Abstract
Horizontal gene transfer (HGT)- is defined as the acquisition of genetic material from another organism. However, recent findings indicate a possible role of HGT in the acquisition of traits with adaptive significance, suggesting that HGT is an important driving force in the evolution of eukaryotes as well as prokaryotes. It has been noted that, in eukaryotes, HGT is more prevalent than originally thought. Mitochondria and chloroplasts lost a large number of genes after their respective endosymbiotic events occurred. Even after this major content loss, organelle genomes still continue to lose their own genes. Many of these are subsequently acquired by intracellular gene transfer from the original plastid. The aim of our review was to elucidate the role of chloroplasts in the transfer of genes. This review also explores gene transfer involving mitochondrial and nuclear genomes, though recent studies indicate that chloroplast genomes are far more active in HGT as compared to these other two DNA-containing cellular compartments.
1. Introduction
Horizontal gene transfer (HGT), also called lateral gene transfer (LGT), is the process of transferring genetic material between organisms by routes other than parent-offspring (vertical gene transfer—VGT). HGT can involve the transfer of genetic material from one cell to another: between different bacterial species by processes of conjugation, transduction, and transformation; between different single-celled organisms, e.g., bacteria and yeasts; or between even more distantly related organisms, e.g., bacteria and insects, fungi and animals, or plants and fungi. In 1984, Syvanen introduced and gradually developed the concept of HGT as interspecies gene flow [1,2]. The phenomenon was first discovered in 1951 in Klebs-Löffler bacillus (Corynebacterium diphtheriae). It was observed that the gene responsible for pathogenicity, tox, was of viral origin and could transfer from pathogenic to non-pathogenic bacteria. In 1959, it was shown that bacterial genes responsible for antibiotic resistance could also be transferred via this route. HGT has further been shown to have a significant role in the evolution of eukaryotes. Above all, the role of this process in protists is to be highlighted. HGT can be considered a universal phenomenon observed in the genomes of bacteria, fungi, plants, or animals [1,2]. HGT is commonplace in prokaryotes [3,4] as demonstrated by studies typing approximately 81% of genes in which transfer was noted [5]. In recent years, the readability of eukaryotic genomes in next generation sequencing (NGS) studies has facilitated the characterization of the occurrence and mechanism of HGT in eukaryotes [6].
Moreover, data on the entire genomes of prokaryotes revealed the presence of HGT between distant species [7,8]. Extensive research on the participation of HGT in prokaryotic evolution (e.g., archaea and bacteria) has shown a possible mechanism to facilitate the acquisition of new features [9]. However, this phenomenon has been speculated to be rare in eukaryotes [10,11].
Transmission and integration of transferred genes can provide several beneficial features, including prokaryotic adaptation during environmental changes [12,13], acquisition of new features/functionalities [14], and evolutionary adaptation in eukaryotes [15,16]. Most cases revealed one transfer of one gene in one species, but recent findings reported a case of massive transfer of at least tens, and probably hundreds, of foreign mitochondrial genes in Amborella trichopoda.
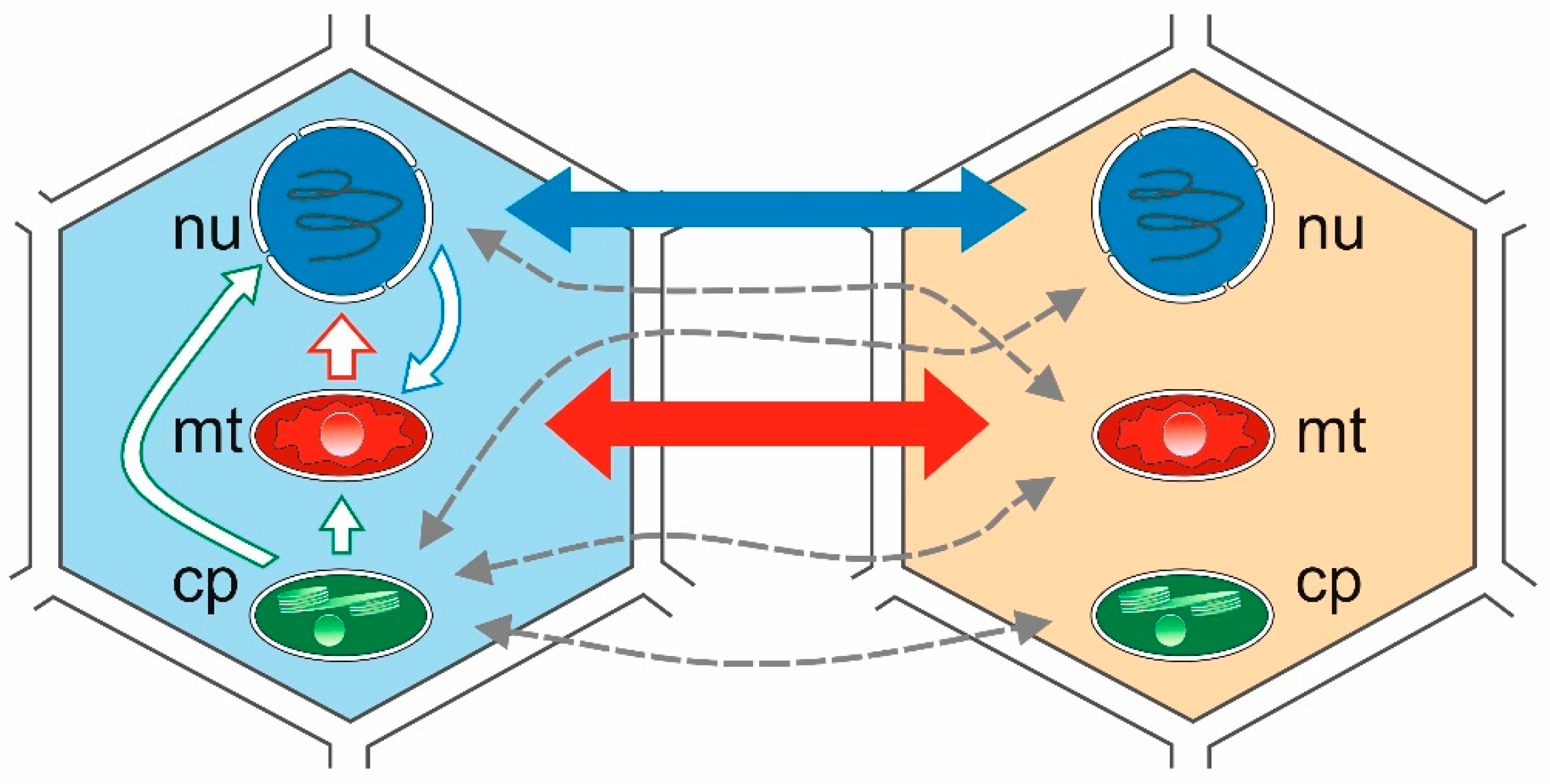
There are also other mechanisms, e.g., intracellular transfer involving cell organelles, such as the nucleus, the mitochondria, and the chloroplasts (intracellular gene transfer—IGT) [17], or the transfer of genetic material to a different location in the genome of the same organism (intragenomic transfer or transposition) (Figure 1). Given the endosymbiotic origin of mitochondria and chloroplasts, many genes of eubacterial origin migrated from these organelles to the nucleus via IGT as well as horizontal transfer. Many recorded transfers occurred relatively recently during evolutionary history and were limited to a single recipient, or to a small number of species within the genus. However, while the IGT rate has dropped significantly since the appearance of eukaryotes, it remains a common process characteristic of the evolution of the nuclear and organelle genomes in plants [18,19,20,21,22,23]. Among the three types of genomes in a plant cell, there are six possible directions for gene transfer. The most prominent are from the organelle (mitochondrial) genome to the nuclear genome [22,23], and from the nuclear and plastid (chloroplast) genome to the mitochondrial genome [18]. Intracellular transfer to the highly compact plastid genome appears to be quite rare; however, it is being reported increasingly frequently. More information can be found in the literature indicating numerous cases of nuclear-to-mitochondrial IGT transfer than from chloroplast to mitochondrion or to cell nucleus [18].
