Horizontal Gene Transfer Involving Chloroplasts
Abstract
1. Introduction
2. Organisms between Which HGT Occurs
3. HGT Routes and Mechanisms
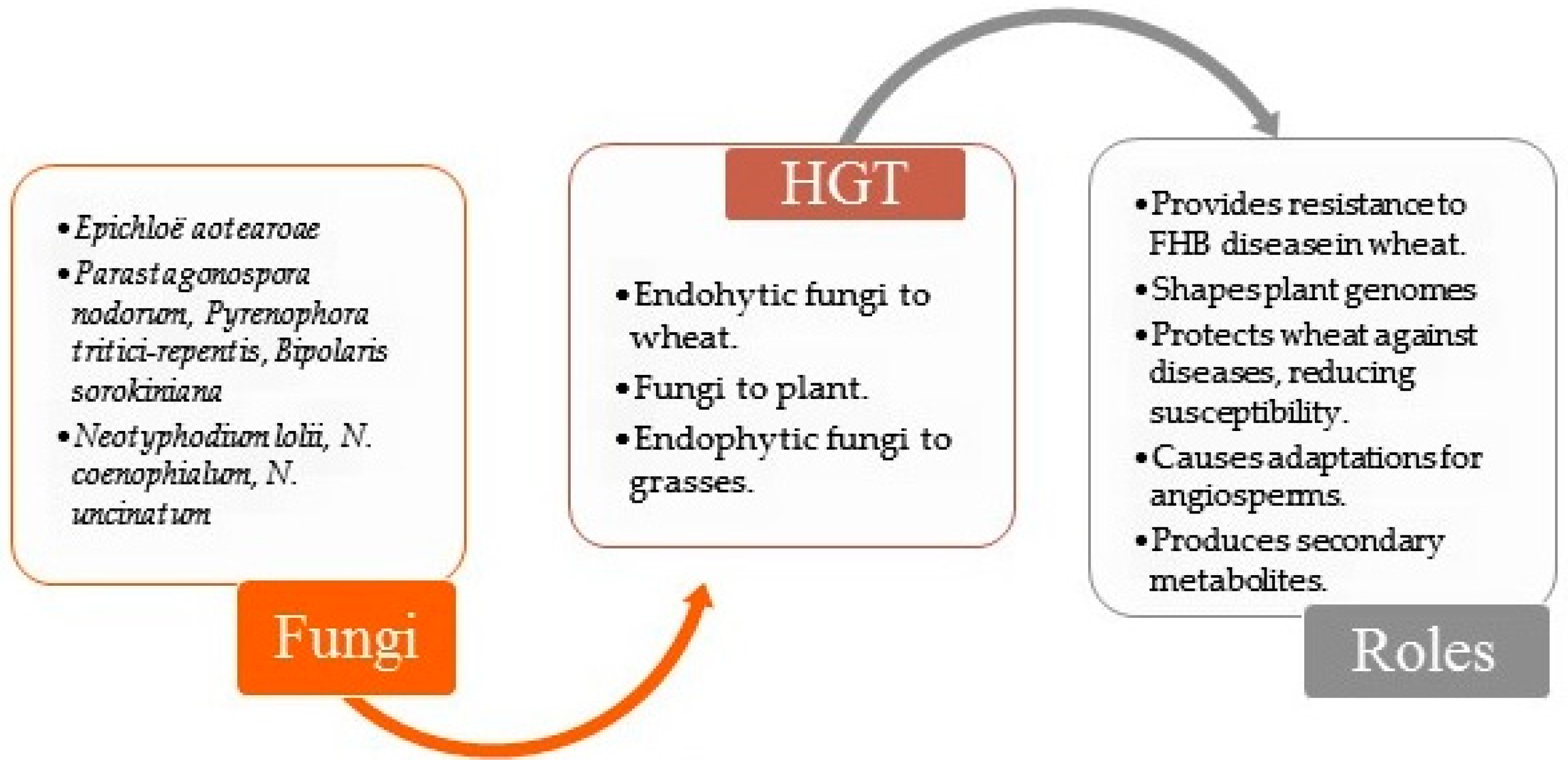
4. HGT and Endophytes
5. Intracellular Gene Transfer
5.1. HGT in Cell Nuclei and Plastids
5.2. Chloroplast-to-Mitochondria Transfer
5.3. Mitochondrion-to-Chloroplast Transfer of Rps16
5.4. Chloroplast-to-Nucleus Transfer
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HGT | Horizontal Gene Transfer |
| LGT | Lateral Gene Transfer |
| VGT | Vertical Gene Transfer |
| IGT | Intracellular Gene Transfer |
| IGT | Intragenomic Gene Transfer |
| cpDNA | chloroplast genome |
| nDNA | nuclear genome |
| mtDNA | mitochondrial genome |
| LTR | LTR-retro retrotransposons |
| FP-HGT | fungus-to-plant HGT |
| FHB | Fusarium head blight |
| IR | inverted repeat cpDNA |
| SSC | small single copy cpDNA |
| LSC | large single copy cpDNA |
Appendix A
| Gene Donor | Recipient of Gene | References |
|---|---|---|
| Bacteria: | Plant: | [108] |
| Agrobacterium rhizogenes | Nicotiana | |
| Plant: | Plant: | [109] |
| Oryza | Zea mays | |
| Plant: | Plant: | [110] |
| Flowering plant: | Gnetum | |
| Asterid | ||
| Plant: | Plant: | [58] |
| Monocot | Aktinidia | |
| Eudicot | Amborella | |
| Unknown | Betulaceae | |
| Ranunculales | Caprifoliaceae | |
| Monocot | Sanguinaria | |
| Bacteria: | Fern: | [111] |
| Nostoc azollae | genus Azolla | |
| Plant: | Plant: | [112] |
| Multiple grass lineages | Alloteropsis semialata | |
| Cyanobacteria: | [113] | |
| Calothrix | Alga: | |
| Alga: | Euglena myxocylindracea | |
| Porphyra purpurea | ||
| Bacteria: | Yeast: | [114] |
| Lactococcus sp. | Saccharomyces cerevisiae | |
| Plant: | Plant: | [115] |
| Flowering plants | Amborella trichopoda | |
| Mosses | ||
| Plant: | Plant: | [82] |
| Eudicot | Phaseolus vulgaris | |
| Plant: | Plant: | [116] |
| Fabales | Apodanthaceae | |
| Plant: | Plant: | [117] |
| Orobanchaceae | Plantago | |
| Bacteria: | Yeast: | [118] |
| Pseudomonas spp. | Saccharomyces cerevisiae | |
| Plant: | Plant: | [119] |
| Flowering plant: | Fern: | |
| Santalales | Botrychium virginianum | |
| Plant: | Plant: | [120] |
| Ericaceae | Ternstroemia | |
| Cyrillaceae | ||
| Plant: | Plant: | [121] |
| Flowering plant: | Flowering plant: | |
| Populus | Populus | |
| Cyanobacteria: | Alga: | [122] |
| Crocosphaera watsonii | Heterocapsa triquetra | |
| Trichodesmium erythraeum | Karlodinium micrum | |
| Oxyrrhis marina | ||
| Plant: | Plant: | [123] |
| Poa palustris | Festuca ovina | |
| Plant: | Plant: | [124] |
| Orobanche | Phelipanche | |
| Fungus: | Plant: | [125] |
| Physcomitrella patens | Oryza sativa | |
| Coprinopsis cinerea | Arabidopsis thaliana | |
| Laccaria bicolor | ||
| Plant: | Plant: | [126] |
| Cuscuta | Plantago | |
| Dinoflagellate: | Dinoflagellate: | [127] |
| Prorocentrum minimum | Prorocentrum minimum nucleus | |
| Alga | Fern: | [128] |
| Bacteria | Mankyua chejuense Helminthostachys zeylanica Botrychium ternatum | |
| Bacteria | Alga: | [129] |
| Ochrophytes | ||
| Bacteria: | Nematode: | [130] |
| Wolbachia | Onchocerca volvulus | |
| Bacteria: | Animal: | [131] |
| Wolbachia | Insect: | |
| Callosobruchus | ||
| Bacteria: | Animal: | [132] |
| Wolbachia | Insect: | |
| Drosophila ananassae | ||
| Alga: | Animal: | [133] |
| Vaucheria litorea | Snail: | |
| Elysia chlorotica | ||
| Bacteria: | Animal: | [134] |
| Wolbachia | Insect: | |
| Aedes aegypti | ||
| Cyanobacteria: | Protozoan: | [135] |
| Synechococcus sp. Prochlo rococcus sp. | Amoeba: | |
| Paulinella chromatophora | ||
| Human: | Protozoan: | [136] |
| Homo sapiens | Malaria parasite: | |
