.707639059 | 2.19 × 10−2 | 14 | Down |
| Hspb7 | −3.272825898 | 1.68 × 10−2 | 4 | Down |
| Dclk2 | −2.383273176 | 4.25 × 10−2 | 3 | Down |
| Slc7a15 | −2.101674471 | 2.79 × 10−2 | 12 | Down |
| Nanos1 | −1.666406569 | 6.68 × 10−2 | 19 | Down |
| Dach1 | −1.613454215 | 1.72 × 10−3 | 14 | Down |
| Srpk3 | −1.591562463 | 6.24 × 10−3 | X | Down |
| Slc6a4 | −1.584727023 | 3.21 × 10−2 | 11 | Down |
| Nptx1 | −1.450738194 | 3.30 × 10−6 | 11 | Down |
| Smpd3 | −1.391289481 | 7.65 × 10−4 | 8 | Down |
| Pgm5 | −1.31912871 | 4.92 × 10−2 | 19 | Down |
| Hmga2 | −1.300711982 | 2.19 × 10−14 | 10 | Down |
| Fam78b | −1.285355042 | 1.93 × 10−3 | 1 | Down |
| Cmbl | −1.256341752 | 2.78 × 10−2 | 15 | Down |
| Chst3 | −1.22498333 | 1.07 × 10−3 | 10 | Down |
| Slco2b1 | −1.20549131 | 1.27 × 10−4 | 7 | Down |
| Klhl3 | −1.173243947 | 1.22 × 10−2 | 13 | Down |
| Slc24a3 | −1.163460987 | 1.40 × 10−4 | 2 | Down |
| Prokr1 | −1.100130971 | 9.67 × 10−4 | 6 | Down |
| Efr3b | −1.063158734 | 5.39 × 10−13 | 12 | Down |
| Sox4 | −1.052753293 | 2.51 × 10−2 | 13 | Down |
| Fabp7 | −1.049413652 | 1.71 × 10−2 | 10 | Down |
| Etl4 | −1.039530137 | 1.92 × 10−3 | 2 | Down |
| Angptl2 | −1.033436675 | 1.59 × 10−23 | 2 | Down |
| Saa3 | 6.725187234 | 1.04 × 10−272 | 7 | Up |
| Cfb | 6.339511013 | 1.73 × 10−5 | 17 | Up |
| Lad1 | 6.049625834 | 8.45 × 10−5 | 1 | Up |
| Iigp1 | 5.288407326 | 5.88 × 10−35 | 18 | Up |
| Cacng8 | 5.269289134 | 1.18 × 10−4 | 7 | Up |
| Ly6i | 5.021584581 | 5.25 × 10−41 | 15 | Up |
| Serpina3g | 4.932160399 | 1.43 × 10−11 | 12 | Up |
| Cdkn2a | 4.865779496 | 9.13 × 10−3 | 4 | Up |
| Ptx3 | 4.863346866 | 8.39 × 10−3 | 3 | Up |
| Acod1 | 4.818905659 | 2.31 × 10−292 | 14 | Up |
| Adgre4 | 4.767373764 | 1.20 × 10−94 | 17 | Up |
| Il1b | 4.68955843 | 1.37 × 10−46 | 2 | Up |
| Ly6a | 4.571707887 | 8.85 × 10−71 | 15 | Up |
| Lpar1 | 4.533771177 | 4.00 × 10−6 | 4 | Up |
| Fpr1 | 4.518260903 | 7.01 × 10−59 | 17 | Up |
| Cxcl10 | 4.51118115 | 8.99 × 10−41 | 5 | Up |
| Ccl5 | 4.470370636 | 2.30 × 10−128 | 11 | Up |
| Gbp4 | 4.398911115 | 4.55 × 10−11 | 5 | Up |
| Marco | 4.320725477 | 6.79 × 10−122 | 1 | Up |
| Fpr2 | 4.318699393 | 5.20 × 10−73 | 17 | Up |
| Ly6c2 | 4.262622986 | 2.41 × 10−27 | 15 | Up |
| Serpina3f | 4.249141119 | 3.35 × 10−5 | 12 | Up |
| Ppm1n | 4.157670609 | 4.40 × 10−16 | 7 | Up |
| Apol9b | 4.156227055 | 1.44 × 10−2 | 15 | Up |
| Gene_Symbol | log2FoldChange | Padj | Gene_Chrosome | Change |
|---|---|---|---|---|
| Gm45820 | −1.686832907 | 2.71 × 10−2 | 8 | Down |
| Gm28800 | −1.563748269 | 2.10 × 10−2 | 1 | Down |
| Dnmt3aos | −1.056605119 | 2.92 × 10−5 | 12 | Down |
| Gm13822 | 4.654103949 | 2.29 × 10−2 | 5 | Up |
| AC113595.1 | 1.919370036 | 7.44 × 10−4 | 15 | Up |
| Gm11772 | 1.832830691 | 1.76 × 10−4 | 11 | Up |
| Gene_Symbol | log2FoldChange | Padj | Gene_Chrosome | Change |
|---|---|---|---|---|
| AW112010 | 2.84032013 | 3.87 × 10−31 | 19 | Up |
| Gm34643 | 2.150264718 | 2.24 × 10−5 | 14 | Up |