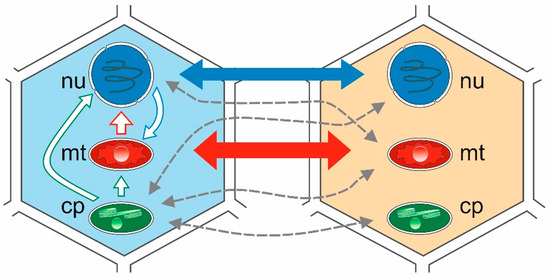
Figure 1.
Intracellular gene transfer (IGT) and horizontal gene transfer (HGT) between two plant species. Intergenomic gene transfer represent lines in the cell from the left. Solid blue, green, and red lines depict the number of reported HGT events between cell compartments. Thicker lines indicate more frequent events than thinner lines. Dashed grey lines depict putative transfers. cp: chloroplast; mt: mitochondria; nu: nucleus.
Recent studies have shown that plant mitochondrial genomes are more likely to transfer DNA to the other two cellular compartments mentioned above. The ever-increasing number of plant mitochondrial genomes (mtDNA) sequenced [20,24,25,26,27] reveals the extent of transfer from both chloroplast (cpDNA) and nuclear genomes (nDNA) [28,29,30,31,32,33,34,35,36,37,38]. In general, plant mtDNAs consist of between 0.56% (Marchantia polymorpha) and 10.85% (Phoenix dactylifera) plastid-derived sequences [33]. The integration of nuclear sequences is usually more complex and more difficult to identify, as it involves retrotransposons and other repetitive sequences. Repetitive sequences are common to mtDNA of various seed-bearing plants, including Oryza sativa [33], Arabidopsis thaliana [35], Cucumis melo [34], and Cucumis sativus [28], as well as Gossypium species [31,32]. Long terminal repeat retrotransposons (LTR-retro) typically constitute the largest proportion of plant nuclear repeats, and often have a dominant influence on nuclear and mitochondrial genome size [38,39,40,41,42,43]. A 2019 study [18] on four cotton species (Gossypium raimondii, Gossypium arboreum, Gossypium hirsutum, and Gossypium barbadense) indicated the presence of multiple nuclear and chloroplast tRNA gene repeats in the plant mtDNA genome. The authors highlighted the discovery of eight chloroplast-derived tRNA genes—trnD, trnH, trnM, trnN, trnP, trnS, trnV, and trnW—in the mitochondrial genomes of all four cotton species [18]. In this paper, we discuss mainly cases of HGT and IGT gene transfer involving the chloroplast genome of plants.
2. Organisms between Which HGT Occurs
In contrast to gene transfer by interspecies crossover, which is restricted to closely related organisms, there appear to be no clear taxonomic boundaries for HGT. As described above, transfers between different species are known, e.g., between bacteria and fungi, bacteria and plants, bacteria and animals, fungi and animals, and fungi and plants. The highest number of HGT cases among Eukaryotes was observed in unicellular organisms [44,45] (Table A1). The number of transferred genes in these organisms ranges from one to several dozen, comprising less than 1% of the total genome. Most were found to have come from bacterial donors. In the plant world, a small number of HGT exchanges have been observed. One of the first described was the exchange of DNA between cells of Agrobacterium and the cells of higher plants [46]. Additional putative cases include the acquisition of aquaglyceroporins from eubacteria (1200 million years ago) [47] and glutathione biosynthesis genes from α-proteobacteria [21].
Viral DNA does not typically integrate into the host’s nuclear genome; this has occurred only sporadically throughout evolutionary history. Random recombination between the viral genome and the plant chromosomal DNA is believed to be the molecular mechanism underlying such events [44].
It is worth noting that chloroplast and mitochondrial RNA polymerases encoded by the nuclear genome are similar to bacteriophage T7 polymerase, which may have contributed to the emergence of HGT transfers during the evolution of eukaryotes [40]. In addition to genes or their fragments, mobile genetic elements can also be transferred. Roulin et al. [48] reported the transfer of retrotransposon LTR Route66 between Sorghum and several Oryza species, remaining active after transfer.
Fungus-to-plant HGT (FP-HGT) plays a significant role in shaping plant genomes. Published literature on this mechanism has deepened our understanding of the genetic evolution of disease resistance, and has opened up a new avenue for the identification of plant resistance genes. It seems, therefore, that there are no genetic barriers “prohibiting” the transfer of genetic material between even evolutionarily distant organisms. Further discussion will focus primarily on plants [49,50].
3. HGT Routes and Mechanisms
The mechanisms responsible for HGT are not sufficiently understood. Transfer of nucleic acids via intermediaries, such as viruses, bacteria, fungi, and transposons, or via the direct uptake of nucleic acids (especially in parasite-host systems) are usually indicated. Genetic material could theoretically be transferred by DNA fragments or by mRNA, which would then be converted back into DNA by reverse transcription. Research tends to point to the former possibility.
Two modes of transfer have been identified—vector and direct [50]. The vector route requires the presence of vectors such as bacteria, fungi, viruses, etc. [51,52], while the direct route can occur through direct DNA exchange [18]. HGT between nuclear and organelle genomes [53] has received particular attention in the literature [54]. HGT involving bacteria or fungi has also been documented [54,55]. Extensive studies suggest the possibility of HGT from plants to other genomes via pathogens, transgenic bacteria (e.g., Agrobacterium tumefaciens), viruses, fungi, and nematodes [56,57,58,59].
Furthermore, plant cells are characterized by the presence of three types of the genome: nuclear, mitochondrial, and chloroplast, which differ in their susceptibility to HGT. It is believed that there is a low level of horizontal transfer of nuclear genes, despite evidence of numerous horizontal movements of their genetic elements [36,60]. Some authors conclude that the nuclear genome arose through HGT by the fusion of archaebacterial and eubacterial genomes [18,60]. Given the endosymbiotic origin of mitochondria and chloroplasts, many genes of eubacterial origin have migrated from these organelles to the nucleus by intracellular modes of HGT [18,61,62,63,64] as well as by intercellular modes.
4. HGT and Endophytes
Recent scientific reports indicate that plant growth under natural conditions is closely linked to the microorganisms accompanying them, known as endophytes—organisms that develop asymptomatically in plant tissues for at least part of their life cycle. These can be either fungi or bacteria, including actinomycetes. Thus far, they have been isolated from numerous species of cultivated, forest, and aquatic plants [44,65]. Endophytes influence inhabited plants in various ways, which can be characterized as direct, indirect, or ecological.
Direct influences are related to the synthesis of compounds by the endophyte that destroy pathogens. Such compounds include terpenoids, alkaloids, aromatic compounds, and also lytic enzymes capable of degrading chitin, proteins, cellulose, hemicellulose, and DNA. Indirect endophyte influences on plant resistance are often related to the induction of plant defense mechanisms, such as the synthesis of secondary metabolites. Another mechanism of indirect influence is the improvement of growth and overall condition, making the host more resistant to stresses. Thus, the relationships between endophyte and plant are economically important due to their potential applications in agriculture, industry, and health, as well as environmental protection.
