Genome-Wide Analysis of the Late Embryogenesis Abundant (LEA) and Abscisic Acid-, Stress-, and Ripening-Induced (ASR) Gene Superfamily from Canavalia rosea and Their Roles in Salinity/Alkaline and Drought Tolerance
Abstract
:1. Introduction
2. Results
2.1. Identification of the C. rosea LEA Family and ASR Family
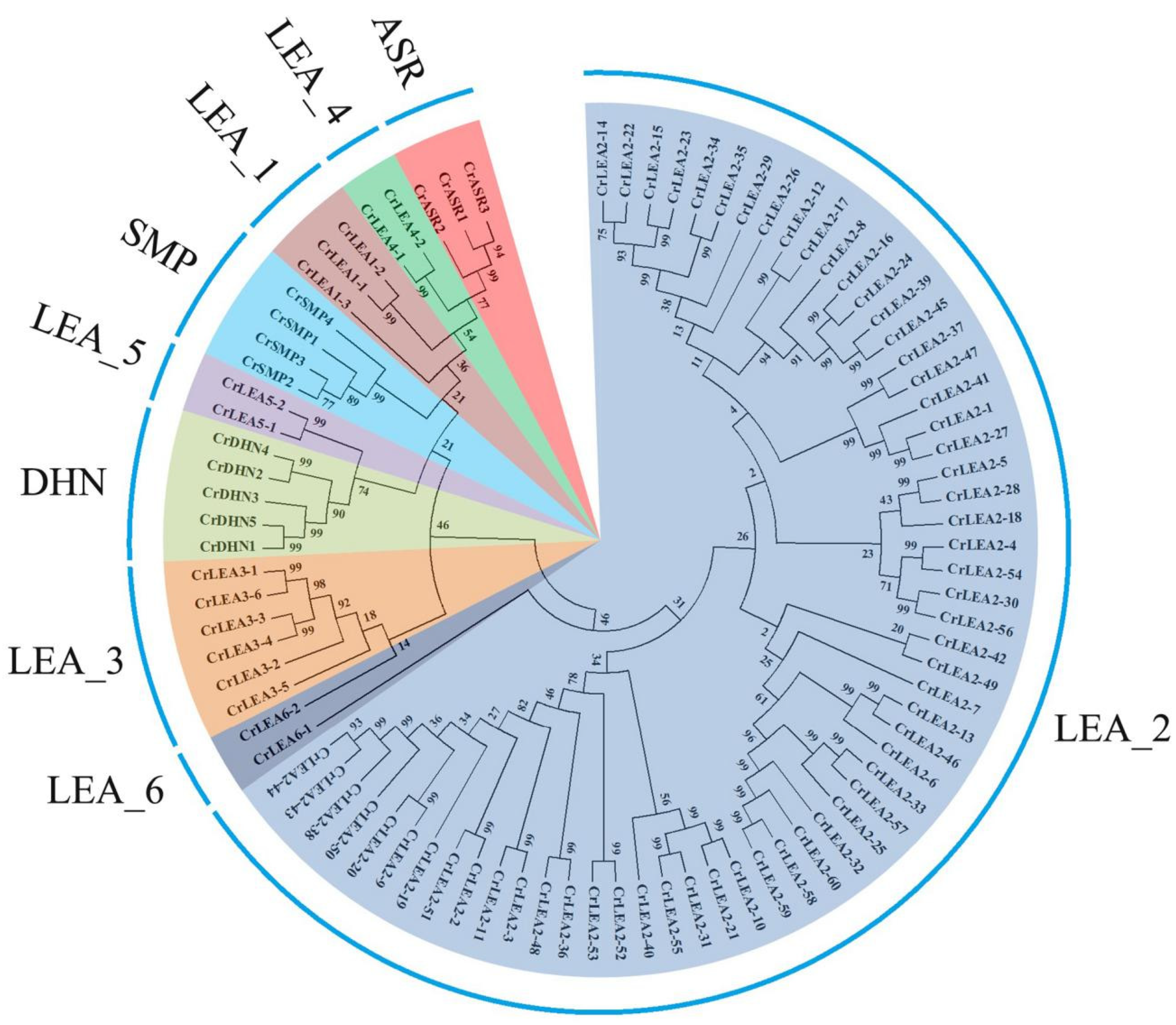
2.2. Phylogenetic Analysis of CrLEA Proteins and CrASR Proteins
2.3. Gene Structures and Conserved Motifs of CrLEAs and CrASRs in C. rosea