| Plasmodium vivax |
References
- Bansal, A.K.; Meyer, T.E. Evolutionary analysis by whole-genome comparisons. J. Bacteriol. 2002, 184, 2260–2272. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.; Hesselman, M.C.; Te Beek, T.A.; van Passel, M.W.J.; Eyre-Walker, A. Rates of Lateral gene transfer in prokaryotes: High but why? Trends Microbiol. 2015, 23, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Neuveglise, C.; Sacerdot, C.; Dujon, B. Insertion of horizontally transferred genes within conserved syntenic regions of yeast genomes. PLoS ONE 2009, 4, e6515. [Google Scholar] [CrossRef] [PubMed]
- Mc Ginty, S.E.; Rankin, D.J.; Brown, S.P. Horizontal gene transfer and the evolution of bacterial cooperation. Evolution 2011, 65, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Dagan, T.; Artzy-Randrup, Y.; Martin, W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 10039–10044. [Google Scholar] [CrossRef] [PubMed]
- Deschavanne, P.; Mallet, L.V.; Becq, J. Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus. BMC Genom. 2010, 11, 1–13. [Google Scholar]
- Kurland, C.G.; Canback, B.; Berg, O.G. Horizontal gene transfer: A critical view. Proc. Natl. Acad. Sci. USA 2003, 100, 9658–9662. [Google Scholar] [CrossRef] [PubMed]
- Tamames, J.; Moya, A. Estimating the extent of horizontal gene transfer in metagenomic sequences. BMC Genom. 2008, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Babić, A.; Lindner, A.B.; Vulić, M.; Stewart, E.J.; Radman, M. Direct visualization of horizontal gene transfer. Science 2008, 319, 1533–1536. [Google Scholar] [CrossRef] [PubMed]
- Bock, R. The give-and-take of DNA: Horizontal gene transfer in plants. Trends Plant Sci. 2010, 15, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Huang, J. Horizontal gene transfer in eukaryotes: The weak-link model. Bioessays 2013, 35, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M. Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 2015, 41, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Popa, O.; Dagan, T. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 2011, 14, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. Transfer of DNA from bacteria to eukaryotes. MBio 2016, 7, e00863. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; McCutcheon, J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2017, 16, 67–79. [Google Scholar] [CrossRef] [PubMed]
- McInerney, J.O. Horizontal gene transfer is less frequent in eukaryotes than prokaryotes but can be important. Bioessays 2017, 39, 1700002. [Google Scholar] [CrossRef] [PubMed]
- Skuza, L.; Demska, K. Horizontal gene transfer in plant mitochondria. Adv. Cell Biol. 2013, 40, 231–252. [Google Scholar]
- Zhao, N.; Grover, C.E.; Chen, Z.; Wendel, J.F.; Hua, J. Intergenomic gene transfer in diploid and allopolyploid Gossypium. BMC Plant Biol. 2019, 19, 1–18. [Google Scholar] [CrossRef]
- Iorizzo, M.; Grzebelus, D.; Senalik, D.; Szklarczyk, M.; Spooner, D.; Simon, P. Against the traffic: The first evidence for mitochondrial DNA transfer into the plastid genome. Mob. Genet. Elem. 2012, 2, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Senalik, D.; Szklarczyk, M.; Grzebelus, D.; Spooner, D.; Simon, P. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Copley, S.D.; Dhillon, J.K. Lateral gene transfer and parallel evolution in the history of gluathione biosynthesis genes. Genome Biol. 2002, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. Mitochondrion-to-plastid DNA transfer: It happens. New Phytol. 2014, 202, 736–738. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, S.; Hartmann, S.; Ruf, S.; Bock, R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. USA 2003, 100, 8828–8833. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Storchova, H.; Palmer, J.D.; Taylor, D.R. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol. Biol. 2010, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; de Pamphilis, C.W.; et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 2013, 342, 1468–1473. [Google Scholar]