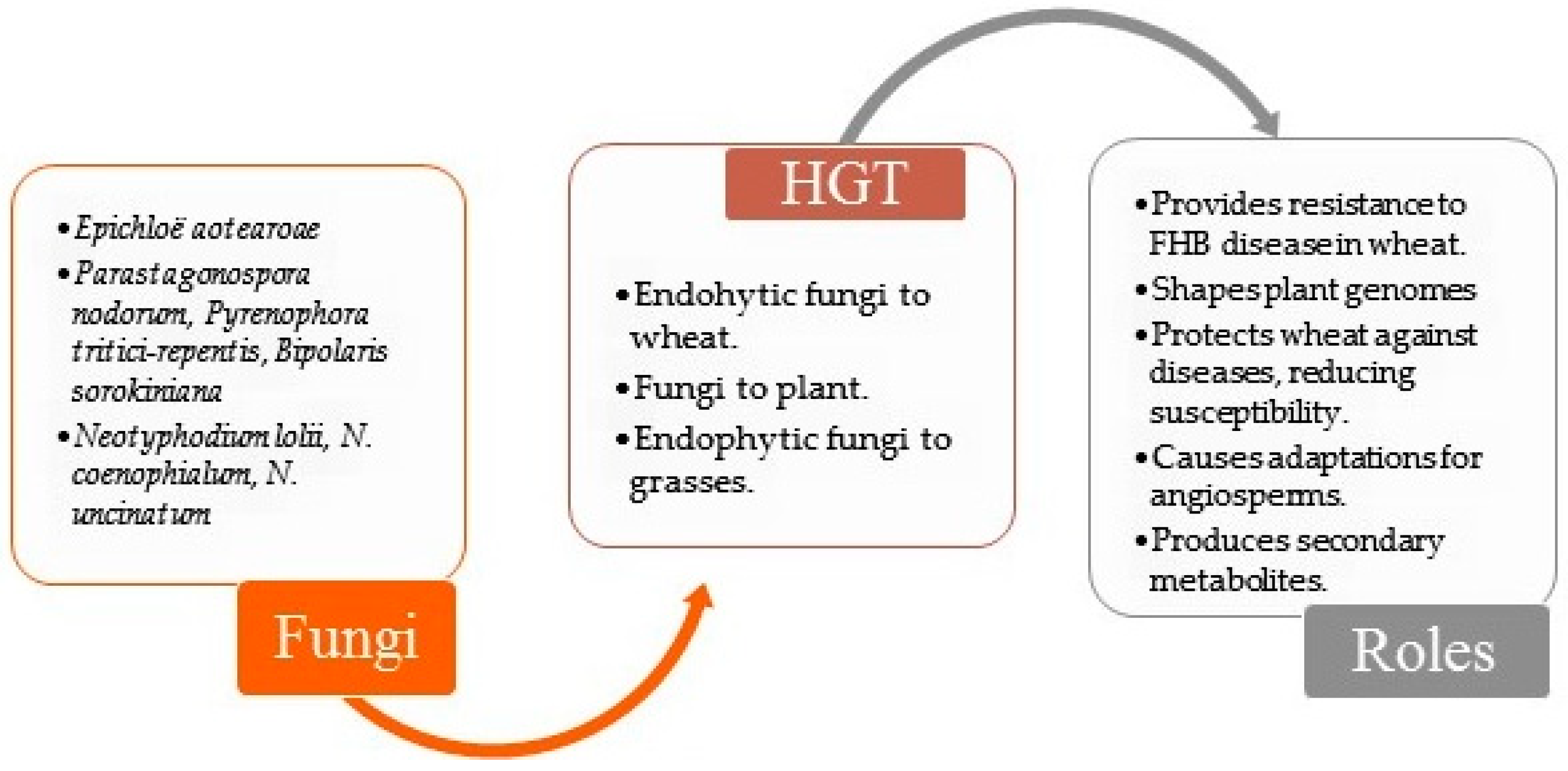
The phenomenon of HGT in endophytes highlights an important biological mechanism for their evolutionary adaptation within the host plant, as they continuously acquire ‘new traits’ of adaptation. It should be noted that this research problem remains unexplored in the literature. There is evidence for the role of endophytic bacteria in the biodegradation of toluene and the reduction of disease in durum wheat (Triticum durum) and maize (Zea mays) [45,49]. Furthermore, researchers have hypothesized that genetic recombination between plants and endophytes may have led to the incorporation of metabolic pathway genes into the host plant [45]. Figure 2 illustrates occurrences of transfer between plant and endophyte (bacterium or fungus).
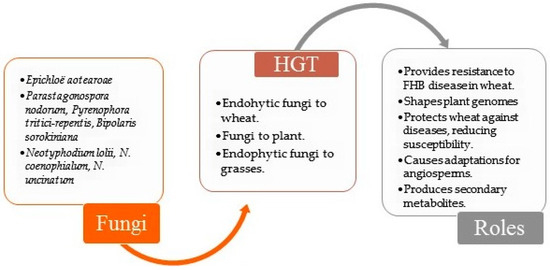
Figure 2.
Examples of HGT cases described in plant and fungi endophyte relationships [43,66,67,68,69,70,71,72,73,74,75]. FHB—Fusarium head blight.
The effects of HGT following a relationship with an endophytic microorganism represent an important process by which adaptation problems may be solved. Attention has been drawn to both the beneficial and detrimental effects of the coexistence of grasses with endophytes. Since few studies are available on this topic, we wish to highlight the importance of HGT in plants in beneficial relationships with endophytes.
Common wheat (Triticum aestivum L.) is a major source of calories for the human population [43,69]. The occurrence and spread of the devastating disease Fusarium head blight (FHB), exacerbated by recent climate change and certain cropping practices, poses a threat to global wheat production and food security. FHB is caused by fungi belonging to the genus Fusarium, and its occurrence is observed in all major cereals grown worldwide (wheat, triticale, rye, oats, barley, maize). FHB has the greatest impact on wheat crops. For example, F. graminea rumist is the main pathogen causing FHB of wheat in China, the United States, Canada, European countries, and many other countries [66,70]. This is due to the high susceptibility of this cereal, the prevalence of the pathogen, and the large area under wheat cultivation. Fusarium produces epoxy-sesquiterpene compounds known as trichothecenes. These compounds are protein synthesis inhibitors and virulent pathogens [70]. Trichothecene contamination of cereal grains results in immunotoxicity and cytotoxicity in humans and animals. Wheat resistance FHB is a quantitative trait [72,73,74,75], and numerous quantitative trait loci (QTL) associated with resistance to this disease have been reported.
In their study, Wang et al. [69] indicated that Fhb7 (encoding glutathione S-transferase—GST) shows similar effects on FHB resistance and confers broad resistance to Fusarium to species. At the same time, studies of wheat genomes have been undertaken in which the Fhb7 reference gene for Th. elongatum has been developed. It was found that Fhb7 can detoxify trichothecenes by catalyzing the conjugation of a glutathione (GSH) unit onto their toxic epoxide moiety, where the active sulfhydryl group neutralizes the toxicity. In the authors’ view, the coding sequence of Fhb7 has no obvious homology with any known sequence across the plant kingdom, but shows 97% sequence identity with a species of endophytic fungus (Epichloë aotearoae) known to infect grasses. This provides evidence that Fhb7 in Th. elongatum probably originates from the fungus, and arrived via HGT. Furthermore, such transfer carried out experimentally has been shown to provide wheat with resistance to FHB [69,70,71].
The analysis of HGT from fungi to plants uses sequenced endophyte genomes to understand how fungal pathogens interact with plants. For some time now, it has been possible to find information in the literature about the ToxA gene that is produced by two fungal pathogens of wheat, Parastagonospora nodorum and Pyrenophora tritici-repentis. It has been hypothesized that these pathogens shared ToxA and part of the surrounding repetitive DNA via HGT, resulting in a sequence of 11,000 bases that is almost identical between the two species [76].
Subsequent genome sequencing results indicated the presence of ToxA in the genome of Bipolaris sorokiniana infecting wheat and barley. Interestingly, the ToxA identification in the pathogen genome has an identical 11,000 kbp fragment. The high identity between these three species of Parastagonospora nodorum, Pyrenophora tritici-repentis, and Bipolaris sorokiniana shows that HGT must have occurred very recently [77,78]. Parastagonospora nodorum and Pyrenophora tritici-repentis are believed to share ToxA and a number of DNA sequences, presumably through HGT, resulting in an 11 kbp transfer which is almost identical between Parastagonospora nodorum and Pyrenophora tritici-repentis. These fungi are the main causes of diseases in cereals such as root rot, leaf spot disease, seedling blight, and black point. The high homology between these three pathogens indicates further evidence of HGT.
This also highlights the importance of ToxA itself, which plays a significant role in wheat diseases. ToxA is responsible for necrosis (cell death) in wheat leaves during infection. It does this in a very specific way, acting in a gene-for-gene relationship with the gene responsible for the susceptibility of wheat to fungal diseases: Tsn1. If both genes are present, ToxA in the fungus and Tsn1 in wheat, the infected leaf dies. If either gene is absent, there are no ToxA-related symptoms. This has been confirmed by studies showing that when the pathogen is a vector, ToxA has a significant effect on disease symptoms in wheat varieties containing the Tsn1 gene. Thus, ToxA plays a key role in the disease caused by these pathogens. Work on this phenomenon further solidifies HGT as a mechanism by which fungal pathogens can share strategies to exploit host vulnerabilities. HGT is, of course, not limited to bacterial plant pathogens. In the case of ToxA, it has been shown to weaken wheat resistance to various fungal diseases. All of these diseases occur worldwide in wheat crops, meaning that ToxA poses a global threat to the yields of common wheat [77,78].