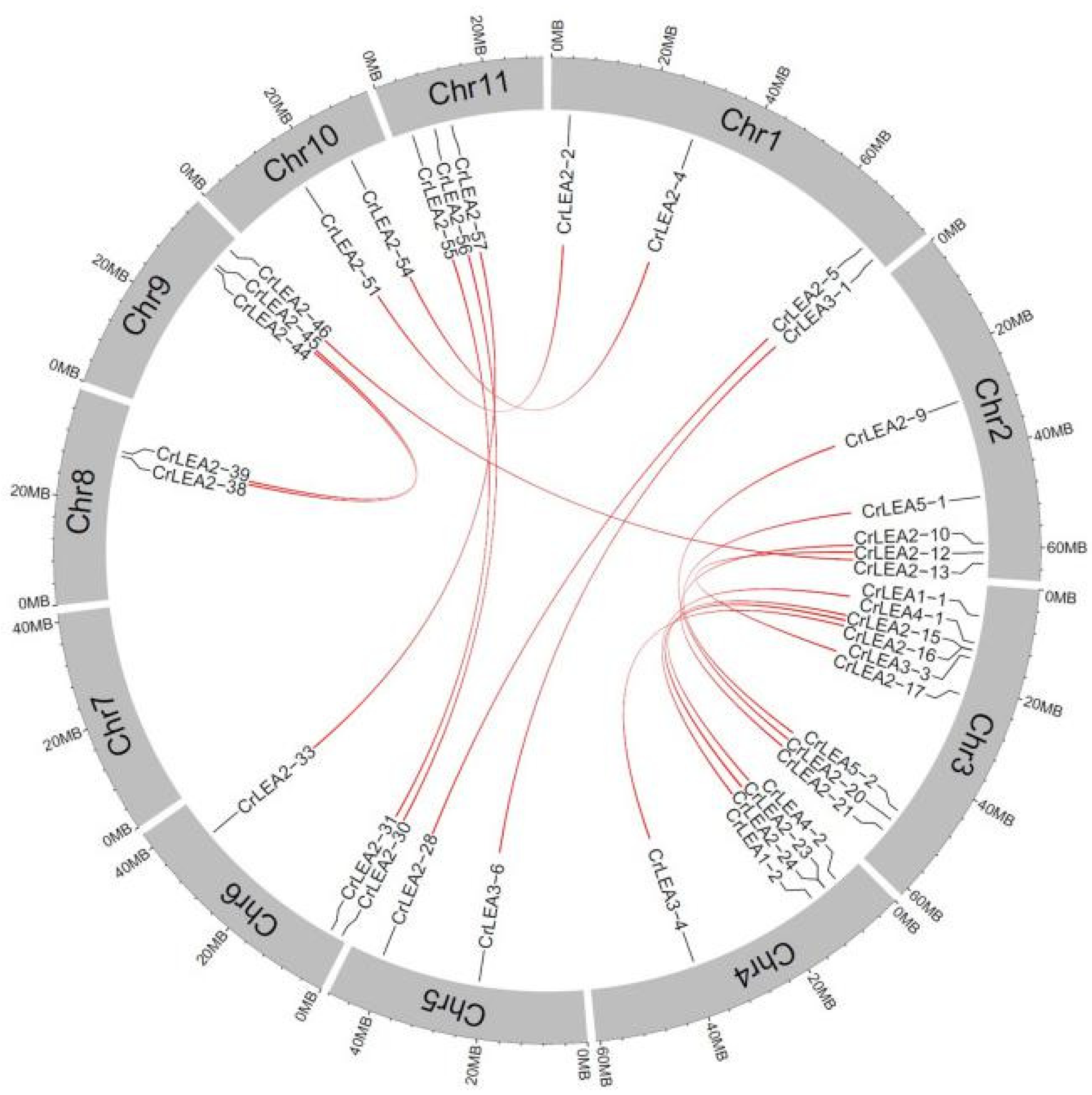
2.4. Chromosomal Locations and Evolutionary Characterization of CrLEAs and CrASRs
2.5. Cis-Regulatory Element Analyses of CrLEAs and CrASRs
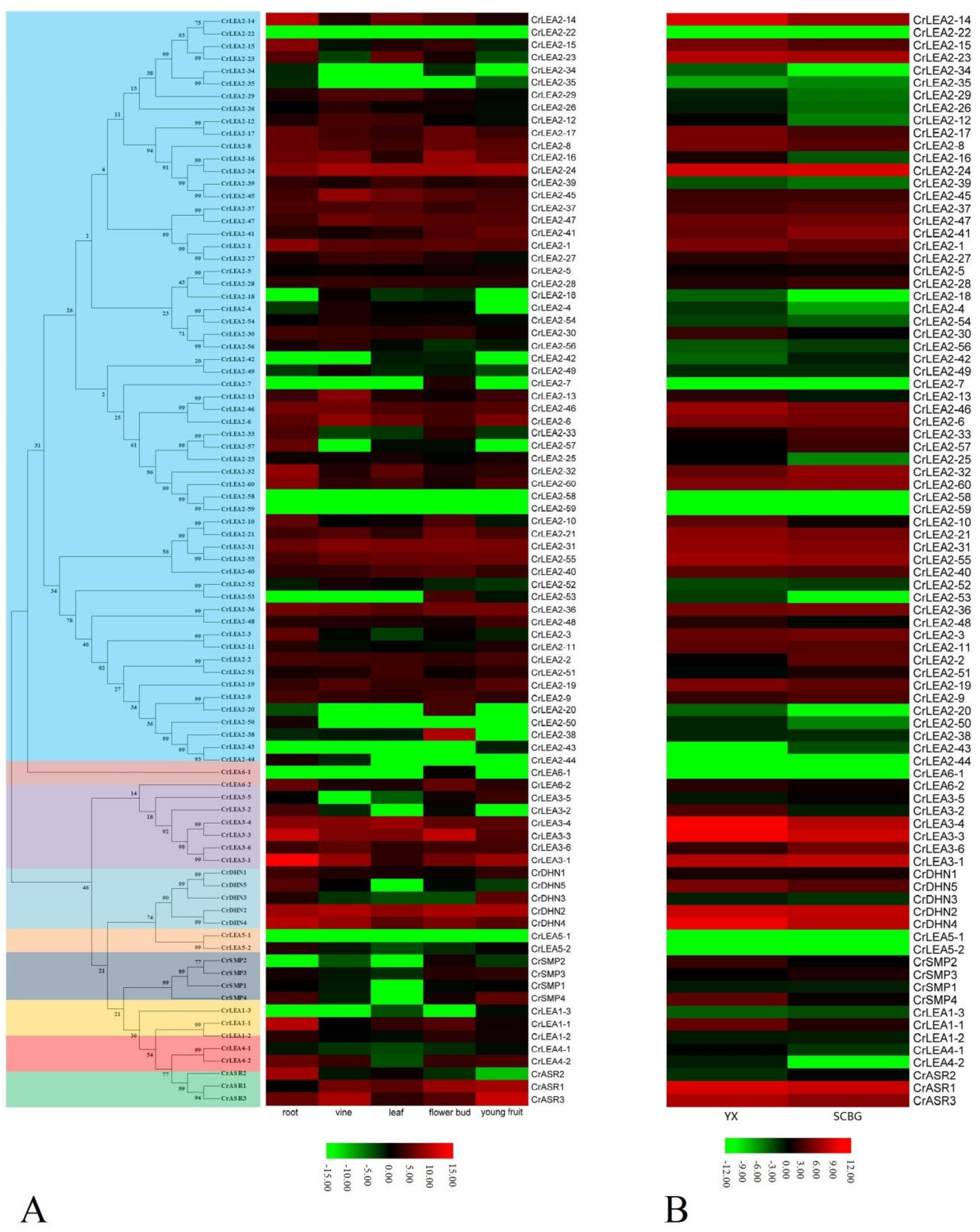
