Back to the Origins: Background and Perspectives of Grapevine Domestication
Abstract
:1. Introduction
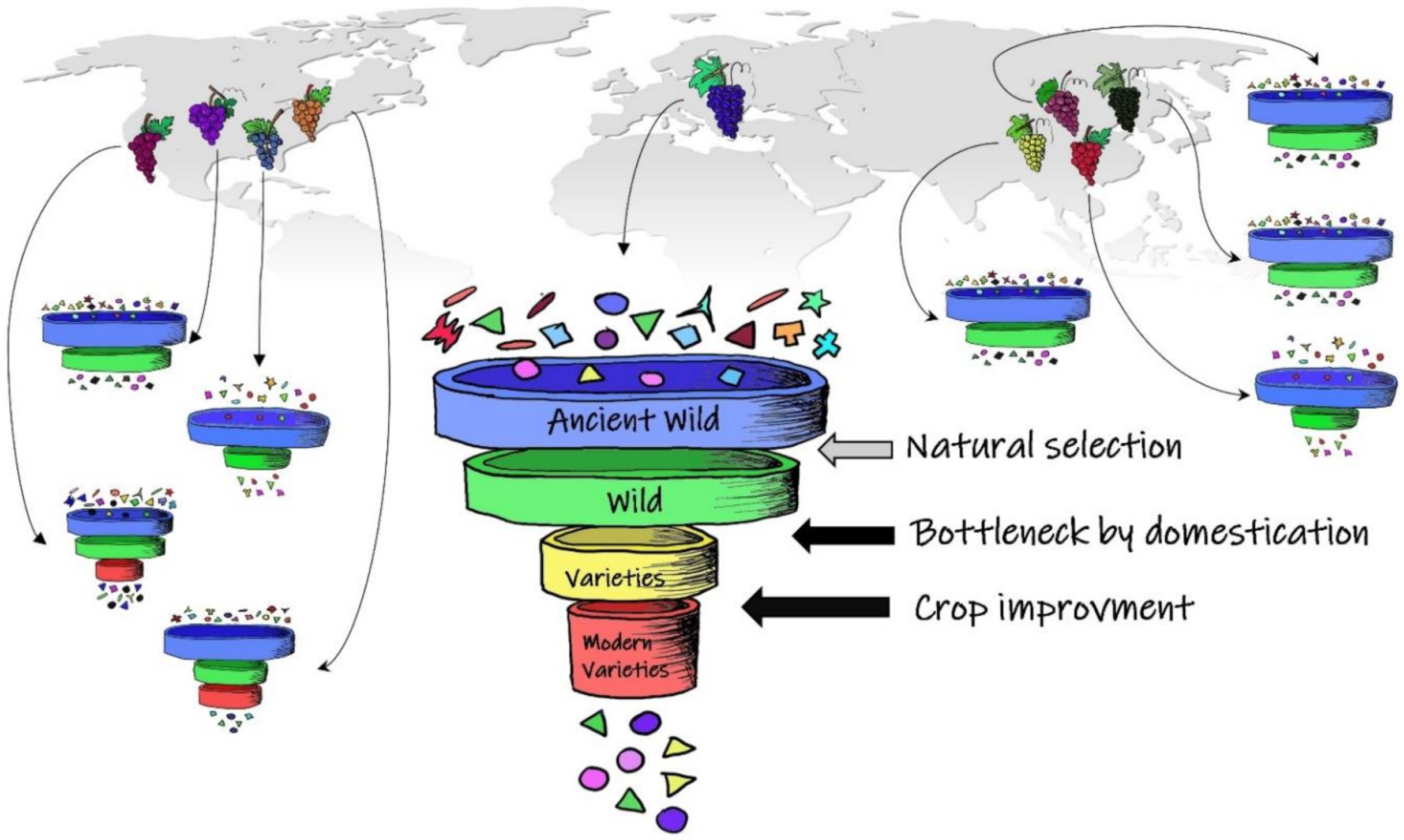
2. What Is Lurking behind the Domestication Process?
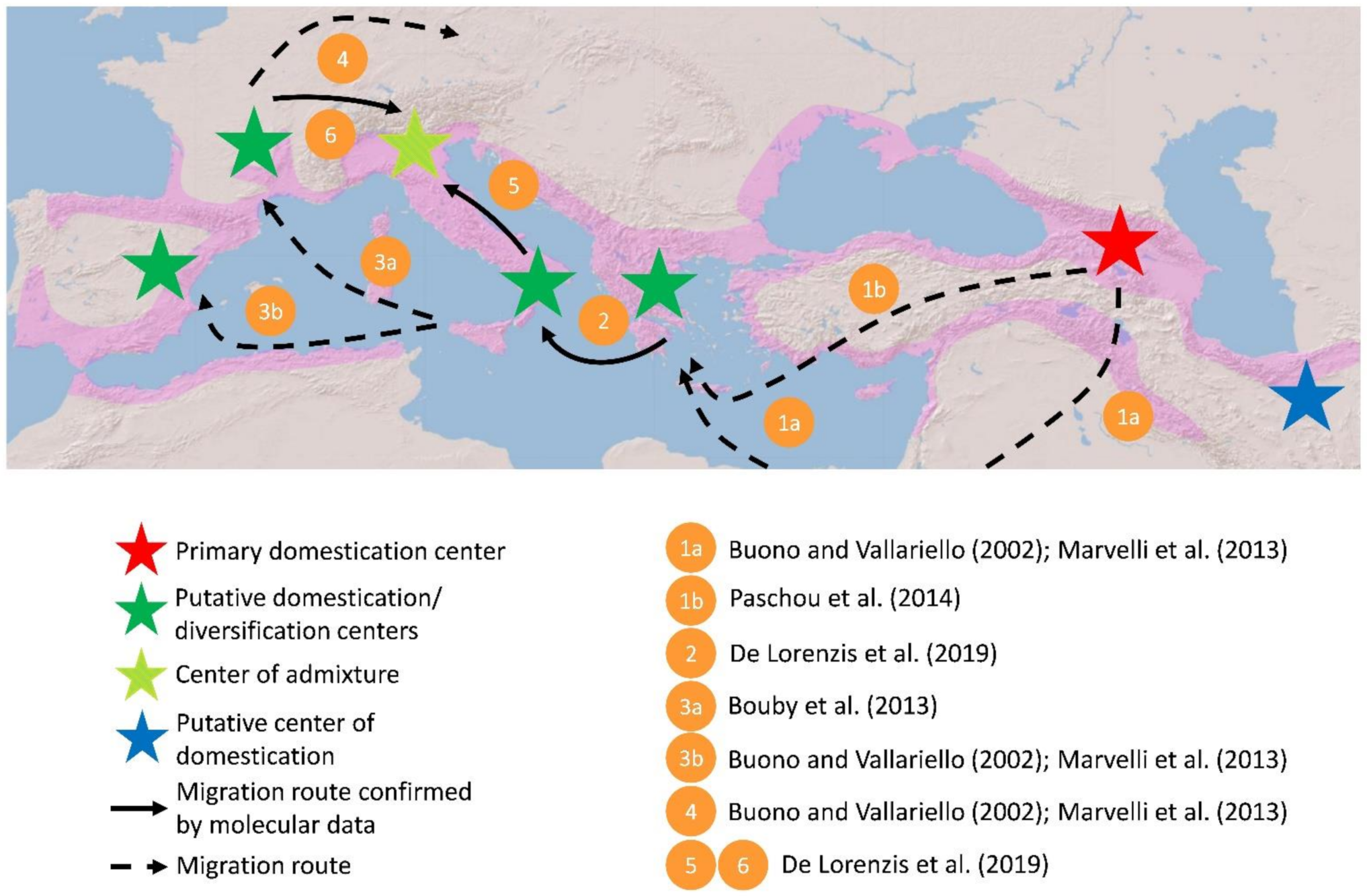
3. Where and How Many Times Has Grapevine Domestication Taken Place?
4. Plastid DNA to Explore the Maternal Lineages
5. When Did Grapevine Domestication Occur?