- Goremykin, V.V.; Salamini, F.; Velasco, R.; Viola, R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2009, 26, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Rousseau-Gueutin, M.; Timmis, J.N. Plastid sequences contribute to some plant mitochondrial genes. Mol. Biol. Evol. 2012, 29, 1707–1711. [Google Scholar] [CrossRef]
- Wang, D.; Wu, Y.W.; Shih, A.C.C.; Wu, C.S.; Wang, Y.N.; Chaw, S.M. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 mya. Mol. Biol. Evol. 2007, 24, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Cao, D.; Li, S.; Su, A.; Geng, J.; Grover, C.E.; Hu, S.; Hua, J. The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS ONE 2013, 8, e69476. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Chen, Z.; Grover, C.E.; Wang, Y.; Li, S.; Liu, G.; Ma, Z.; Wendel, J.F.; Hua, J. Rapid evolutionary divergence of Gossypium barbadense and G hirsutum mitochondrial genomes. BMC Genom. 2015, 16, 770. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Wang, Y.; Hua, J. The roles of mitochondrion in intergenomic gene transfer in plants: A source and a pool. Int. J. Mol. Sci. 2018, 19, 547. [Google Scholar] [CrossRef] [PubMed]
- Cusimano, N.; Wicke, S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2016, 210, 680–693. [Google Scholar] [CrossRef]
- Knoop, V.; Unseld, M.; Marienfeld, J.; Brandt, P.; Sünkel, S.; Ullrich, H.; Brennicke, A. Copia-, gypsy- and line-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics 1996, 142, 579–585. [Google Scholar] [CrossRef]
- Rodriguez-Moreno, L.; Gonzalez, V.M.; Benjak, A.; Marti, M.C.; Puigdomenech, P.; Aranda, M.A.; Garcia-Mas, J. Determination of melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genom. 2011, 12, 424. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Feng, K.; Grover, C.E.; Li, P.; Liu, F.; Wang, Y.; Xu, Q.; Shang, M.; Zhou, Z.; Cai, X.; et al. Chloroplast DNA structural variation, phylogeny, and age of divergence among diploid cotton species. PLoS ONE 2016, 11, e0157183. [Google Scholar] [CrossRef]
- Feschotte, C.; Jiang, N.; Wessler, S.R. Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 2002, 3, 329–341. [Google Scholar] [CrossRef]
- Cossu, R.M.; Casola, C.; Giacomello, S.; Vidalis, A.; Scofield, D.G.; Zuccolo, A. LTR retrotransposons show low levels of unequal recombination and high rates of intraelement gene conversion in large plant genomes. Genome Biol. Evol. 2017, 9, 3449–3462. [Google Scholar] [CrossRef]
- SanMiguel, P.; Gaut, B.S.; Tikhonov, A.; Nakajima, Y.; Bennetzen, J.L. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998, 20, 43–45. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O.; Quesneville, H. Ltr retrotransposons in rice (Oryza sativa, L. ): Recent burst amplifications followed by rapid DNA loss. BMC Genom. 2007, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, J.S. LTR retrotransposon landscape in Medicago truncatula: More rapid removal than in rice. BMC Genom. 2008, 9, 382. [Google Scholar] [CrossRef]
- Hawkins, J.S.; Proulx, S.R.; Rapp, R.A.; Wendel, J.F. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc. Natl. Acad. Sci. USA 2009, 106, 17811–17816. [Google Scholar] [CrossRef]