5. Intracellular Gene Transfer
As described above, DNA fragments can migrate from the cell nucleus of one organism to the cell nucleus of another organism. This process can also occur between all the elements of a cell containing genetic material: nucleus, mitochondria, and plastids. The mitochondria of most seed-bearing plants examined have both nuclear and plastid sequences. Mitochondrial genes are also found in plastids, but rarely. The difference is probably due to the fact that mitochondria, unlike plastids, have efficient mechanisms to take up foreign DNA. Many genes of mitochondrial origin have been found in nuclei. In such cases, prokaryotic genes are converted into eukaryotic genes, which are related, among other things, to the fact that they undergo recombination during sexual reproduction. Presumably, RNA is involved as a mediator in this type of IGT [18,19,20,21,22].
5.1. HGT in Cell Nuclei and Plastids
Many traces of HGT have also been found in the nuclei of angiosperms. These concern nuclear genes as well as transposons. One interesting case is the parasitic plant Rafflesia cattleyi, in which more than 30 genes have been found that have been transferred from the host. At least some of these are functional. Plastids, on the other hand, are thought to be highly resistant to incoming processes of HGT or IGT. Plastid sequences, rather, are found in other genomes of the cell—mitochondrial and nuclear—having transferred from a starting point in the plastid [79,80,81].
5.2. Chloroplast-to-Mitochondria Transfer
Today, HGT into mitochondria occurring between distantly related higher plant species is a well-known phenomenon, and is no longer as controversial as when the work by Woloszynska et al. was released [82], which was the first to demonstrate horizontal transfer of DNA sequences from chloroplasts to mitochondria. The subject of the study was a fragment of chloroplast trnA gene intron, named pvs-trnA because it is part of the pvs sequence that determines male sterility. The pvs-trnA sequence was identified in only three species of the genus Phaseolus and it was found that, although it contains only 190 bp, it differs from the chloroplast trnA sequence of beans in as many as 10 positions and is most similar to the chloroplast genes of Philodendron scandens and Magnolia grandiflora, showing only three differences. In view of this, the phylogenetic trees generated placed pvs-trnA between plants from the class of monocotyledons and the order of magnoliales, at a position isolated from the leguminous plants to which beans belong. The results of this study demonstrated that the pvs-trnA sequence did not arise from intracellular transfer from chloroplasts to mitochondria of the same plant, but from horizontal transfer of a trnA intron fragment from the chloroplasts of a non-dicotyledons plant to mitochondria of a plant of the genus Phaseolus [82].
Among the cases of horizontal gene transfer from plant to plant that have been studied, in about 1 in 40 cases, a gene of mitochondrial origin encoding respiration-associated proteins or ribosomal proteins had been transferred. To date, only one exception has been described: the horizontal transfer of a fragment of the intron pvs-trnA described above. Subsequently, in Isoetes engelmannii, an insert of chloroplast origin containing trnA and 23S rRNA gene sequences was identified in the mtDNA. These sequences are highly similar to their counterparts in the cpDNA of I. malinverniana, indicating inter-organelle transfer rather than HGT [83].
Recent data indicate that between 1.1% and 6.3% of mitochondrial DNA has been transferred via the IGT pathway from chloroplasts [27]. Chloroplast sequences are more often transferred to the mitochondrial genome, and also by pathways other than HGT, e.g., via IGT [84,85,86,87,88,89,90,91,92,93,94,95,96], which is why we find traces of chloroplast-derived sequences in the mtDNA of genera such as Arabidopsis, Beta, Brassica, and Oryza [87]. This is not surprising since the homology of certain chloroplast and mitochondrial genome sequences had already been noted in the 1980s [89]. According to Hao and Palmer [89], the mitochondrial genome of flowering plants tends to adopt chloroplast sequences that have no effect on its function [92] as these were progressively degraded to pseudogenes.
It was, therefore, decided to look for homologous recombination (and gene conversion) events between the sequences of the two genomes. Consequently, it was possible to observe a repeated conversion—the replacement of short mitochondrial sequences of the atp1 gene by chloroplast homologous atp1 sequences. Both homologues encode an α subunit of ATP synthase. It is likely that, after integration of the atp1 gene into the mitochondrial genome, intra-mitochondrial recombination occurred. The chimeric atp1 genes in the mitochondrion are presumed to be functional, to have ORFs and to not exhibit the characteristics of pseudogenes, and it is likely that substitutions in the recombinant region are synonymous [89]. Analysis of HGT cases in plants has also led to the introduction of the term “duplicative” HGT, which is linked to conversion. Duplicative HGT involves the transfer and integration of foreign genes (single genes, their fragments, whole assemblies) which, in the recipient mitochondrial genome, do not immediately replace their counterparts. HGT, together with recombination, contributes to the genetic diversity of mtDNA [90].
5.3. Mitochondrion-to-Chloroplast Transfer of Rps16
An interesting case is the transfer of the rps16 gene, which is present in the chloroplast genome of most higher plants, while it is absent in the cpDNA of Medicago truncatula and Populus alba. In these species, the rps16 gene is present in the mitochondrial genome. The product of mitochondrial gene expression may be directed to both mitochondria and chloroplasts. Such targeting of the mitochondrial product of the rps16 gene is also characteristic of Arabidopsis thaliana, Lycopersicon esculentum, and Oryza sativa, in which the gene is present in both organellar genomes (Table 1) [94].

Table 1.
Plastid genes in HGT involved in HGT in plants and type of the transfer.
5.4. Chloroplast-to-Nucleus Transfer
The structure and gene content of the plastid genome are well conserved among different land plant species [101]. The chloroplast genome of land plants has a highly conserved organization. It is a circularly shaped, double-stranded DNA molecule that contains two inverted repeat (IR) copies that separate a small and a large single-copy region (small single copy—SSC; large single copy—LSC). The cpDNA contains genes that encode proteins responsible for the mechanisms of photosynthesis, and biosynthesis of fatty acids, amino acids, pigment, and vitamins, among others [102]. The region of inverted repeats usually contains three rRNA genes (rns, rnl, rrn5) and two tRNAs (trnA, trnI), but there may be more as a result of contraction or by the expansion of this region. Nevertheless, the gene repeats are rarely identical. Plastids are characterized by the presence of proteins that are encoded by nuclear genes and post-traumatically imported into their region. Moreover, genes in the plastid genome show a high degree of similarity among a wide variety of plants. Therefore, it is believed that most genes were transferred from the original endosymbiont to the nucleus at an early stage of plant evolution [103,104]. However, evidence for the ongoing occurrence of chloroplast gene transfer to the nucleus has been demonstrated. It is believed that the tufA gene, encoding the chloroplast elongation factor Tu, was transferred from the chloroplast to the nucleus in the algae line [105,106].
During research on sequencing of the tobacco chloroplast genome, a homologue of the Escherichia coli rp122 gene, encoding the ribosomal protein L22, was found [95]. This gene was subsequently found in the cpDNA of monocotyledons and dicotyledons, vascular land plants, and in algae [96,97]. However, rp122, which is part of a large region of ribosomal protein genes in rice, tobacco, and liverwort, is not found in its normal position in the cpDNA of soybean and subterranean clover (Trifolium subterraneum) [95]. Thus, it can be concluded that the plastid genomes of legumes do not contain rp122, which raises the question of whether this gene has been completely lost from legumes or has been transferred to the nucleus. There is evidence that in pea, rp122 is located in the cell nucleus, which has been confirmed by detailed studies involving the structure of this gene and clarifying its transfer mechanism [95]. Studies on this case, initiated in the 20th century, showed that sequences corresponding to the exon and intron of pea rp122 had not been found in any characterized chloroplast gene. Thus, long-term studies on the evolutionary transfer of pea rp122 have completely placed it outside the angiosperm chloroplast gene clade. This demonstrates that this gene was transferred to the nucleus long before its subsequent loss from the chloroplast genome of its legume ancestor. Gantt et al. [95], based on the best phylogenetic tree they have obtained in the course of their study, showed that nuclear transfer probably preceded the loss of chloroplasts by at least 100 million years.