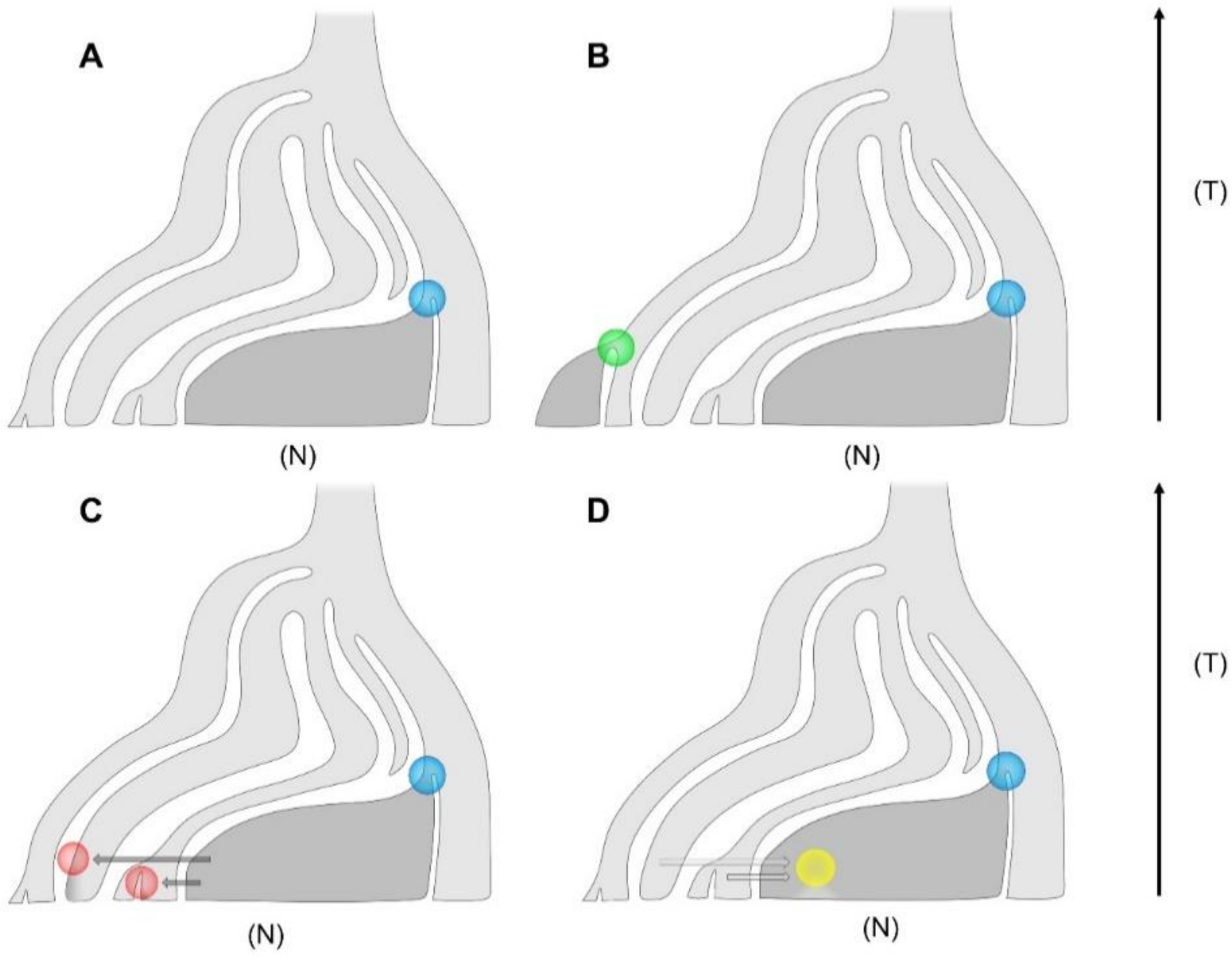
6. Introgression between Wild and Domesticated Grapevine
7. Ancient DNA to Investigate Grapevine Domestication
8. From Genome to Super-Pangenome
9. Concluding Remarks and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Organisation of Vine and Wine. Available online: https://www.oiv.int/en/ (accessed on 1 April 2021).