- Petrinini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Andrews, J.H., Hirano, S.S., Eds.; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- Wang, Y.; Li, H.; Zhao, W.; He, X.; Chen, J.; Geng, X.; Xiao, M. Induction of toluene degradation and growth promotion in corn and wheat by horizontal gene transfer within endophytic bacteria. Soil Biol. Biochem. 2010, 42, 1051–1057. [Google Scholar] [CrossRef]
- Ziemienowicz, A. Transfer plazmidów między bakteriami a komórkami eukariotycznymi. Kosmos 2002, 51, 343–351. [Google Scholar]
- Zardoya, R.; Ding, X.D.; Kitagawa, Y.; Chrispeels, M.J. Origin of plant glycerol transporters by horizontal gene transfer and functional recruitment. Proc. Natl. Acad. Sci. USA 2002, 99, 14893–14896. [Google Scholar] [CrossRef]
- Roulin, A.; Piegu, B.; Fortune, P.M.; Sabot, F.; D’hont, A.; Manicacci, D.; Panaud, O. Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in Poaceae. BMC Evol. Biol. 2009, 9, 58. [Google Scholar] [CrossRef]
- Haile, J.K.; N’Diaye, A.; Walkowiak, S.; Nilsen, K.T.; Clarke, J.M.; Kutcher, H.R.; Steiner, B.; Buerstmayr, H.; Pozniak, C.J. Fusarium head blight in durum wheat: Recent status, breeding directions, and future research prospects. Phytopathology 2019, 109, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Bae, H. Horizontal gene transfer and endophytes: An implication for the acquisition of novel traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ren, X.; Mason, A.S.; Liu, H.; Xiao, M.; Li, J.; Fu, D. Horizontal gene transfer in plants. Funct. Integr. Genom. 2014, 14, 23–29. [Google Scholar] [CrossRef]
- Trobridge, G.D. Foamy virus vectors for gene transfer. Expert Opin. Biol. Ther. 2009, 9, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.G.J.; Haegeman, A.; Jones, J.T. Horizontal gene transfer in nematodes: A catalyst for plant parasitism? Mol. Plant Microbe Interact. 2011, 24, 879–887. [Google Scholar]
- Thyssen, G.; Svab, Z.; Maliga, P. Cell-to-cell movement of plastids in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 2439–2443. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M.; Richards, T.A. Gene transfer: Anything goes in plant mitochondria. BMC Biol. 2010, 8, 147. [Google Scholar] [CrossRef] [PubMed]
- Pal, C. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 2005, 37, 1372–1375. [Google Scholar] [CrossRef]
- Broothaerts, W.; Mitchell, H.J.; Weir, B.; Kaines, S.; Smith, L.M.A.; Yang, W.; Mayer, J.E.; Roa-Rodriguez, C.; Jefferson, R.A. Gene transfer to plants by diverse species of bacteria. Nature 2005, 433, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef]
- Sommer, R.J.; Mayer, W.E.; Schuster, L.N.; Bartelmes, G.; Dieterich, C. Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol. Biol. 2011, 11, 13. [Google Scholar]
- Smith, D.R. Extending the limited transfer window hypothesis to interorganelle DNA migration. Genome Biol. Evol. 2011, 3, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Chaw, S.M.; Shih, A.C.-C.; Wang, D.; Wu, Y.W.; Liu, S.M.; Chou, T.Y. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 2008, 25, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef]
- Kleine, T.; Maier, U.G.; Leister, D. DNA transfer from organelles to the nucleus: The idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 2009, 60, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA 2015, 112, e3515–e3524. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.X.; Zou, W.X. Endophytes: A rich source of functional metabolites. Nat. Prod. Rep. 2001, 18, 448–459. [Google Scholar] [CrossRef]
- Shinozuka, H.; Hettiarachchige, I.K.; Shinozuka, M.; Cogan, N.O.I.; Spangenberg, G.C.; Cocks, B.G.; Forster, J.W.; Sawbridge, T.I. Horizontaltransferofaß-1,6glucanase gene from anancestral species of fungal endophyte to a cool-season grass host. Sci. Rep. 2017, 7, 9024. [Google Scholar] [CrossRef] [PubMed]