| Gm36161 | 1.365143899 | 6.92 × 10−26 | 13 | Up |
| Gm17705 | 1.333355523 | 7.90 × 10−3 | 17 | Up |
| Gm14221 | −1.62639326 | 2.98 × 10−4 | 2 | Down |
| Gm37168 | −1.332031628 | 4.17 × 10−5 | 1 | Down |
| Gm16907 | −1.027914414 | 7.90 × 10−3 | 13 | Down |
| Gene Name Ensembl ID | Species Gene Type | Location Length | Expression Changes (PrRP vs. Control) | Function | Refs. |
|---|---|---|---|---|---|
| Ifit1 (Interferon Induced Protein with Tetratricopeptide Repeats 1) (ENSEMBL: ENSG00000185745) | Mus musculus Protein coding | Chr 19 (2638 bp) | Up-regulated | Activities: Inhibit the translational initiation and replication of virus. Diseases: Hepatitis and Hepatitis C. Pathways: innate immune system and interferon gamma signaling. | [17] |
| Oasl2 (2’-5’ oligoadenylate synthetase-like 2) (ENSMUSG00000029561) | Mus musculus Protein coding | Chr 5 (3136 bp) | Up-regulated | Activities: is involved in the innate immune response to viral infection. Diseases: microphthalmia with limb anomalies and tick-Borne encephalitis. Pathways: innate immune system and interferon gamma signaling. | [18] |
| Irf7 (Interferon Regulatory Factor 7) (ENSG00000185507) | Mus musculus Protein coding | Chr 7 (1876 bp) | Up-regulated | Activities: regulates the transcriptional activation of virus-inducible cellular genes such as interferon beta chain genes. Diseases: influenza and immunodeficiency 39. Pathways: activated TLR4 signaling and apoptosis modulation and signaling. | [19] |
| Ifit3 (Interferon Induced Protein with Tetratricopeptide Repeats 3) (ENSG00000119917) | Mus musculus Protein coding | Chr 19 (1998 bp) | Up-regulated | Activities: inhibits cellular events, including viral processes, signaling, proliferation, cell migration, and viral replication. Diseases: systemic lupus erythematosus and lupus erythematosus. Pathways: innate immune system and interferon gamma signaling. | [17] |
| Ifit2 (Interferon Induced Protein with Tetratricopeptide Repeats 2) (ENSG00000119922) | Mus musculus Protein coding | Chr 19 (3949 bp) | Up-regulated | Activities: inhibits the expression of viral mRNAs. Diseases: microphthalmia with limb anomalies. Pathways: innate immune system and interferon gamma signaling. | [20] |
| Usp18 (Ubiquitin Specific Peptidase 18) (ENSG00000184979) | Mus musculus Protein coding | Chr 6 (1778 bp) | Up-regulated | Activities: inhibits interferon responses. Diseases: torch syndrome and pseudo-torch syndrome 2. Pathways: interferon gamma signaling and immune response IFN alpha/beta signaling pathway. | [21] |
| Ifi44 (Interferon Induced Protein 44) (ENSG00000137965) | Mus musculus Protein coding | Chr 3 (2860 bp) | Up-regulated | Activities: aggregates to form microtubular structures. Diseases: Potocki-Shaffer syndrome and hepatitis D. Pathways: interferon gamma signaling. | [22] |