- This, P.; Lacombe, T.; Thomas, M.R. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006, 22, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Caporali, E.; Spada, A.; Marziani, G.; Failla, O.; Scienza, A. The arrest of development of abortive reproductive organs in the unisexual flower of Vitis vinifera ssp. silvestris. Sex. Plant Reprod. 2003, 15, 291–300. [Google Scholar] [CrossRef]
- Massonnet, M.; Cochetel, N.; Minio, A.; Vondras, A.M.; Lin, J.; Muyle, A.; Garcia, J.F.; Zhou, Y.; Delledonne, M.; Riaz, S.; et al. The genetic basis of sex determination in grapes. Nat. Commun. 2020, 11, 2902. [Google Scholar] [CrossRef] [PubMed]
- Ekhvaia, J.; Akhalkatsi, M. Morphological variation and relationships of Georgian populations of Vitis vinifera L. subsp. sylvestris (C.C. Gmel.) Hegi. Flora Morphol. Distrib. Funct. Ecol. Plants 2010, 205, 608–617. [Google Scholar]
- Naqinezhad, A.; Ramezani, E.; Djamali, M.; Schnitzler, A.; Arnold, C. Wild grapevine (Vitis vinifera subsp. sylvestris) in the Hyrcanian relict forests of northern Iran: An overview of current taxonomy, ecology and palaeorecords. J. For. Res. 2018, 29, 1757–1768. [Google Scholar] [CrossRef] [Green Version]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Zohary, D., Hopf, M., Weiss, E., Eds.; Oxford University Press: Oxford, UK, 2012; ISBN 9780199549061. [Google Scholar]
- McGovern, P.; Jalabadze, M.; Batiuk, S.; Callahan, M.P.; Smith, K.E.; Hall, G.R.; Kvavadze, E.; Maghradze, D.; Rusishvili, N.; Bouby, L.; et al. Early Neolithic wine of Georgia in the South Caucasus. Proc. Natl. Acad. Sci. USA. 2017, 114, 10309–10318. [Google Scholar] [CrossRef] [Green Version]
- Fuller, D.Q.; Stevens, C.J. Between domestication and civilization: The role of agriculture and arboriculture in the emergence of the first urban societies. Veg. Hist. Archaeobot. 2019, 28, 263–282. [Google Scholar] [CrossRef] [Green Version]
- Hegi, G. Illustrierte Flora von Mitteleuropa; Karl Hans Verlag: München, Germany, 1925. [Google Scholar]
- Galet, P. Dictionnaire Encyclopédique des Cepages; Hachette Pratique: Paris, France, 2000. [Google Scholar]
- Negrul, A.M. Evolution of cultivated forms of grapes. Acad. Sci. URSS 1938, 18, 585–588. [Google Scholar]
- Laucou, V.; Lacombe, T.; Dechesne, F.; Siret, R.; Bruno, J.-P.; Dessup, M.; Dessup, T.; Ortigosa, P.; Parra, P.; Roux, C.; et al. High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor. Appl. Genet. 2011, 122, 1233–1245. [Google Scholar] [CrossRef]
- Laucou, V.; Launay, A.; Bacilieri, R.; Lacombe, T.; Adam-Blondon, A.-F.; Bérard, A.; Chauveau, A.; de Andrés, M.T.; Hausmann, L.; Ibáñez, J.; et al. Extended diversity analysis of cultivated grapevine Vitis vinifera with 10K genome-wide SNPs. PLoS ONE 2018, 13, e0192540. [Google Scholar] [CrossRef]
- Riaz, S.; De Lorenzis, G.; Velasco, D.; Koehmstedt, A.; Maghradze, D.; Bobokashvili, Z.; Musayev, M.; Zdunic, G.; Laucou, V.; Walker, M.A.; et al. Genetic diversity analysis of cultivated and wild grapevine (Vitis vinifera L.) accessions around the Mediterranean basin and Central Asia. BMC Plant Biol. 2018, 18, 137. [Google Scholar] [CrossRef]
- Grassi, F.; Arroyo-Garcia, R. Origins and Domestication of the Grape. Front. Plant Sci. 2020, 11, 1176. [Google Scholar] [CrossRef]
- Fernie, A.R.; Yan, J. De Novo Domestication: An Alternative Route toward New Crops for the Future. Mol. Plant 2019, 12, 615–631. [Google Scholar] [CrossRef] [Green Version]
- Gross, B.L.; Olsen, K.M. Genetic perspectives on crop domestication. Trends Plant Sci. 2010, 15, 529–537. [Google Scholar] [CrossRef] [Green Version]
- Warschefsky, E.; Penmetsa, R.V.; Cook, D.R.; von Wettberg, E.J.B. Back to the wilds: Tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am. J. Bot. 2014, 101, 1791–1800. [Google Scholar] [CrossRef]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytol. 2012, 196, 29–48. [Google Scholar] [CrossRef]
- Gepts, P.; Famula, T.R.; Bettinger, R.L.; Brush, S.B.; Damania, A.B.; McGuire, P.E.; Qualset, C.O. Biodiversity in Agriculture; Gepts, P., Famula, T.R., Bettinger, R.L., Brush, S.B., Damania, A.B., McGuire, P.E., Qualset, C.O., Eds.; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Kantar, M.B.; Nashoba, A.R.; Anderson, J.E.; Blackman, B.K.; Rieseberg, L.H. The genetics and genomics of plant domestication. Bioscience 2017, 67, 971–982. [Google Scholar] [CrossRef] [Green Version]
- Vavilov, N.I. Origin and Geography of Cultivated Plants; Cambridge University Press: Cambridge, UK, 1926. [Google Scholar]
- Krishnan, S.G.; Waters, D.L.E.; Henry, R.J. Australian wild rice reveals pre-domestication origin of polymorphism deserts in rice genome. PLoS ONE 2014, 9, e98843. [Google Scholar] [CrossRef]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef]
- Olmo, H.P. Grapes. In Evolution of Crop Plants; Simmonds, N.W., Ed.; Longman: New York, NY, USA, 1976; pp. 86–90. [Google Scholar]
- McGovern, P.E.; Fleming, S.J.; Katz, S.H. The Origins and Ancient History of Wine; McGovern, P.E., Fleming, S.J., Katz, S.H., Eds.; Routledge: London, UK, 1996. [Google Scholar]
- Forni, G. The origin of “Old World” viticulture. In Caucasus and Northern Black Sea Region; Maghradze, D., Rustioni, L., Scienza, A., Turok, J., Failla, O., Eds.; Julius Kuhn-Institut: Albersweiler, Germany, 2012; pp. 27–38. [Google Scholar]
- Buono, R.; Vallariello, G. Introduzione e diffusione della vite (Vitis vinifera L.) in Italia. Delpinoa 2002, 44, 39–51. [Google Scholar]
- Marvelli, S.; De’ Siena, S.; Rizzoli, E.; Marchesini, M. The origin of grapevine cultivation in Italy: The archaeobotanical evidence. Ann. Bot. 2013, 3, 155–163. [Google Scholar]