A 2001 publication [96] showed that among the six fully sequenced chloroplast genomes of angiosperm plants (with the exception of the non-photosynthesizing plant Epifagus virginiana), most protein-coding genes are universal. In contrast, other genes are species-specific, e.g., accD, ycf1, and ycf2 (pseudogenes in rice and maize), rpl23 (pseudogene in spinach), and infA (pseudogene in tobacco, Arabidopsis, and Oenothera elata) [106]. Chloroplast genes have been lost in angiosperms, including rpl22, rps16, and ycf4 (ORF 184), which have been lost in some or all of the legumes [96]. Furthermore, ycf2 and ndhF were repeatedly lost in various angiosperm species [96]. Another example of the loss of the chloroplast genes, i.e., infA, rps16, ycf1, ycf2, and ycf4, has not been elucidated, as it is not known whether they reflect successful gene transfer to the nucleus or complete loss of the gene from the cell. A generally better-understood case is rpl22 in legumes, as it represents a classic chloroplast-to-nucleus gene transfer where the protein is imported back into the chloroplast via a transit peptide [96]. In the case of analysis of the chloroplast locus rpl23 in spinach, it was shown to be a pseudogene and has been functionally replaced with a nuclear gene similar to the homologous cytosolic ribosomal protein gene.
Further confirming the case for DNA sequence transfer is the chloroplast infA gene, which has been repeatedly lost in the evolution of angiosperms. Studies have shown that the four nuclear infA genes characterized so far probably arose by independent gene transfer processes from the chloroplast to the nucleus. The changes in infA in angiosperm cpDNA have been shown to be similar to the evolution of rps10 in mitochondrial DNA of angiosperms [96], as evidenced by loss and for nuclear transfer in both cases (Table 1) [94,97,98,99,100,101,102,103,104,105,106,107].
6. Conclusions
In conclusion, it is important to note the importance of HGT in plant evolution. Currently, extensive literature shows that HGT plays a powerful role in eukaryotes. Remnants of this process are found in all major groups of organisms. For example, the important role that transfer has played in plant evolution has been demonstrated by the transformation of the intracellular prokaryotic endosymbiont into a chloroplast. A very interesting form of evolution occurs when pathogens exchange DNA between species, which, as described above, is a biological process known as horizontal gene transfer (HGT). The HGT events that have been observed by detecting genes or DNA sequences which are almost identical between distantly related species are very interesting. In this paper, we have only highlighted a few of the many such cases where a single horizontally transferred gene can contribute to the acquisition of many important traits for plants. In particular, we have highlighted cases of fungus-to-plant transfer that confer adaptive plasticity in plants to new and extreme conditions, efficient stress responses, more efficient DNA repair, and the ability to neutralize toxicity.
Still, little is known about the functional and implications of HGT between plants, other than the confirmed presence of nuclear and mitochondrial HGT in the plant kingdom. This phenomenon raises a number of questions regarding the scope and mechanism of the exchange of genetic material between plants. There is a lack of information on the consequences for the storage of a given transgene and the flow of information between the organelle (cpDNA, mtDNA) and the nuclear genome. It should be emphasized that few studies indicate cases of sequence transfer of chloroplast origin. Current advances in generating genomic comparative data will contribute to the rapid discovery of evolutionary aspects of HGT between plants, including confirmation of the involvement of cpDNA.
Author Contributions
Discussed the concept of the manuscript, E.F. and L.S.; contributed to literature collection and writing of the manuscript, E.F.; critically reviewed and revised the manuscript, L.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| HGT | Horizontal Gene Transfer |
| LGT | Lateral Gene Transfer |
| VGT | Vertical Gene Transfer |
| IGT | Intracellular Gene Transfer |
| IGT | Intragenomic Gene Transfer |
| cpDNA | chloroplast genome |
| nDNA | nuclear genome |
| mtDNA | mitochondrial genome |
| LTR | LTR-retro retrotransposons |
| FP-HGT | fungus-to-plant HGT |
| FHB | Fusarium head blight |
| IR | inverted repeat cpDNA |
| SSC | small single copy cpDNA |
| LSC | large single copy cpDNA |
Appendix A

Table A1.
Selected examples of organisms in which horizontal gene transfer was found.
Table A1.
Selected examples of organisms in which horizontal gene transfer was found.
| Gene Donor | Recipient of Gene | References |
|---|---|---|
| Bacteria: | Plant: | [108] |
| Agrobacterium rhizogenes | Nicotiana | |
| Plant: | Plant: | [109] |
| Oryza | Zea mays | |
| Plant: | Plant: | [110] |
| Flowering plant: | Gnetum | |
| Asterid | ||
| Plant: | Plant: | [58] |
| Monocot | Aktinidia | |
| Eudicot | Amborella | |
| Unknown | Betulaceae | |
| Ranunculales | Caprifoliaceae | |
| Monocot | Sanguinaria | |
| Bacteria: | Fern: | [111] |
| Nostoc azollae | genus Azolla | |
| Plant: | Plant: | [112] |
| Multiple grass lineages | Alloteropsis semialata | |
| Cyanobacteria: | [113] | |
| Calothrix | Alga: | |
| Alga: | Euglena myxocylindracea | |
| Porphyra purpurea | ||
| Bacteria: | Yeast: | [114] |
| Lactococcus sp. | Saccharomyces cerevisiae | |
| Plant: | Plant: | [115] |
| Flowering plants | Amborella trichopoda | |
| Mosses | ||
| Plant: | Plant: | [82] |
| Eudicot | Phaseolus vulgaris | |
| Plant: | Plant: | [116] |
| Fabales | Apodanthaceae | |
| Plant: | Plant: | [117] |
| Orobanchaceae | Plantago | |
| Bacteria: | Yeast: | [118] |
| Pseudomonas spp. | Saccharomyces cerevisiae | |
| Plant: | Plant: | [119] |
| Flowering plant: | Fern: | |
| Santalales | Botrychium virginianum | |
| Plant: | Plant: | [120] |
| Ericaceae | Ternstroemia | |
| Cyrillaceae | ||
| Plant: | Plant: | [121] |
| Flowering plant: | Flowering plant: | |
| Populus | Populus | |
| Cyanobacteria: | Alga: | [122] |
| Crocosphaera watsonii | Heterocapsa triquetra | |
| Trichodesmium erythraeum | Karlodinium micrum | |
| Oxyrrhis marina | ||
| Plant: | Plant: | [123] |
| Poa palustris | Festuca ovina | |
| Plant: | Plant: | [124] |
| Orobanche | Phelipanche | |
| Fungus: | Plant: | [125] |
| Physcomitrella patens | Oryza sativa | |
| Coprinopsis cinerea | Arabidopsis thaliana | |
| Laccaria bicolor | ||
| Plant: | Plant: | [126] |
| Cuscuta | Plantago | |
| Dinoflagellate: | Dinoflagellate: | [127] |
| Prorocentrum minimum | Prorocentrum minimum nucleus | |
| Alga | Fern: | [128] |
| Bacteria | Mankyua chejuense Helminthostachys zeylanica Botrychium ternatum | |
| Bacteria | Alga: | [129] |
| Ochrophytes | ||
| Bacteria: | Nematode: | [130] |
| Wolbachia | Onchocerca volvulus | |
| Bacteria: | Animal: | [131] |
| Wolbachia | Insect: | |
| Callosobruchus | ||
| Bacteria: | Animal: | [132] |
| Wolbachia | Insect: | |
| Drosophila ananassae | ||
| Alga: | Animal: | [133] |
| Vaucheria litorea | Snail: | |
| Elysia chlorotica | ||
| Bacteria: | Animal: | [134] |
| Wolbachia | Insect: | |
| Aedes aegypti | ||
| Cyanobacteria: | Protozoan: | [135] |
| Synechococcus sp. Prochlo rococcus sp. | Amoeba: | |
| Paulinella chromatophora | ||
| Human: | Protozoan: | [136] |
| Homo sapiens | Malaria parasite: | |
| Plasmodium vivax |
References
- Bansal, A.K.; Meyer, T.E. Evolutionary analysis by whole-genome comparisons. J. Bacteriol. 2002, 184, 2260–2272. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.; Hesselman, M.C.; Te Beek, T.A.; van Passel, M.W.J.; Eyre-Walker, A. Rates of Lateral gene transfer in prokaryotes: High but why? Trends Microbiol. 2015, 23, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Neuveglise, C.; Sacerdot, C.; Dujon, B. Insertion of horizontally transferred genes within conserved syntenic regions of yeast genomes. PLoS ONE 2009, 4, e6515. [Google Scholar] [CrossRef] [PubMed]