- Sachin, N.; Manjunatha, B.L.; Kumara, P.M.; Ravikanth, G.; Shweta, S.; Suryanarayanan, T.S.; Ganeshaiah, K.N.; Shaanker, R.U. Do endophytic fungi possess pathway genes for plant secondary metabolites? Curr. Sci. 2013, 104, 178–182. [Google Scholar]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Trail, F. For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiol. 2009, 149, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Hathout, A.S.; Aly, S.E. Biological detoxification of mycotoxins: A review. Ann. Microbiol. 2014, 64, 905–919. [Google Scholar] [CrossRef]
- Hao, Y.; Rasheed, A.; Zhu, Z.; Wulff, B.B.H.; He, Z. Harnessing wheat Fhb1 for Fusarium resistance. Trends Plant Sci. 2020, 25, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Lagudah, E.S.; Krattinger, S.G. A new player contributing to durable Fusarium resistance. Nat. Genet. 2019, 51, 1070–1071. [Google Scholar] [CrossRef]
- Li, G.; Zhou, J.; Jia, H.; Gao, Z.; Fan, M.; Luo, Y.; Zhao, P.; Xue, S.; Li, N.; Yuan, Y.; et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat.Genet. 2019, 51, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Navathe, S.; Yadav, P.S.; Chand, R.; Mishra, V.K.; Vasistha, N.K.; Meher, P.K.; Joshi, A.K.; Gupta, P.K. ToxA- Tsn1 Interaction for Spot Blotch Susceptibility in Indian Wheat: An Example of Inverse Gene-for-Gene Relationship. Plant Dis. 2020, 104, 71–81. [Google Scholar] [CrossRef]
- Friesen, T.L.; Holmes, D.J.; Bowden, R.L.; Faris, J.D. ToxA Is Present in the U.S. Bipolaris sorokiniana Population and Is a Significant Virulence Factor on Wheat Harboring Tsn1. Plant Dis. 2018, 102, 2446–2452. [Google Scholar]
- Oliver, R.P.; Lord, M.; Rybak, K.; Faris, J.D.; Solomon, P.S. Emergence of tan spot disease caused by toxigenic Pyrenophora tritici-repentis in Australia is not associated with increased deployment of toxin-sensitive cultivars. Phytopathology 2008, 98, 488–491. [Google Scholar] [CrossRef]
- Xi, Z.; Bradley, R.K.; Wurdack, K.J.; Wong, K.; Sugumaran, M.; Bomblies, K.; Rest, J.S.; Davis, C.C. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genom. 2012, 13, 227. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Wang, Y.; Bradley, R.K.; Sugumaran, M.; Marx, C.J.; Rest, J.S.; Davis, C.C. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013, 9, e1003265. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.C.; Wurdack, K.J. Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science 2004, 305, 676–678. [Google Scholar] [CrossRef] [PubMed]
- Woloszyńska, M.; Bocer, T.; Mackiewicz, P.; Janska, H. A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots, to mitochondrial genome of Phaseolus. Plant Mol. Biol. 2004, 56, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Grewe, F.; Viehoever, P.; Weisshaar, B.; Knoop, V. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2009, 37, 5093–5104. [Google Scholar] [CrossRef] [PubMed]
- Clifton, S.W.; Minx, P.; Fauron, C.M.; Gibson, M.; Allen, J.O.; Sun, H.; Thompson, M.; Barbazuk, W.B.; Kanuganti, S.; Tayloe, C.; et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004, 136, 3486–3503. [Google Scholar] [CrossRef] [PubMed]
- Handa, H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): Comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003, 31, 5907–5916. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA (Cys) (GCA). Nucleic Acids Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Notsu, Y.; Masood, S.; Nishikawa, T.; Kubo, N.; Akiduki, G.; Nakazono, M.; Hirai, A.; Kadowaki, K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: Frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genom. 2002, 268, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.B.; Lonsdale, D.M. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc. Natl. Acad. Sci. USA 1984, 81, 1946–1950. [Google Scholar] [CrossRef]