- Sefc, K.M.; Lopes, M.S.; Lefort, F.; Botta, R.; Roubelakis-Angelakis, K.A.; Ibáñez, J.; Pejić, I.; Wagner, H.W.; Glössl, J.; Steinkellner, H. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor. Appl. Genet. 2000, 100, 498–505. [Google Scholar] [CrossRef]
- Aradhya, M.K.; Dangl, G.S.; Prins, B.H.; Boursiquot, J.-M.; Walker, M.A.; Meredith, C.P.; Simon, C.J. Genetic structure and differentiation in cultivated grape, Vitis vinifera L. Genet. Res. 2003, 81, 179–192. [Google Scholar] [CrossRef] [Green Version]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef]
- Imazio, S.; Maghradze, D.; Lorenzis, G.; Bacilieri, R.; Laucou, V.; This, P.; Scienza, A.; Failla, O. From the cradle of grapevine domestication: Molecular overview and description of Georgian grapevine (Vitis vinifera L.) germplasm. Tree Genet. Genomes 2013, 9, 641–658. [Google Scholar] [CrossRef]
- De Lorenzis, G.; Las Casas, G.; Brancadoro, L.; Scienza, A. Genotyping of Sicilian grapevine germplasm resources (V. vinifera L.) and their relationships with Sangiovese. Sci. Hortic. 2018, 169, 189–198. [Google Scholar] [CrossRef]
- Grassi, F.; Labra, M.; Imazio, S.; Spada, A.; Sgorbati, S.; Scienza, A.; Sala, F. Evidence of a secondary grapevine domestication centre detected by SSR analysis. Theor. Appl. Genet. 2003, 107, 1315–1320. [Google Scholar] [CrossRef]
- Snoussi, H.; Ben Slimane, M.H.; Ruiz-García, L.; Martínez-Zapater, J.M.; Arroyo-García, R. Genetic relationship among cultivated and wild grapevine accessions from Tunisia. Genome 2004, 47, 1211–1219. [Google Scholar] [CrossRef]
- Vouillamoz, J.; Maigre, D.; Meredith, C.P. Microsatellite analysis of ancient alpine grape cultivars: Pedigree reconstruction of Vitis vinifera L. “Cornalin du Valais”. Theor. Appl. Genet. 2003, 107, 448–454. [Google Scholar] [CrossRef]
- Lacombe, T.; Boursiquot, J.-M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.-P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 2012, 126, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Labra, M.; Imazio, S.; Grassi, F.; Rossoni, M.; Citterio, S.; Sgorbati, S.; Scienza, A.; Failla, O. Molecular approach to assess the origin of cv. Marzemino. Vitis J. Grapevine Res. 2003, 42, 137–140. [Google Scholar]
- Crespan, M.; Cabello, F.; Giannetto, S.; Ibanez, J.; Karoglan Kontic, J.; Maletic, E.; Pejic, I.; Rodrìgez-Torres, I.; Antonacci, D. Malvasia delle Lipari, Malvasia di Sardegna, Greco di Gerace, Malvasia de Sitges and Malvasia dubrovačka—Synonyms of an old and famous grape cultivar. Vitis Geilweilerhof 2006, 45, 69–73. [Google Scholar]
- Crespan, M.; Cancellier, S.; Chies, R.; Giannetto, S.; Meneghetti, S.; Costacurta, A.; Sperimentale, I.; Sezione, A.M.G. Molecular contribution to the knowledge of two ancient varietal populations: ‘Rabosi’ and ‘Glere’. Acta Hortic. 2009, 217–220. [Google Scholar] [CrossRef]
- De Andrés, M.T.; Benito, A.; Pérez-Rivera, G.; Ocete, R.; Lopez, M.; Gaforio, L.; Muñoz, G.; Cabello, F.; Zapater, J.M.M.; Arroyo-García, R. Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol. Ecol. 2012, 21, 800–816. [Google Scholar] [CrossRef]
- Zdunić, G.; Lukšić, K.; Nagy, Z.A.; Mucalo, A.; Hančević, K.; Radić, T.; Butorac, L.; Jahnke, G.G.; Kiss, E.; Ledesma-Krist, G.; et al. Genetic structure and relationships among wild and cultivated grapevines from Central Europe and part of the Western Balkan Peninsula. Genes 2020, 11, 962. [Google Scholar] [CrossRef]
- Lopes, M.S.; Mendonça, D.; dos Santos, M.R.; Eiras-Dias, J.E.; da Machado, A.C. New insights on the genetic basis of Portuguese grapevine and on grapevine domestication. Genome 2009, 52, 790–800. [Google Scholar] [CrossRef] [Green Version]
- De Michele, R.; La Bella, F.; Gristina, A.S.; Fontana, I.; Pacifico, D.; Garfi, G.; Motisi, A.; Crucitti, D.; Abbate, L.; Carimi, F. Phylogenetic relationship among wild and cultivated grapevine in sicily: A hotspot in the middle of the Mediterranean Basin. Front. Plant Sci. 2019, 10, 1506. [Google Scholar] [CrossRef] [Green Version]
- Cunha, J.; Ibáñez, J.; Teixeira-Santos, M.; Brazão, J.; Fevereiro, P.; Martínez-Zapater, J.M.; Eiras-Dias, J.E. Genetic Relationships among portuguese cultivated and wild Vitis vinifera L. Germplasm. Front. Plant Sci. 2020, 11, 127. [Google Scholar] [CrossRef] [Green Version]
- Marrano, A.; Grzeskowiak, L.; Moreno Sanz, P.; Lorenzi, S.; Prazzoli, M.L.; Arzumanov, A.; Amanova, M.; Failla, O.; Maghradze, D.; Grando, M.S. Genetic diversity and relationships in the grapevine germplasm collection from Central Asia. Vitis Geilweilerhof 2005, 54, 233–237. [Google Scholar]
- Bacilieri, R.; Lacombe, T.; Le Cunff, L.; Di Vecchi-Staraz, M.; Laucou, V.; Genna, B.; Péros, J.-P.; This, P.; Boursiquot, J.-M. Genetic structure in cultivated grapevines is linked to geography and human selection. BMC Plant Biol. 2013, 13, 25. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, D.B.; Pollock, D.D. Launching Microsatellites: A review of mutation processes and methods of phylogenetic inference. J. Hered. 1997, 88, 335–342. [Google Scholar] [CrossRef]
- Estoup, A.; Jarne, P.; Cornuet, J.-M. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 2002, 11, 1591–1604. [Google Scholar] [CrossRef]
- Estoup, A.; Angers, B. Microsatellites and minisatellites for molecular ecology: Theoretical and empirical considerations. In Advances in Molecular Ecology; Carvalho, G., Ed.; NATO Press: Amsterdam, The Netherlands, 1998; pp. 55–86. [Google Scholar]
- Barkley, N.A.; Krueger, R.R.; Federici, C.T.; Roose, M.L. What phylogeny and gene genealogy analyses reveal about homoplasy in citrus microsatellite alleles. Plant Syst. Evol. 2009, 282, 71–86. [Google Scholar] [CrossRef] [Green Version]