| Rtp4 (Receptor Transporter Protein 4) (ENSG00000136514) | Mus musculus Protein coding | Chr 16 (1573 bp) | Up-regulated | Activities: enhances functional expression of the opioid receptor heterodimer OPRM1-OPRD1. Diseases: pain-related diseases. Pathways: olfactory transduction and signaling by GPCR. | [23] |
| Gene | GO Analysis [24,25] |
|---|---|
| Ifit1 | MF: RNA binding |
| BP: immune system process; response to stimulus | |
| CC: cytosol | |
| Oasl2 | MF: carbohydrate derivative binding; RNA binding; transferase |
| BP: immune system process; response to stimulus | |
| CC: cytosol; nucleus; organelle lumen | |
| Irf7 | MF: DNA binding; transcription |
| BP: immune system process; nucleic acid-templated transcription; response to stimulus; signaling; system development | |
| CC: cytosol; nucleus; organelle lumen | |
| Ifit3 | MF: RNA binding |
| BP: immune system process; response to stimulus | |
| CC: cytosol; mitochondrion | |
| Ifit2 | MF: RNA binding |
| BP: cell death; immune system process; response to stimulus | |
| CC: cytosol; endoplasmic reticulum | |
| Usp18 | MF: hydrolase |
| BP: protein metabolic process; response to stimulus | |
| CC: cytosol; nucleus | |
| Ifi44 | MF: none |
| BP: immune system process; response to stimulus | |
| CC: none | |
| Rtp4 | MF: signaling receptor binding |
| BP: cellular component organization; establishment of localization; immune system process; response to stimulus | |
| CC: none |
| # | Key TF | Description | # of Overlapped Genes | p Value | FDR |
|---|---|---|---|---|---|
| 1 | Irf1 | interferon regulatory factor 1 | 5 | 2.16 × 10−9 | 3.46 × 10−8 |
| 2 | Stat1 | signal transducer and activator of transcription 1 | 5 | 1.12 × 10−8 | 8.92 × 10−8 |
| 3 | Nfkb1 | nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 | 6 | 4.95 × 10−6 | 2.64 × 10−5 |
| 4 | Jun | jun proto-oncogene | 5 | 1.02 × 10−5 | 4.07 × 10−5 |
| 5 | Irf8 | interferon regulatory factor 8 | 3 | 1.74 × 10−5 | 5.56 × 10−5 |
| 6 | Rel | reticuloendotheliosis oncogene | 3 | 3.19 × 10−5 | 8.50 × 10−5 |
| 7 | Rela | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | 4 | 1.47 × 10−4 | 0.000336 |
| 8 | Foxo3 | forkhead box O3 | 2 | 1.71 × 10−4 | 0.000341 |
| 9 | Irf4 | interferon regulatory factor 4 | 2 | 2.60 × 10−4 | 0.000462 |
| 10 | Ikbkb | inhibitor of kappaB kinase beta | 2 | 3.11 × 10−4 | 0.000498 |
| 11 | Foxm1 | forkhead box M1 | 2 | 1.18 × 10−3 | 0.00171 |
| 12 | Hdac1 | histone deacetylase 1 | 2 | 1.74 × 10−3 | 0.00232 |
| 13 | Spi1 | spleen focus forming virus (SFFV) proviral integration oncogene | 2 | 3.54 × 10−3 | 0.00435 |
| 14 | Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 2 | 3.89 × 10−3 | 0.00445 |
| 15 | Fos | FBJ osteosarcoma oncogene | 2 | 6.83 × 10−3 | 0.00729 |
| 16 | Stat3 | signal transducer and activator of transcription 3 | 2 | 1.32 × 10−2 | 0.0132 |
| Proteins | Species | Protein Length (aa) | Model Templates (Query Cover, Identify) |
|---|---|---|---|
| Ifit1 | Mus musculus | 461 | 5W5H_A (2.79 Å) (99%, 52.98%) 6C6K_A (2.54 Å) (97%, 53.25%) 5UDI_A (1.58 Å) (95%, 53.22%) |