- Hao, W.; Palmer, J.D. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl. Acad. Sci. USA 2009, 106, 16728–16733. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Richardson, A.O.; Zheng, Y.; Palmer, J.D. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. USA 2010, 107, 21576–21581. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.O.; Palmer, J.D. Horizontal gene transfer in plants. J. Exp. Bot. 2007, 58, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Nakazono, M.; Hirai, A. Transcription of plastid-derived tRNA genes in rice mitochondria. Curr. Genet. 1998, 34, 216–220. [Google Scholar] [CrossRef]
- Nakazono, M.; Nishiwaki, S.; Tsutsumi, N.; Hirai, A. A chloroplast-derived sequence is utilized as a source of promoter sequences for the gene for subunit 9 of NADH dehydrogenase (nad9) in rice mitochondria. Mol. Gen. Genet. 1996, 252, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Nishikawa, T.; Fujimoto, M.; Takanashi, H.; Arimura, S.; Tsutsumi, N.; Kadowaki, K. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008, 25, 1566–1575. [Google Scholar] [CrossRef] [PubMed]
- Gantt, J.S.; Baldauf, S.L.; Calie, P.J.; Weeden, N.F.; Palmer, J.D. Transfer of rp122 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991, 10, 3073–3078. [Google Scholar] [CrossRef] [PubMed]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Gray, J.C.; Morden, C.W.; et al. Many Parallel Losses of infA from Chloroplast DNA during Angiosperm Evolution with Multiple Independent Transfers to the Nucleus. Plant Cell 2001, 13, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Fujimoto, M.; Arimura, S.; Murata, J.; Tsutsumi, N.; Kadowaki, K. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 2007, 402, 51–56. [Google Scholar] [CrossRef]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366 924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Hupfer, H.; Swiatek, M.; Hornung, S.; Herrmann, R.G.; Maier, R.M.; Chiu, W.L.; Sears, B. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol. Gen. Genet. 2000, 263, 581–585. [Google Scholar] [CrossRef]
- Thomas, F.; Massenet, O.; Dorne, A.M.; Briat, J.F.; Mache, R. Expression of the rpl23, rpl2 and rps19 genes in spinach chloroplasts. Nucleic Acids Res. 1988, 16, 2461–2472. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Liu, T.; Wang, X.; Qu, J.; Jia, X. The complete chloroplast genome of Sargassum horneri and its phylogenetic analysis. Mitochondrial DNA B Resour. 2019, 4, 3312–3313. [Google Scholar] [CrossRef]
- Cui, Y.; Liang, R. The chloroplast genome sequence of Commelina communis (Commelinaceae). Mitochondrial DNA B Resour. 2019, 4, 2631–2632. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Delwiche, C.F. The origin and evolution of plastids and their genomes. In Molecular Systematics of Plants. II; DNA Sequencing; Soltis, D.E., Soltis, P.S., Doyle, J.J., Eds.; Kluwer Academic Publishing: Boston, MA, USA, 1998; pp. 375–409. [Google Scholar]
- Palmer, J.D.; Adams, K.L.; Cho, Y.; Parkinson, C.L.; Qiu, Y.L.; Song, K. Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. USA 2000, 97, 6960–6966. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Daley, D.O.; Qiu, Y.L.; Whelan, J.; Palmer, J.D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 2000, 408, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Morden, C.W.; Palmer, J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 1992, 89, 10648–10652. [Google Scholar] [CrossRef]