- Zarouri, B.; Vargas, A.M.; Gaforio, L.; Aller, M.; de Andrés, M.T.; Cabezas, J.A. Whole-genome genotyping of grape using a panel of microsatellite multiplex PCRs. Tree Genet. Genomes 2015, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Jarne, P.; Lagoda, P.J.L. Microsatellites, from molecules to populations and back. Trends Ecol. Evol. 1996, 11, 424–429. [Google Scholar] [CrossRef]
- Myles, S.; Chia, J.-M.; Hurwitz, B.; Simon, C.; Zhong, G.Y.; Buckler, E.; Ware, D. Rapid genomic characterization of the genus vitis. PLoS ONE 2010, 5, e8219. [Google Scholar] [CrossRef] [Green Version]
- Myles, S.; Boyko, A.R.; Owens, C.L.; Brown, P.J.; Grassi, F.; Aradhya, M.K.; Prins, B.; Reynolds, A.; Chia, J.-M.; Ware, D.; et al. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 2011, 108, 3530–3535. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.J.; Gross, B.L. From forest to field: Perennial fruit crop domestication. Am. J. Bot. 2011, 98, 1389–1414. [Google Scholar] [CrossRef] [Green Version]
- Grassi, F.; Imazio, S.; Failla, O.; Scienza, A.; Ocete Rubio, R.; Lopez, M.A.; Sala, F.; Labra, M. Genetic Isolation and diffusion of wld grapevine Italian and Spanish populations as estimated by nuclear and chloroplast SSR Analysis. Plant Biol. 2003, 5, 608–614. [Google Scholar] [CrossRef]
- Grassi, F.; Labra, M.; Imazio, S.; Rubio, R.O.; Failla, O.; Scienza, A.; Sala, F. Phylogeographical structure and conservation genetics of wild grapevine. Conserv. Genet. 2006, 7, 837–845. [Google Scholar] [CrossRef]
- Marrano, A.; Birolo, G.; Prazzoli, M.L.; Lorenzi, S.; Valle, G.; Grando, M.S. SNP-Discovery by RAD-sequencing in a germplasm collection of wild and cultivated grapevines (V. vinifera L.). PLoS ONE 2017, 12, e170655. [Google Scholar] [CrossRef]
- De Lorenzis, G.; Chipashvili, R.; Failla, O.; Maghradze, D. Study of genetic variability in Vitis vinifera L. germplasm by high-throughput Vitis18kSNP array: The case of Georgian genetic resources. BMC Plant Biol. 2015, 15, 154. [Google Scholar] [CrossRef] [Green Version]
- Mercati, F.; De Lorenzis, G.; Brancadoro, L.; Lupini, A.; Abenavoli, M.R.; Barbagallo, M.G.; Di Lorenzo, R.; Scienza, A.; Sunseri, F. High-throughput 18K SNP array to assess genetic variability of the main grapevine cultivars from Sicily. Tree Genet. Genomes 2016, 12, 59. [Google Scholar] [CrossRef] [Green Version]
- Sunseri, F.; Lupini, A.; Mauceri, A.; De Lorenzis, G.; Araniti, F.; Brancadoro, L.; Dattola, A.; Gullo, G.; Zappia, R.; Mercati, F. Single nucleotide polymorphism profiles reveal an admixture genetic structure of grapevine germplasm from Calabria, Italy, uncovering its key role for the diversification of cultivars in the Mediterranean Basin. Aust. J. Grape Wine Res. 2018, 24, 345–359. [Google Scholar] [CrossRef]
- De Lorenzis, G.; Mercati, F.; Bergamini, C.; Cardone, M.F.; Lupini, A.; Mauceri, A.; Caputo, A.R.; Abbate, L.; Barbagallo, M.G.; Antonacci, D.; et al. SNP genotyping elucidates the genetic diversity of Magna Graecia grapevine germplasm and its historical origin and dissemination. BMC Plant Biol. 2019, 19, 7. [Google Scholar] [CrossRef]
- D’Onofrio, C.; Tumino, G.; Gardiman, M.; Crespan, M.; Schneider, A.; Bignami, C.; de Palma, L.; Barbagallo, M.; Morcia, C.; Novello, V.; et al. Parentage atlas of widely and locally cultivated Italian grapevine varieties as inferred from SNP Genotyping. Front. Plant Sci. 2021, 11, 605934. [Google Scholar] [CrossRef]
- Özdoğan, M. A new look at the introduction of the Neolithic way of life in Southeastern Europe. Changing paradigms of the expansion of the Neolithic way of life. Doc. Praehist. 2014, 41, 33–49. [Google Scholar] [CrossRef] [Green Version]
- Perlès, C. From the Near East to Greece: Let’s reverse the focus—Cultural elements that didn’t transfer. In How Did Farming Reach Europe? Anatolian- European Relations from the Second Half of the 7th Through the First Half of the 6th Millennium cal BC; Lichter, C., Ed.; Ege Yayınları: Instanbul, Turkey, 2005; pp. 275–290. [Google Scholar]
- Paschou, P.; Drineas, P.; Yannaki, E.; Razou, A.; Kanaki, K.; Tsetsos, F.; Padmanabhuni, S.S.; Michalodimitrakis, M.; Renda, M.C.; Pavlovic, S.; et al. Maritime route of colonization of Europe. Proc. Natl. Acad. Sci. USA 2014, 111, 9211–9216. [Google Scholar] [CrossRef] [Green Version]
- Rogalski, M.; do Vieira, L.N.; Fraga, H.P.; Guerra, M.P. Plastid genomics in horticultural species: Importance and applications for plant population genetics, evolution, and biotechnology. Front. Plant Sci. 2015, 6, 586. [Google Scholar] [CrossRef] [Green Version]
- Grassi, F.; Labra, M.; Scienza, A.; Imazio, S. Chloroplast SSR markers to assess DNA diversity in wild and cultivated grapevines. Vitis Geilweilerhof 2002, 41, 157–158. [Google Scholar]
- Arroyo-García, R.; Lefort, F.; de Andrés, M.T.; Ibáñez, J.; Borrego, J.; Jouve, N.; Cabello, F.; Martínez-Zapater, J.M. Chloroplast microsatellite polymorphisms in Vitis species. Genome 2002, 45, 1142–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Mattia, F.; Imazio, S.; Grassi, F.; Baneh, H.D.; Scienza, A.; Labra, M. Study of genetic relationships between wild and domesticated grapevine distributed from Middle East Regions to European countries. Rend. LINCEI 2008, 19, 223–240. [Google Scholar] [CrossRef]
- Imazio, S.; Labra, M.; Grassi, F.; Scienza, A.; Failla, O. Chloroplast microsatellites to investigate the origin of grapevine. Genet. Resour. Crops Evol. 2006, 53, 1003–1011. [Google Scholar] [CrossRef]
- Arroyo-García, R.; Ruiz-García, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.; Ergul, A.; Söylemezoğlu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms . Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar]