| Oasl2 | Mus musculus | 439 | 4XQ7_A (1.60 Å) (68%, 49%) 4IG8_A (2.70 Å) (65%, 45.40%) 1PX5_A (1.74 Å) (66%, 43.02%) |
| Irf7 | Mus musculus | 330 | 1QWT_A (2.10 Å) (49%, 28.57%) 1J2F_A (2.30 Å) (49%, 28.57%) 5JEJ_A (2.00 Å) (46%, 28.97%) |
| Ifit3 | Mus musculus | 391 | 4G1T_A (2.80 Å) (99%, 51.33%) 6C6K_A (2.54 Å) (92%, 41.58%) 5W5H_A (2.79 Å) (92%, 41.58%) |
| Ifit2 | Mus musculus | 468 | 4G1T_A (2.80 Å) (98%, 63.03%) 5UDI_A (1.58 Å) (92%, 41.03%) 5W5H_A (2.79 Å) (92%, 41.03%) |
| Usp18 | Mus musculus | 308 | 5CHV_A (3.00 Å) (87%, 100%) 5CHT_A (2.80 Å) (87%, 100%) 2F1Z_A (3.20 Å) (87%, 26.89%) |
| Ifi44 | Mus musculus | 420 | De novo by Rosetta |
| Rtp4 | Mus musculus | 247 | De novo by Rosetta |
| Proteins | Number of Residues in Favoured Regions | Number of Residues in Allowed Region | Number of Outliers |
|---|---|---|---|
| Ifit1 | 452/461 (98%) | 459/461 (99.6%) | 2 (0.004%) |
| Oasl2 | 408/439 (92.9%) | 433/439 (98.6%) | 6 (0.014%) |
| Irf7 | 295/330 (89.4%) | 316/330 (95.8%) | 14 (0.042%) |
| Ifit3 | 372/391 (95.1%) | 386/391 (98.7%) | 5 (0.013%) |
| Ifit2 | 450/468 (96.2%) | 459/468 (98.1%) | 9 (0.019%) |
| Usp18 | 292/308 (94.8%) | 304/308 (98.7%) | 4 (0.013%) |
| Ifi44 | 399/420 (95.0%) | 420/420 (100.0%) | 0 (0.000%) |
| Rtp4 | 237/247 (96.0%) | 247/247 (100.0%) | 0 (0.000%) |
| Genes | Primers | Sequences (5′ to 3′) | Products (bp) |
|---|---|---|---|
| Gapdh | Forward | TGTGTCCGTCGTGGATCTGA | 150 |
| Reverse | TTGCTGTTGAAGTCGCAGGAG | ||
| Ifit1 | Forward | TTGTTGTTGTTGTTGTTC | 127 |
| Reverse | GTGAGTATGTATCCTTGG | ||
| Oasl2 | Forward | TGTTGGATGATGAGGAGTTG | 75 |
| Reverse | GTATGATGGTGTCGCAGTC | ||
| Irf7 | Forward | AATCTACACTGAGTTCTG | 154 |
| Reverse | GACCAAGTTTCACAAATG | ||
| Ifit3 | Forward | GTCCTTTGAACTCCTACTC | 80 |
| Reverse | GCTCTCCTTACTGATGAC | ||
| Ifit2 | Forward | TATATGACACAGACAGAG | 163 |
| Reverse | TCTAACTTCTTCCTATCC | ||
| Usp18 | Forward | CTTAGGTGACAGAACTTG | 94 |
| Reverse | AACAGGAAGAAGAACTATTAG | ||
| Ifi44 | Forward | TAGTTCTGCTTGCTTCTC | 180 |
| Reverse | TCTGTGCCTTCTTCATTC | ||
| Rtp4 | Forward | TCAGAAGTGCCAGAAGTG | 123 |
| Reverse | TTCCTGTGTCCATAGTATCTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Zuo, Z.; Kuang, Y. Prolactin-Releasing Peptide Differentially Regulates Gene Transcriptomic Profiles in Mouse Bone Marrow-Derived Macrophages. Int. J. Mol. Sci. 2021, 22, 4456. https://doi.org/10.3390/ijms22094456
Sun Y, Zuo Z, Kuang Y. Prolactin-Releasing Peptide Differentially Regulates Gene Transcriptomic Profiles in Mouse Bone Marrow-Derived Macrophages. International Journal of Molecular Sciences. 2021; 22(9):4456. https://doi.org/10.3390/ijms22094456
Chicago/Turabian StyleSun, Yulong, Zhuo Zuo, and Yuanyuan Kuang. 2021. "Prolactin-Releasing Peptide Differentially Regulates Gene Transcriptomic Profiles in Mouse Bone Marrow-Derived Macrophages" International Journal of Molecular Sciences 22, no. 9: 4456. https://doi.org/10.3390/ijms22094456