- Cusack, B.P.; Wolfe, K.H. When gene marriages don’t work out: Divorce by subfunctionalization. Trends Genet. 2007, 23, 270–272. [Google Scholar] [CrossRef]
- Aoki, S.; Syno, K. Horizontal gene transfer and mutation: Ngrol genes in the genome of Nicotiana glauca. Proc. Natl. Acad. Sci. USA 1999, 96, 13229–13234. [Google Scholar] [CrossRef]
- Adams, K.L.; Ong, H.C.; Palmer, J.D. Mitochondrial gene transfer in pieces: Fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 2001, 18, 2289–2297. [Google Scholar] [CrossRef]
- Won, H.; Renner, S.S. Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl. Acad. Sci. USA 2003, 100, 10824–10829. [Google Scholar] [CrossRef]
- Li, F.W.; Brouwer, P.; Carretero-Paulet, L.; Cheng, S.; De Vries, J.; Delaux, P.M.; Eily, A.; Koppers, N.; Kuo, L.Y.; Li, Z.; et al. Fern genomes alicidate land plant evolution and cyanobacterial symbioses. Nat. Plants 2018, 4, 460–472. [Google Scholar] [CrossRef]
- Dunning, L.T.; Olofsson, J.K.; Parisod, C.; Choudhury, R.R.; Moreno-Villena, J.J.; Yang, Y.; Dionora, J.; Quick, W.P.; Park, M.; Bennetzen, J.L.; et al. Lateral transfer of large DNA fragments spread functional genes among grasses. Proc. Natl. Acad. Sci. USA 2019, 116, 4416–4425. [Google Scholar] [CrossRef]
- Sheveleva, E.V.; Hallick, R.B. Recent horizontal intron transfer to a chloroplast genome. Nucleic Acids Res. 2004, 32, 803–810. [Google Scholar] [CrossRef]
- Gojković, Z.; Knecht, W.; Zameitat, E.; Warneboldt, J.; Coutelis, J.B.; Pynyaha, Y.; Neuveglise, C.; Moller, K.; Loffler, M.; Piskur, J. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Gen. Genom. 2004, 271, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Bergthorsson, U.; Richardson, A.O.; Young, G.J.; Goertzen, L.R.; Palmer, J.D. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl. Acad. Sci. USA 2004, 101, 17747–17752. [Google Scholar] [CrossRef] [PubMed]
- Nickrent, D.L.; Blarer, A.; Qiu, Y.L.; Vidal-Russell, R.; Anderson, F.E. Phylogenetic inference in Rafflesiales: The influence of rate heterogeneity and horizontal gene transfer. BMC Evol. Biol. 2004, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Stefanovic, S.; Young, G.J.; Palmer, J.D. Gene transfer from parasitic to host plants. Nature 2004, 432, 165–166. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Brachat, S.; Dietrich, F.S. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1102–1115. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.C.; Anderson, W.R.; Wurdack, K.J. Gene transfer from a parasitic flowering plant to a fern. Proc. R. Soc. Lond. B Biol. Sci. 2005, 272, 2237–2242. [Google Scholar] [CrossRef] [PubMed]
- Schönenberger, J.; Anderberg, A.A.; Sytsma, K.J. Molecular phylogenetics and patterns of floral evolution in the Ericales. Int. J. Plant Sci. 2005, 166, 265–288. [Google Scholar] [CrossRef]
- Choi, C.; Liu, Z.; Adams, K.L. Evolutionary transfers of mitochondrial genes to the nucleus in the Populus lineage and coexpression of nuclear and mitochondrial Sdh4 genes. New Phytol. 2006, 172, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.F.; Slamovits, C.H.; Keeling, P.J. Lateral gene transfer of a multigene region from Cyanobacteria to Dinoflagellates resulting in a novel plastid-targeted fusion protein. Mol. Biol. Evol. 2006, 23, 1437–1443. [Google Scholar] [CrossRef]
- Vallenback, P.; Jaarola, M.; Ghatnekar, L.; Bengtsson, B.O. Origin and timing of the horizontal transfer of a PgiC gene from Poa to Festuca ovina. Mol. Phylogenet. Evol. 2008, 46, 890–896. [Google Scholar] [CrossRef]
- Park, J.M.; Manen, J.F.; Schneeweiss, G.M. Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae). Mol. Phylogenet. Evol. 2007, 43, 974–985. [Google Scholar] [CrossRef]