- Mc Ginty, S.E.; Rankin, D.J.; Brown, S.P. Horizontal gene transfer and the evolution of bacterial cooperation. Evolution 2011, 65, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Dagan, T.; Artzy-Randrup, Y.; Martin, W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 10039–10044. [Google Scholar] [CrossRef] [PubMed]
- Deschavanne, P.; Mallet, L.V.; Becq, J. Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus. BMC Genom. 2010, 11, 1–13. [Google Scholar]
- Kurland, C.G.; Canback, B.; Berg, O.G. Horizontal gene transfer: A critical view. Proc. Natl. Acad. Sci. USA 2003, 100, 9658–9662. [Google Scholar] [CrossRef] [PubMed]
- Tamames, J.; Moya, A. Estimating the extent of horizontal gene transfer in metagenomic sequences. BMC Genom. 2008, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Babić, A.; Lindner, A.B.; Vulić, M.; Stewart, E.J.; Radman, M. Direct visualization of horizontal gene transfer. Science 2008, 319, 1533–1536. [Google Scholar] [CrossRef] [PubMed]
- Bock, R. The give-and-take of DNA: Horizontal gene transfer in plants. Trends Plant Sci. 2010, 15, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Huang, J. Horizontal gene transfer in eukaryotes: The weak-link model. Bioessays 2013, 35, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M. Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 2015, 41, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Popa, O.; Dagan, T. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 2011, 14, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. Transfer of DNA from bacteria to eukaryotes. MBio 2016, 7, e00863. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; McCutcheon, J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2017, 16, 67–79. [Google Scholar] [CrossRef] [PubMed]
- McInerney, J.O. Horizontal gene transfer is less frequent in eukaryotes than prokaryotes but can be important. Bioessays 2017, 39, 1700002. [Google Scholar] [CrossRef] [PubMed]
- Skuza, L.; Demska, K. Horizontal gene transfer in plant mitochondria. Adv. Cell Biol. 2013, 40, 231–252. [Google Scholar]
- Zhao, N.; Grover, C.E.; Chen, Z.; Wendel, J.F.; Hua, J. Intergenomic gene transfer in diploid and allopolyploid Gossypium. BMC Plant Biol. 2019, 19, 1–18. [Google Scholar] [CrossRef]
- Iorizzo, M.; Grzebelus, D.; Senalik, D.; Szklarczyk, M.; Spooner, D.; Simon, P. Against the traffic: The first evidence for mitochondrial DNA transfer into the plastid genome. Mob. Genet. Elem. 2012, 2, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Senalik, D.; Szklarczyk, M.; Grzebelus, D.; Spooner, D.; Simon, P. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Copley, S.D.; Dhillon, J.K. Lateral gene transfer and parallel evolution in the history of gluathione biosynthesis genes. Genome Biol. 2002, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. Mitochondrion-to-plastid DNA transfer: It happens. New Phytol. 2014, 202, 736–738. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, S.; Hartmann, S.; Ruf, S.; Bock, R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. USA 2003, 100, 8828–8833. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Storchova, H.; Palmer, J.D.; Taylor, D.R. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol. Biol. 2010, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; de Pamphilis, C.W.; et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 2013, 342, 1468–1473. [Google Scholar]
- Goremykin, V.V.; Salamini, F.; Velasco, R.; Viola, R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2009, 26, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Rousseau-Gueutin, M.; Timmis, J.N. Plastid sequences contribute to some plant mitochondrial genes. Mol. Biol. Evol. 2012, 29, 1707–1711. [Google Scholar] [CrossRef]
- Wang, D.; Wu, Y.W.; Shih, A.C.C.; Wu, C.S.; Wang, Y.N.; Chaw, S.M. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 mya. Mol. Biol. Evol. 2007, 24, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Cao, D.; Li, S.; Su, A.; Geng, J.; Grover, C.E.; Hu, S.; Hua, J. The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS ONE 2013, 8, e69476. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Chen, Z.; Grover, C.E.; Wang, Y.; Li, S.; Liu, G.; Ma, Z.; Wendel, J.F.; Hua, J. Rapid evolutionary divergence of Gossypium barbadense and G hirsutum mitochondrial genomes. BMC Genom. 2015, 16, 770. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Wang, Y.; Hua, J. The roles of mitochondrion in intergenomic gene transfer in plants: A source and a pool. Int. J. Mol. Sci. 2018, 19, 547. [Google Scholar] [CrossRef] [PubMed]
- Cusimano, N.; Wicke, S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2016, 210, 680–693. [Google Scholar] [CrossRef]
- Knoop, V.; Unseld, M.; Marienfeld, J.; Brandt, P.; Sünkel, S.; Ullrich, H.; Brennicke, A. Copia-, gypsy- and line-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics 1996, 142, 579–585. [Google Scholar] [CrossRef]
- Rodriguez-Moreno, L.; Gonzalez, V.M.; Benjak, A.; Marti, M.C.; Puigdomenech, P.; Aranda, M.A.; Garcia-Mas, J. Determination of melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genom. 2011, 12, 424. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Feng, K.; Grover, C.E.; Li, P.; Liu, F.; Wang, Y.; Xu, Q.; Shang, M.; Zhou, Z.; Cai, X.; et al. Chloroplast DNA structural variation, phylogeny, and age of divergence among diploid cotton species. PLoS ONE 2016, 11, e0157183. [Google Scholar] [CrossRef]
- Feschotte, C.; Jiang, N.; Wessler, S.R. Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 2002, 3, 329–341. [Google Scholar] [CrossRef]
- Cossu, R.M.; Casola, C.; Giacomello, S.; Vidalis, A.; Scofield, D.G.; Zuccolo, A. LTR retrotransposons show low levels of unequal recombination and high rates of intraelement gene conversion in large plant genomes. Genome Biol. Evol. 2017, 9, 3449–3462. [Google Scholar] [CrossRef]
- SanMiguel, P.; Gaut, B.S.; Tikhonov, A.; Nakajima, Y.; Bennetzen, J.L. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998, 20, 43–45. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O.; Quesneville, H. Ltr retrotransposons in rice (Oryza sativa, L. ): Recent burst amplifications followed by rapid DNA loss. BMC Genom. 2007, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, J.S. LTR retrotransposon landscape in Medicago truncatula: More rapid removal than in rice. BMC Genom. 2008, 9, 382. [Google Scholar] [CrossRef]
- Hawkins, J.S.; Proulx, S.R.; Rapp, R.A.; Wendel, J.F. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc. Natl. Acad. Sci. USA 2009, 106, 17811–17816. [Google Scholar] [CrossRef]
- Petrinini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Andrews, J.H., Hirano, S.S., Eds.; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- Wang, Y.; Li, H.; Zhao, W.; He, X.; Chen, J.; Geng, X.; Xiao, M. Induction of toluene degradation and growth promotion in corn and wheat by horizontal gene transfer within endophytic bacteria. Soil Biol. Biochem. 2010, 42, 1051–1057. [Google Scholar] [CrossRef]
- Ziemienowicz, A. Transfer plazmidów między bakteriami a komórkami eukariotycznymi. Kosmos 2002, 51, 343–351. [Google Scholar]
- Zardoya, R.; Ding, X.D.; Kitagawa, Y.; Chrispeels, M.J. Origin of plant glycerol transporters by horizontal gene transfer and functional recruitment. Proc. Natl. Acad. Sci. USA 2002, 99, 14893–14896. [Google Scholar] [CrossRef]
- Roulin, A.; Piegu, B.; Fortune, P.M.; Sabot, F.; D’hont, A.; Manicacci, D.; Panaud, O. Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in Poaceae. BMC Evol. Biol. 2009, 9, 58. [Google Scholar] [CrossRef]
- Haile, J.K.; N’Diaye, A.; Walkowiak, S.; Nilsen, K.T.; Clarke, J.M.; Kutcher, H.R.; Steiner, B.; Buerstmayr, H.; Pozniak, C.J. Fusarium head blight in durum wheat: Recent status, breeding directions, and future research prospects. Phytopathology 2019, 109, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Bae, H. Horizontal gene transfer and endophytes: An implication for the acquisition of novel traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ren, X.; Mason, A.S.; Liu, H.; Xiao, M.; Li, J.; Fu, D. Horizontal gene transfer in plants. Funct. Integr. Genom. 2014, 14, 23–29. [Google Scholar] [CrossRef]