- Cunha, J.; Santos, M.T.; Carneiro, L.C.; Fevereiro, P.; Eiras-Dias, J.E. Portuguese traditional grapevine cultivars and wild vines (Vitis vinifera L.) share morphological and genetic traits. Genet. Resour. Crop Evol. 2009, 56, 975–989. [Google Scholar] [CrossRef]
- Pipia, I.; Gogniashvili, M.; Tabidze, V.; Beridze, T.; Gamkrelidze, M.; Gotsiridze, V.; Melyan, G.; Musayev, M.; Salimov, V.; Beck, J.B.; et al. Plastid DNA sequence diversity in wild grapevine samples (Vitis vinifera subsp. sylvestris) from the Caucasus region. Vitis Geilweilerhof 2012, 51, 119–124. [Google Scholar]
- Zecca, G.; Abbott, J.R.; Sun, W.-B.; Spada, A.; Sala, F.; Grassi, F. The timing and the mode of evolution of wild grapes (Vitis). Mol. Phylogenet. Evol. 2012, 62, 736–747. [Google Scholar] [CrossRef]
- Grassi, F.; Mattia, F.D.E.; Zecca, G.; Sala, F.; Labra, M. Historical isolation and Quaternary range expansion of divergent lineages in wild grapevine. Biol. J. Linn. Soc. 2008, 79, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Zecca, G.; Grassi, F.; Tabidze, V.; Pipia, I.; Kotorashvili, A.; Kotaria, N.; Beridze, T. Dates and rates in grape’s plastomes: Evolution in slow motion. Curr. Genet. 2020, 66, 123–140. [Google Scholar] [CrossRef]
- Péros, J.-P.; Berger, G.; Portemont, A.; Boursiquot, J.-M.; Lacombe, T. Genetic variation and biogeography of the disjunct Vitis subg. Vitis (Vitaceae). J. Biogeogr. 2011, 38, 471–486. [Google Scholar] [CrossRef]
- Riaz, S.; Pap, D.; Uretsky, J.; Laucou, V.; Boursiquot, M.J.; Kocsis, L.; Walker, M.A. Genetic diversity and parentage analysis of grape rootstocks. Theor. Appl. Genet. 2019, 132, 1847–1860. [Google Scholar] [CrossRef]
- Poczai, P.; Hyvönen, J.; Taller, J.; Jahnke, G.; Kocsis, L. Phylogenetic analyses of Teleki grapevine rootstocks using three chloroplast DNA markers. Plant Mol. Biol. Rep. 2013, 31, 371–386. [Google Scholar] [CrossRef]
- Wen, J.; Herron, S.A.; Yang, X.; Liu, B.-B.; Zuo, Y.-J.; Harris, A.; Kalburgi, Y.; Johnson, G.; Zimmer, E.A. Nuclear and Chloroplast sequences resolve the enigmatic origin of the concord grape. Front. Plant Sci. 2020, 11, 263. [Google Scholar] [CrossRef]
- Tabidze, V.; Baramidze, G.; Pipia, I.; Gogniashvili, M.; Ujmajuridze, L.; Beridze, T.; Hernandez, A.G.; Schaal, B. The complete chloroplast DNA sequence of eleven grape cultivars. Simultaneous resequencing methodology. OENO One 2014, 48, 99. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Harris, A.; Kalburgi, Y.; Zhang, N.; Xu, Y.; Zheng, W.; Ickert-Bond, S.M.; Johnson, G.; Zimmer, E.A. Chloroplast phylogenomics of the New World grape species (Vitis, Vitaceae). J. Syst. Evol. 2018, 56, 297–308. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Schwaninger, H.R.; Baldo, A.M.; Labate, J.A.; Zhong, G.-Y.; Simon, C.J. A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during neogene and quaternary climate change. BMC Evol. Biol. 2013, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Zeller, U.; Göttert, T. The relations between evolution and domestication reconsidered—Implications for systematics, ecology, and nature conservation. Glob. Ecol. Conserv. 2019, 20, e00756. [Google Scholar] [CrossRef]
- Zhang, L.-B.; Zhu, Q.; Wu, Z.-Q.; Ross-Ibarra, J.; Gaut, B.S.; Ge, S.; Sang, T. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009, 184, 708–720. [Google Scholar] [CrossRef]
- Allaby, R.G.; Fuller, D.Q.; Brown, T.A. The genetic expectations of a protracted model for the origins of domesticated crops. Proc. Natl. Acad. Sci. USA 2008, 105, 13982–13986. [Google Scholar] [CrossRef] [Green Version]
- Purugganan, M.D.; Fuller, D.Q. Archaeological data reveal slow rates of evolution during plant domestication. Evolution 2011, 65, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Purugganan, M.D. Evolutionary Insights into the Nature of Plant Domestication. Curr. Biol. 2019, 29, R705–R714. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Massonnet, M.; Sanjak, J.S.; Cantu, D.; Gaut, B.S. Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc. Natl. Acad. Sci. USA 2017, 114, 11715–11720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, R.S.; Choi, J.Y.; Sanches, M.; Plessis, A.; Flowers, J.M.; Amas, J.; Dorph, K.; Barretto, A.; Gross, B.; Fuller, D.Q.; et al. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat. Genet. 2016, 48, 1083–1088. [Google Scholar] [CrossRef]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and Related Genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.A.; Donoghue, M.J. Rates of molecular evolution are linked to life history in flowering plants. Science 2008, 322, 86–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bromham, L. Why do species vary in their rate of molecular evolution? Biol. Lett. 2009, 5, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Z.; Duan, S.; Sheng, J.; Zhu, S.; Ni, X.; Shao, J.; Liu, C.; Nick, P.; Du, F.; Fan, P.; et al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gepts, P. The contribution of genetic and genomic approaches to plant domestication studies. Curr. Opin. Plant Biol. 2014, 18, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Schröder, S.; Kortekamp, A.; Heene, E.; Daumann, J.; Valea, I.; Nick, P. Crop wild relatives as genetic resources—The case of the European wild grape. Can. J. Plant Sci. 2015, 95, 905–912. [Google Scholar] [CrossRef] [Green Version]
- Aradhya, M.; Wang, Y.; Walker, M.A.; Prins, B.H.; Koehmstedt, A.M.; Velasco, D.; Gerrath, J.M.; Dangl, G.S.; Preece, J.E. Genetic diversity, structure, and patterns of differentiation in the genus Vitis. Plant Syst. Evol. 2013, 299, 317–330. [Google Scholar] [CrossRef]
- Zecca, G.; Labra, M.; Grassi, F. Untangling the evolution of American wild grapes: Admixed species and how to find them. Front. Plant Sci. 2020, 10, 1814. [Google Scholar] [CrossRef]
- Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005, 20, 229–237. [Google Scholar] [CrossRef]
- Levadoux, L. Les populations sauvages et cultivées de Vitis vinifera L. Ann. L’amélioration des Plantes 1956, 1, 59–118. [Google Scholar]
- Zecca, G.; De Mattia, F.; Lovicu, G.; Labra, M.; Sala, F.; Grassi, F. Wild grapevine: Silvestris, hybrids or cultivars that escaped from vineyards? Molecular evidence in Sardinia. Plant Biol. 2010, 12, 558–562. [Google Scholar] [CrossRef]
- Arrigo, N.; Arnold, C. Naturalised Vitis rootstocks in Europe and consequences to native wild grapevine. PLoS ONE 2007, 2, e521. [Google Scholar] [CrossRef] [Green Version]
- Di Vecchi-Staraz, M.; Laucou, V.; Bruno, G.; Lacombe, T.; Gerber, S.; Bourse, T.; Boselli, M.; This, P. Low level of pollen-mediated gene flow from cultivated to wild grapevine: Consequences for the evolution of the endangered subspecies Vitis vinifera L. subsp. silvestris. J. Hered. 2009, 100, 66–75. [Google Scholar] [CrossRef]
- D’Onofrio, C. Introgression among cultivated and wild grapevine in Tuscany. Front. Plant Sci. 2020, 11, 202. [Google Scholar] [CrossRef]
- Arnold, C.; Bachmann, O.; Schnitzler, A. Insights into the Vitis complex in the Danube floodplain (Austria). Ecol. Evol. 2017, 7, 7796–7806. [Google Scholar] [CrossRef]
- Zecca, G.; Grassi, F. RPB2 gene reveals a phylodemographic signal in wild and domesticated grapevine (Vitis vinifera). J. Syst. Evol. 2013, 51, 205–211. [Google Scholar] [CrossRef]
- Arnold, C.; Schnitzler, A.; Douard, A.; Peter, R.; Gillet, F. Is there a future for wild grapevine (Vitis vinifera subsp. silvestris) in the Rhine Valley? Biodivers. Conserv. 2005, 14, 1507–1523. [Google Scholar] [CrossRef]
- Ocete Rubio, R.; Ocete Rubio, E.; Ocete Pérez, C.; Ángeles Pérez Izquierdo, M.; Rustioni, L.; Failla, O.; Chipashvili, R.; Maghradze, D. Ecological and sanitary characteristics of the Eurasian wild grapevine (Vitis vinifera L. ssp. sylvestris (Gmelin) Hegi) in Georgia (Caucasian region). Plant Genet. Resour. 2012, 10, 155–162. [Google Scholar] [CrossRef]
- Arnold, C.; Gilett, F.; Gobat, J.M. Situation de la vigne sauvage Vitis vinifera L sp. sylvestris (Gmelin) Hegi en Europe. Vitis Geilweilerhof 1998, 37, 159–170. [Google Scholar]
- Butorac, L.; Hančević, K.; Lukšić, K.; Škvorc, Ž.; Leko, M.; Maul, E.; Zdunić, G. Assessment of wild grapevine (Vitis vinifera ssp. sylvestris) chlorotypes and accompanying woody species in the Eastern Adriatic region. PLoS ONE 2018, 13, e0199495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from Genome-Wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [Green Version]
- Flowers, J.M.; Hazzouri, K.M.; Gros-Balthazard, M.; Mo, Z.; Koutroumpa, K.; Perrakis, A.; Ferrand, S.; Khierallah, H.S.M.; Fuller, D.Q.; Aberlenc, F.; et al. Cross-species hybridization and the origin of North African date palms. Proc. Natl. Acad. Sci. USA 2019, 116, 1651–1658. [Google Scholar] [CrossRef] [Green Version]
- Helmstetter, A.J.; Oztolan-Erol, N.; Lucas, S.J.; Buggs, R.J.A. Genetic diversity and domestication of hazelnut (Corylus avellana L.) in Turkey. Plants People Planet 2020, 2, 326–339. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.-Y.; Wen, J.; Tian, J.-P.; Jamal, A.; Chen, L.-Q.; Liu, X.-Q. Testing reticulate evolution of four Vitis species from East Asia using restriction-site associated DNA sequencing. J. Syst. Evol. 2018, 56, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Bouby, L.; Figueiral, I.; Bouchette, A.; Rovira, N.; Ivorra, S.; Lacombe, T.; Pastor, T.; Picq, S.; Marinval, P.; Terral, J.-F. Bioarchaeological insights into the process of domestication of grapevine (Vitis vinifera L.) during Roman times in Southern France. PLoS ONE 2013, 8, e63195. [Google Scholar] [CrossRef] [Green Version]
- Bouby, L.; Wales, N.; Jalabadze, M.; Rusishvili, N.; Bonhomme, V.; Ramos-Madrigal, J.; Evin, A.; Ivorra, S.; Lacombe, T.; Pagnoux, C.; et al. Tracking the history of grapevine cultivation in Georgia by combining geometric morphometrics and ancient DNA. Veg. Hist. Archaeobot. 2021, 30, 63–76. [Google Scholar] [CrossRef]
- Pagnoux, C.; Bouby, L.; Valamoti, S.M.; Bonhomme, V.; Ivorra, S.; Gkatzogia, E.; Karathanou, A.; Kotsachristou, D.; Kroll, H.; Terral, J.-F. Local domestication or diffusion? Insights into viticulture in Greece from Neolithic to Archaic times, using geometric morphometric analyses of archaeological grape seeds. J. Archaeol. Sci. 2021, 125, 105263. [Google Scholar] [CrossRef]
- Terral, J.F.; Tabard, E.; Bouby, L.; Ivorra, S.; Pastor, T.; Figueiral, I.; Picq, S.; Chevance, J.B.; Jung, C.; Fabre, L.; et al. Evolution and history of grapevine (Vitis vinifera) under domestication: New morphometric perspectives to understand seed domestication syndrome and reveal origins of ancient European cultivars. Ann. Bot. 2010, 105, 443–455. [Google Scholar] [CrossRef]
- Pagnoux, C.; Bouby, L.; Ivorra, S.; Petit, C.; Valamoti, S.-M.; Pastor, T.; Picq, S.; Terral, J.-F. Inferring the agrobiodiversity of Vitis vinifera L. (grapevine) in ancient Greece by comparative shape analysis of archaeological and modern seeds. Veg. Hist. Archaeobot. 2015, 24, 75–84. [Google Scholar] [CrossRef]
- Bouby, L.; Bonhomme, V.; Ivorra, S.; Pastor, T.; Rovira, N.; Tillier, M.; Pagnoux, C.; Terral, J.F. Back from burn out: Are experimentally charred grapevine pips too distorted to be characterized using morphometrics? Archaeol. Anthropol. Sci. 2018, 10, 943–954. [Google Scholar] [CrossRef]
- Wales, N.; Andersen, K.; Cappellini, E.; Ávila-Arcos, M.C.; Gilbert, M.T.P. Optimization of DNA Recovery and Amplification from Non-Carbonized Archaeobotanical Remains. PLoS ONE 2014, 9, e86827. [Google Scholar] [CrossRef] [Green Version]