- Richards, T.A.; Soanes, D.M.; Foster, P.G.; Leonard, G.; Thornton, C.R.; Talbot, N.J. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell 2009, 21, 1897–1911. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Stefanović, S.; Hao, W.; Gummow, J.S.; Jain, K.; Ahmed, D.; Palmer, J.D. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010, 8, 150. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Abassi, S.; Ki, J.-S. Origin and roles of a novel copper-zinc superoxide dismutase (CuZnSOD) gene from the harmful dinoflagellate Prorocentrum minimum. Gene 2019, 683, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kim, K.J. Evolution of six novel ORFs in the plastome of Mankyua chejuense and phylogeny of eusporangiate ferns. Sci. Rep. 2018, 8, 16466. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, R.G.; Villain, A.; Perez-Lamarque, B.; Audren de Kerdrel, G.; McCallum, G.; Watson, A.K.; Ait-Mohamed, O.; Alberti, A.; Corre, E.; Frischkorn, K.R.; et al. Phylogenomic fingerprinting of tempo and functions of horizontal gene transfer within ochrophytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2009974118. [Google Scholar] [CrossRef]
- Fenn, K.; Conlon, C.; Jones, M.; Quail, M.A.; Holroyd, N.E.; Parkhill, J.; Blaxter, M. Phylogenetic relationships of the Wolbachia of Nematodes and Arthropods. PLoS Pathog. 2006, 2, 887–899. [Google Scholar] [CrossRef]
- Kondo, N.; Ijichi, N.; Shimada, M.; Fukatsu, T. Prevailing triple infection with Wolbachia in Callosobruchus chinensis (Coleoptera: Bruchidae). Mol. Ecol. 2002, 11, 167–180. [Google Scholar] [CrossRef]
- Hotopp, J.C.D.; Clark, M.E.; Oliveira, D.C.S.G.; Foster, J.M.; Fischer, P.; MuzonTorres, M.C.; Giebel, J.D.; Kumar, N.; Ishmael, N.; Wang, S.; et al. Widespread latelar gene transfer from intracellular bacteria to multicellular eukaryotes. Science 2007, 317, 1753–1756. [Google Scholar] [CrossRef] [PubMed]
- Rumpho, M.E.; Pelletreau, K.N.; Moustafa, A.; Bhattacharya, D. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. USA 2008, 105, 17867–17871. [Google Scholar] [CrossRef] [PubMed]
- Klasson, L.; Kambris, Z.; Cook, P.E.; Walker, T.; Sinkins, S.P. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genom. 2009, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Ishida, K.I. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr. Biol. 2009, 19, 284–285. [Google Scholar] [CrossRef] [PubMed]
- Bar, D.Z. Evidence of massive horizontal gene transfer between humans and Plasmodium vivax. Nat. Preced. 2011, 1–17. [Google Scholar] [CrossRef]

| Gen(s) | Transfer Type | Ref. |
|---|---|---|
| rpl22 | chloroplast to nucleus | [95] |
| infA | chloroplast to nucleus | [96] |
| tufA | chloroplast to nucleus | [95] |
| rpl32, rps122 | chloroplast to nucleus | [91,92,93,94,95,96,97,98,99,100] |
| pvs-trnA | chloroplast to mitochondrion | [83] |
| rps16 | chloroplast to mitochondrion | [94] |
| rps16 | mitochondrion to mitochondrion | [94] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filip, E.; Skuza, L. Horizontal Gene Transfer Involving Chloroplasts. Int. J. Mol. Sci. 2021, 22, 4484. https://doi.org/10.3390/ijms22094484
Filip E, Skuza L. Horizontal Gene Transfer Involving Chloroplasts. International Journal of Molecular Sciences. 2021; 22(9):4484. https://doi.org/10.3390/ijms22094484
Chicago/Turabian StyleFilip, Ewa, and Lidia Skuza. 2021. "Horizontal Gene Transfer Involving Chloroplasts" International Journal of Molecular Sciences 22, no. 9: 4484. https://doi.org/10.3390/ijms22094484
APA StyleFilip, E., & Skuza, L. (2021). Horizontal Gene Transfer Involving Chloroplasts. International Journal of Molecular Sciences, 22(9), 4484. https://doi.org/10.3390/ijms22094484