- Trobridge, G.D. Foamy virus vectors for gene transfer. Expert Opin. Biol. Ther. 2009, 9, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.G.J.; Haegeman, A.; Jones, J.T. Horizontal gene transfer in nematodes: A catalyst for plant parasitism? Mol. Plant Microbe Interact. 2011, 24, 879–887. [Google Scholar]
- Thyssen, G.; Svab, Z.; Maliga, P. Cell-to-cell movement of plastids in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 2439–2443. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M.; Richards, T.A. Gene transfer: Anything goes in plant mitochondria. BMC Biol. 2010, 8, 147. [Google Scholar] [CrossRef] [PubMed]
- Pal, C. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 2005, 37, 1372–1375. [Google Scholar] [CrossRef]
- Broothaerts, W.; Mitchell, H.J.; Weir, B.; Kaines, S.; Smith, L.M.A.; Yang, W.; Mayer, J.E.; Roa-Rodriguez, C.; Jefferson, R.A. Gene transfer to plants by diverse species of bacteria. Nature 2005, 433, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef]
- Sommer, R.J.; Mayer, W.E.; Schuster, L.N.; Bartelmes, G.; Dieterich, C. Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol. Biol. 2011, 11, 13. [Google Scholar]
- Smith, D.R. Extending the limited transfer window hypothesis to interorganelle DNA migration. Genome Biol. Evol. 2011, 3, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Chaw, S.M.; Shih, A.C.-C.; Wang, D.; Wu, Y.W.; Liu, S.M.; Chou, T.Y. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 2008, 25, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef]
- Kleine, T.; Maier, U.G.; Leister, D. DNA transfer from organelles to the nucleus: The idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 2009, 60, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA 2015, 112, e3515–e3524. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.X.; Zou, W.X. Endophytes: A rich source of functional metabolites. Nat. Prod. Rep. 2001, 18, 448–459. [Google Scholar] [CrossRef]
- Shinozuka, H.; Hettiarachchige, I.K.; Shinozuka, M.; Cogan, N.O.I.; Spangenberg, G.C.; Cocks, B.G.; Forster, J.W.; Sawbridge, T.I. Horizontaltransferofaß-1,6glucanase gene from anancestral species of fungal endophyte to a cool-season grass host. Sci. Rep. 2017, 7, 9024. [Google Scholar] [CrossRef] [PubMed]
- Sachin, N.; Manjunatha, B.L.; Kumara, P.M.; Ravikanth, G.; Shweta, S.; Suryanarayanan, T.S.; Ganeshaiah, K.N.; Shaanker, R.U. Do endophytic fungi possess pathway genes for plant secondary metabolites? Curr. Sci. 2013, 104, 178–182. [Google Scholar]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Trail, F. For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiol. 2009, 149, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Hathout, A.S.; Aly, S.E. Biological detoxification of mycotoxins: A review. Ann. Microbiol. 2014, 64, 905–919. [Google Scholar] [CrossRef]
- Hao, Y.; Rasheed, A.; Zhu, Z.; Wulff, B.B.H.; He, Z. Harnessing wheat Fhb1 for Fusarium resistance. Trends Plant Sci. 2020, 25, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Lagudah, E.S.; Krattinger, S.G. A new player contributing to durable Fusarium resistance. Nat. Genet. 2019, 51, 1070–1071. [Google Scholar] [CrossRef]
- Li, G.; Zhou, J.; Jia, H.; Gao, Z.; Fan, M.; Luo, Y.; Zhao, P.; Xue, S.; Li, N.; Yuan, Y.; et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat.Genet. 2019, 51, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Navathe, S.; Yadav, P.S.; Chand, R.; Mishra, V.K.; Vasistha, N.K.; Meher, P.K.; Joshi, A.K.; Gupta, P.K. ToxA- Tsn1 Interaction for Spot Blotch Susceptibility in Indian Wheat: An Example of Inverse Gene-for-Gene Relationship. Plant Dis. 2020, 104, 71–81. [Google Scholar] [CrossRef]
- Friesen, T.L.; Holmes, D.J.; Bowden, R.L.; Faris, J.D. ToxA Is Present in the U.S. Bipolaris sorokiniana Population and Is a Significant Virulence Factor on Wheat Harboring Tsn1. Plant Dis. 2018, 102, 2446–2452. [Google Scholar]
- Oliver, R.P.; Lord, M.; Rybak, K.; Faris, J.D.; Solomon, P.S. Emergence of tan spot disease caused by toxigenic Pyrenophora tritici-repentis in Australia is not associated with increased deployment of toxin-sensitive cultivars. Phytopathology 2008, 98, 488–491. [Google Scholar] [CrossRef]
- Xi, Z.; Bradley, R.K.; Wurdack, K.J.; Wong, K.; Sugumaran, M.; Bomblies, K.; Rest, J.S.; Davis, C.C. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genom. 2012, 13, 227. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Wang, Y.; Bradley, R.K.; Sugumaran, M.; Marx, C.J.; Rest, J.S.; Davis, C.C. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013, 9, e1003265. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.C.; Wurdack, K.J. Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science 2004, 305, 676–678. [Google Scholar] [CrossRef] [PubMed]
- Woloszyńska, M.; Bocer, T.; Mackiewicz, P.; Janska, H. A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots, to mitochondrial genome of Phaseolus. Plant Mol. Biol. 2004, 56, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Grewe, F.; Viehoever, P.; Weisshaar, B.; Knoop, V. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2009, 37, 5093–5104. [Google Scholar] [CrossRef] [PubMed]
- Clifton, S.W.; Minx, P.; Fauron, C.M.; Gibson, M.; Allen, J.O.; Sun, H.; Thompson, M.; Barbazuk, W.B.; Kanuganti, S.; Tayloe, C.; et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004, 136, 3486–3503. [Google Scholar] [CrossRef] [PubMed]
- Handa, H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): Comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003, 31, 5907–5916. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA (Cys) (GCA). Nucleic Acids Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Notsu, Y.; Masood, S.; Nishikawa, T.; Kubo, N.; Akiduki, G.; Nakazono, M.; Hirai, A.; Kadowaki, K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: Frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genom. 2002, 268, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.B.; Lonsdale, D.M. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc. Natl. Acad. Sci. USA 1984, 81, 1946–1950. [Google Scholar] [CrossRef]
- Hao, W.; Palmer, J.D. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl. Acad. Sci. USA 2009, 106, 16728–16733. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Richardson, A.O.; Zheng, Y.; Palmer, J.D. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. USA 2010, 107, 21576–21581. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.O.; Palmer, J.D. Horizontal gene transfer in plants. J. Exp. Bot. 2007, 58, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Nakazono, M.; Hirai, A. Transcription of plastid-derived tRNA genes in rice mitochondria. Curr. Genet. 1998, 34, 216–220. [Google Scholar] [CrossRef]
- Nakazono, M.; Nishiwaki, S.; Tsutsumi, N.; Hirai, A. A chloroplast-derived sequence is utilized as a source of promoter sequences for the gene for subunit 9 of NADH dehydrogenase (nad9) in rice mitochondria. Mol. Gen. Genet. 1996, 252, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Nishikawa, T.; Fujimoto, M.; Takanashi, H.; Arimura, S.; Tsutsumi, N.; Kadowaki, K. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008, 25, 1566–1575. [Google Scholar] [CrossRef] [PubMed]
- Gantt, J.S.; Baldauf, S.L.; Calie, P.J.; Weeden, N.F.; Palmer, J.D. Transfer of rp122 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991, 10, 3073–3078. [Google Scholar] [CrossRef] [PubMed]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Gray, J.C.; Morden, C.W.; et al. Many Parallel Losses of infA from Chloroplast DNA during Angiosperm Evolution with Multiple Independent Transfers to the Nucleus. Plant Cell 2001, 13, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Fujimoto, M.; Arimura, S.; Murata, J.; Tsutsumi, N.; Kadowaki, K. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 2007, 402, 51–56. [Google Scholar] [CrossRef]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366 924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Hupfer, H.; Swiatek, M.; Hornung, S.; Herrmann, R.G.; Maier, R.M.; Chiu, W.L.; Sears, B. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol. Gen. Genet. 2000, 263, 581–585. [Google Scholar] [CrossRef]