- Cappellini, E.; Gilbert, M.T.P.; Geuna, F.; Fiorentino, G.; Hall, A.; Thomas-Oates, J.; Ashton, P.D.; Ashford, D.A.; Arthur, P.; Campos, P.F.; et al. A multidisciplinary study of archaeological grape seeds. Naturwissenschaften 2010, 97, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Foley, B.P.; Hansson, M.C.; Kourkoumelis, D.P.; Theodoulou, T.A. Aspects of ancient Greek trade re-evaluated with amphora DNA evidence. J. Archaeol. Sci. 2012, 39, 389–398. [Google Scholar] [CrossRef]
- Hansson, M.C.; Foley, B.P. Ancient DNA fragments inside Classical Greek amphoras reveal cargo of 2400-year-old shipwreck. J. Archaeol. Sci. 2008, 35, 1169–1176. [Google Scholar] [CrossRef]
- Manen, J.-F.; Bouby, L.; Dalnoki, O.; Marinval, P.; Turgay, M.; Schlumbaum, A. Microsatellites from archaeological Vitis vinifera seeds allow a tentative assignment of the geographical origin of ancient cultivars. J. Archaeol. Sci. 2003, 30, 721–729. [Google Scholar] [CrossRef]
- Bacilieri, R.; Bouby, L.; Figueiral, I.; Schaal, C.; Terral, J.-F.; Breton, C.; Picq, S.; Weber, A.; Schlumbaum, A. Potential of combining morphometry and ancient DNA information to investigate grapevine domestication. Veg. Hist. Archaeobot. 2017, 26, 345–356. [Google Scholar] [CrossRef]
- Rizzi, E.; Lari, M.; Gigli, E.; De Bellis, G.; Caramelli, D. Ancient DNA studies: New perspectives on old samples. Genet. Sel. Evol. 2012, 44, 21. [Google Scholar] [CrossRef] [Green Version]
- Knapp, M.; Hofreiter, M. Next Generation Sequencing of Ancient DNA: Requirements, Strategies and Perspectives. Genes 2010, 1, 227–243. [Google Scholar] [CrossRef] [Green Version]
- Wales, N.; Madrigal, J.R.; Cappellini, E.; Baez, A.C.; Castruita, J.A.S.; Romero-Navarro, J.A.; Carøe, C.; Ávila-Arcos, M.C.; Peñaloza, F.; Moreno-Mayar, J.V.; et al. The limits and potential of paleogenomic techniques for reconstructing grapevine domestication. J. Archaeol. Sci. 2016, 72, 57–70. [Google Scholar] [CrossRef]
- Ramos-Madrigal, J.; Runge, A.K.W.; Bouby, L.; Lacombe, T.; Castruita, J.A.S.; Adam-Blondon, A.-F.; Figueiral, I.; Hallavant, C.; Martínez-Zapater, J.M.; Schaal, C.; et al. Palaeogenomic insights into the origins of French grapevine diversity. Nat. Plants 2019, 5, 595–603. [Google Scholar] [CrossRef]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S. Retrotransposon-Induced Mutations in Grape Skin Color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Fernandez, L.; Torregrosa, L.; Segura, V.; Bouquet, A.; Martinez-Zapater, J.M. Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. Plant J. 2010, 61, 545–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Minio, A.; Massonnet, M.; Solares, E.; Lv, Y.; Beridze, T.; Cantu, D.; Gaut, B.S. The population genetics of structural variants in grapevine domestication. Nat. Plants 2019, 5, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Barrera-Redondo, J.; Piñero, D.; Eguiarte, L.E. Genomic, transcriptomic and epigenomic tools to study the domestication of plants and animals: A field guide for beginners. Front. Genet. 2020, 11, 742. [Google Scholar] [CrossRef]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef]
- Moore, M.O.; Wen, J. Vitaceae. In Flora of North America North of Mexico; Flora of North America Editorial Committee, Ed.; Flora of North America Association: New York, NY, USA; Oxford, UK, 2016; pp. 3–23. [Google Scholar]
- Callen, S.T.; Klein, L.L.; Miller, A.J. Climatic niche characterization of 13 North American Vitis species. Am. J. Enol. Vitic. 2016, 67, 339–349. [Google Scholar] [CrossRef]
- Töpfer, R.; Hausmann, L.; Harst, M.; Maul, E.; Zyprian, E.; Eibach, R. New horizons for grapevine breeding. In Methods in Temperate Fruit Breeding; Flachowsky, H., Hanke, M.V., Eds.; Gloval Science Books: Isleworth, UK, 2011; pp. 79–100. [Google Scholar]
- Bavaresco, L.; Gardiman, M.; Brancadoro, L.; Espen, L.; Failla, O.; Scienza, A.; Vezzulli, S.; Zulini, L.; Velasco, R.; Stefanini, M.; et al. Grapevine breeding programs in Italy. In Grapevine Breeding Programs for the Wine Industry; Reynolds, A., Ed.; Elsevier: Cambridge, UK, 2015; pp. 135–155. [Google Scholar]
- Meggio, F.; Prinsi, B.; Negri, A.S.; Simone Di Lorenzo, G.; Lucchini, G.; Pitacco, A.; Failla, O.; Scienza, A.; Cocucci, M.; Espen, L. Biochemical and physiological responses of two grapevine rootstock genotypes to drought and salt treatments. Aust. J. Grape Wine Res. 2014, 20, 310–323. [Google Scholar] [CrossRef]
- Bianchi, D.; Caramanico, L.; Grossi, D.; Brancadoro, L.; De Lorenzis, G. How do novel M-rootstock (Vitis spp.) genotypes cope with drought? Plants 2020, 9, 1385. [Google Scholar] [CrossRef]
- Khoury, C.K.; Greene, S.L.; Krishnan, S.; Miller, A.J.; Moreau, T. A road map for conservation, use, and public engagement around North America’s crop wild relatives and wild utilized plants. Crops Sci. 2019, 59, 2302–2307. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grassi, F.; De Lorenzis, G. Back to the Origins: Background and Perspectives of Grapevine Domestication. Int. J. Mol. Sci. 2021, 22, 4518. https://doi.org/10.3390/ijms22094518
Grassi F, De Lorenzis G. Back to the Origins: Background and Perspectives of Grapevine Domestication. International Journal of Molecular Sciences. 2021; 22(9):4518. https://doi.org/10.3390/ijms22094518
Chicago/Turabian StyleGrassi, Fabrizio, and Gabriella De Lorenzis. 2021. "Back to the Origins: Background and Perspectives of Grapevine Domestication" International Journal of Molecular Sciences 22, no. 9: 4518. https://doi.org/10.3390/ijms22094518
APA StyleGrassi, F., & De Lorenzis, G. (2021). Back to the Origins: Background and Perspectives of Grapevine Domestication. International Journal of Molecular Sciences, 22(9), 4518. https://doi.org/10.3390/ijms22094518