- Thomas, F.; Massenet, O.; Dorne, A.M.; Briat, J.F.; Mache, R. Expression of the rpl23, rpl2 and rps19 genes in spinach chloroplasts. Nucleic Acids Res. 1988, 16, 2461–2472. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Liu, T.; Wang, X.; Qu, J.; Jia, X. The complete chloroplast genome of Sargassum horneri and its phylogenetic analysis. Mitochondrial DNA B Resour. 2019, 4, 3312–3313. [Google Scholar] [CrossRef]
- Cui, Y.; Liang, R. The chloroplast genome sequence of Commelina communis (Commelinaceae). Mitochondrial DNA B Resour. 2019, 4, 2631–2632. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Delwiche, C.F. The origin and evolution of plastids and their genomes. In Molecular Systematics of Plants. II; DNA Sequencing; Soltis, D.E., Soltis, P.S., Doyle, J.J., Eds.; Kluwer Academic Publishing: Boston, MA, USA, 1998; pp. 375–409. [Google Scholar]
- Palmer, J.D.; Adams, K.L.; Cho, Y.; Parkinson, C.L.; Qiu, Y.L.; Song, K. Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. USA 2000, 97, 6960–6966. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Daley, D.O.; Qiu, Y.L.; Whelan, J.; Palmer, J.D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 2000, 408, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Morden, C.W.; Palmer, J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 1992, 89, 10648–10652. [Google Scholar] [CrossRef]
- Cusack, B.P.; Wolfe, K.H. When gene marriages don’t work out: Divorce by subfunctionalization. Trends Genet. 2007, 23, 270–272. [Google Scholar] [CrossRef]
- Aoki, S.; Syno, K. Horizontal gene transfer and mutation: Ngrol genes in the genome of Nicotiana glauca. Proc. Natl. Acad. Sci. USA 1999, 96, 13229–13234. [Google Scholar] [CrossRef]
- Adams, K.L.; Ong, H.C.; Palmer, J.D. Mitochondrial gene transfer in pieces: Fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 2001, 18, 2289–2297. [Google Scholar] [CrossRef]
- Won, H.; Renner, S.S. Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl. Acad. Sci. USA 2003, 100, 10824–10829. [Google Scholar] [CrossRef]
- Li, F.W.; Brouwer, P.; Carretero-Paulet, L.; Cheng, S.; De Vries, J.; Delaux, P.M.; Eily, A.; Koppers, N.; Kuo, L.Y.; Li, Z.; et al. Fern genomes alicidate land plant evolution and cyanobacterial symbioses. Nat. Plants 2018, 4, 460–472. [Google Scholar] [CrossRef]
- Dunning, L.T.; Olofsson, J.K.; Parisod, C.; Choudhury, R.R.; Moreno-Villena, J.J.; Yang, Y.; Dionora, J.; Quick, W.P.; Park, M.; Bennetzen, J.L.; et al. Lateral transfer of large DNA fragments spread functional genes among grasses. Proc. Natl. Acad. Sci. USA 2019, 116, 4416–4425. [Google Scholar] [CrossRef]
- Sheveleva, E.V.; Hallick, R.B. Recent horizontal intron transfer to a chloroplast genome. Nucleic Acids Res. 2004, 32, 803–810. [Google Scholar] [CrossRef]
- Gojković, Z.; Knecht, W.; Zameitat, E.; Warneboldt, J.; Coutelis, J.B.; Pynyaha, Y.; Neuveglise, C.; Moller, K.; Loffler, M.; Piskur, J. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Gen. Genom. 2004, 271, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Bergthorsson, U.; Richardson, A.O.; Young, G.J.; Goertzen, L.R.; Palmer, J.D. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl. Acad. Sci. USA 2004, 101, 17747–17752. [Google Scholar] [CrossRef] [PubMed]
- Nickrent, D.L.; Blarer, A.; Qiu, Y.L.; Vidal-Russell, R.; Anderson, F.E. Phylogenetic inference in Rafflesiales: The influence of rate heterogeneity and horizontal gene transfer. BMC Evol. Biol. 2004, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Stefanovic, S.; Young, G.J.; Palmer, J.D. Gene transfer from parasitic to host plants. Nature 2004, 432, 165–166. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Brachat, S.; Dietrich, F.S. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1102–1115. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.C.; Anderson, W.R.; Wurdack, K.J. Gene transfer from a parasitic flowering plant to a fern. Proc. R. Soc. Lond. B Biol. Sci. 2005, 272, 2237–2242. [Google Scholar] [CrossRef] [PubMed]
- Schönenberger, J.; Anderberg, A.A.; Sytsma, K.J. Molecular phylogenetics and patterns of floral evolution in the Ericales. Int. J. Plant Sci. 2005, 166, 265–288. [Google Scholar] [CrossRef]
- Choi, C.; Liu, Z.; Adams, K.L. Evolutionary transfers of mitochondrial genes to the nucleus in the Populus lineage and coexpression of nuclear and mitochondrial Sdh4 genes. New Phytol. 2006, 172, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.F.; Slamovits, C.H.; Keeling, P.J. Lateral gene transfer of a multigene region from Cyanobacteria to Dinoflagellates resulting in a novel plastid-targeted fusion protein. Mol. Biol. Evol. 2006, 23, 1437–1443. [Google Scholar] [CrossRef]
- Vallenback, P.; Jaarola, M.; Ghatnekar, L.; Bengtsson, B.O. Origin and timing of the horizontal transfer of a PgiC gene from Poa to Festuca ovina. Mol. Phylogenet. Evol. 2008, 46, 890–896. [Google Scholar] [CrossRef]
- Park, J.M.; Manen, J.F.; Schneeweiss, G.M. Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae). Mol. Phylogenet. Evol. 2007, 43, 974–985. [Google Scholar] [CrossRef]
- Richards, T.A.; Soanes, D.M.; Foster, P.G.; Leonard, G.; Thornton, C.R.; Talbot, N.J. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell 2009, 21, 1897–1911. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Stefanović, S.; Hao, W.; Gummow, J.S.; Jain, K.; Ahmed, D.; Palmer, J.D. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010, 8, 150. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Abassi, S.; Ki, J.-S. Origin and roles of a novel copper-zinc superoxide dismutase (CuZnSOD) gene from the harmful dinoflagellate Prorocentrum minimum. Gene 2019, 683, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kim, K.J. Evolution of six novel ORFs in the plastome of Mankyua chejuense and phylogeny of eusporangiate ferns. Sci. Rep. 2018, 8, 16466. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, R.G.; Villain, A.; Perez-Lamarque, B.; Audren de Kerdrel, G.; McCallum, G.; Watson, A.K.; Ait-Mohamed, O.; Alberti, A.; Corre, E.; Frischkorn, K.R.; et al. Phylogenomic fingerprinting of tempo and functions of horizontal gene transfer within ochrophytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2009974118. [Google Scholar] [CrossRef]
- Fenn, K.; Conlon, C.; Jones, M.; Quail, M.A.; Holroyd, N.E.; Parkhill, J.; Blaxter, M. Phylogenetic relationships of the Wolbachia of Nematodes and Arthropods. PLoS Pathog. 2006, 2, 887–899. [Google Scholar] [CrossRef]
- Kondo, N.; Ijichi, N.; Shimada, M.; Fukatsu, T. Prevailing triple infection with Wolbachia in Callosobruchus chinensis (Coleoptera: Bruchidae). Mol. Ecol. 2002, 11, 167–180. [Google Scholar] [CrossRef]
- Hotopp, J.C.D.; Clark, M.E.; Oliveira, D.C.S.G.; Foster, J.M.; Fischer, P.; MuzonTorres, M.C.; Giebel, J.D.; Kumar, N.; Ishmael, N.; Wang, S.; et al. Widespread latelar gene transfer from intracellular bacteria to multicellular eukaryotes. Science 2007, 317, 1753–1756. [Google Scholar] [CrossRef] [PubMed]
- Rumpho, M.E.; Pelletreau, K.N.; Moustafa, A.; Bhattacharya, D. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. USA 2008, 105, 17867–17871. [Google Scholar] [CrossRef] [PubMed]
- Klasson, L.; Kambris, Z.; Cook, P.E.; Walker, T.; Sinkins, S.P. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genom. 2009, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Ishida, K.I. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr. Biol. 2009, 19, 284–285. [Google Scholar] [CrossRef] [PubMed]
- Bar, D.Z. Evidence of massive horizontal gene transfer between humans and Plasmodium vivax. Nat. Preced. 2011, 1–17. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).