2.6. Expression Profiles of CrLEAs and CrASRs in Different Tissues and Plants Residing in Different Habitats
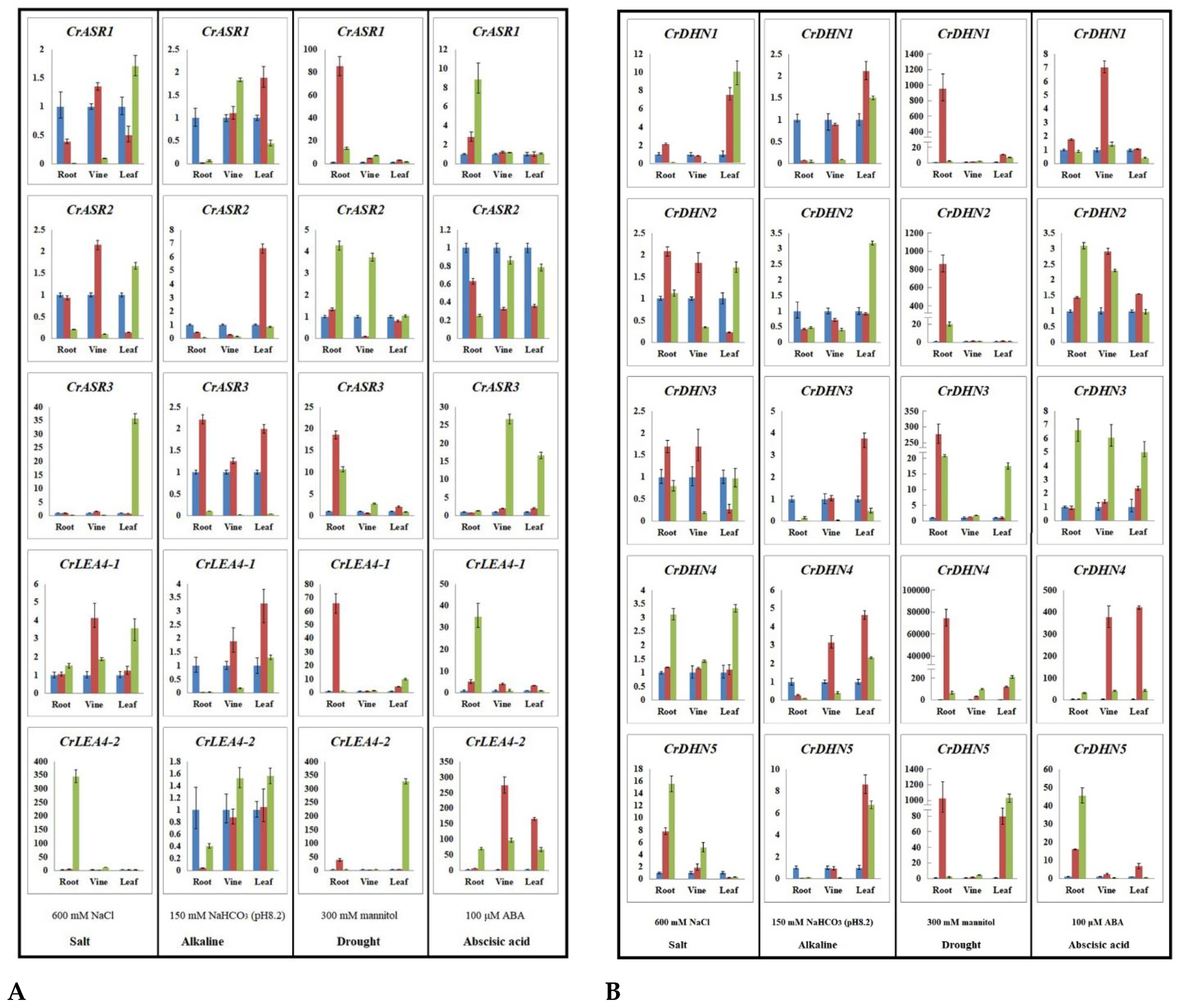
2.7. Expression Profiles of CrLEAs and CrASRs in Response to Different Stressors and the ABA Treatment
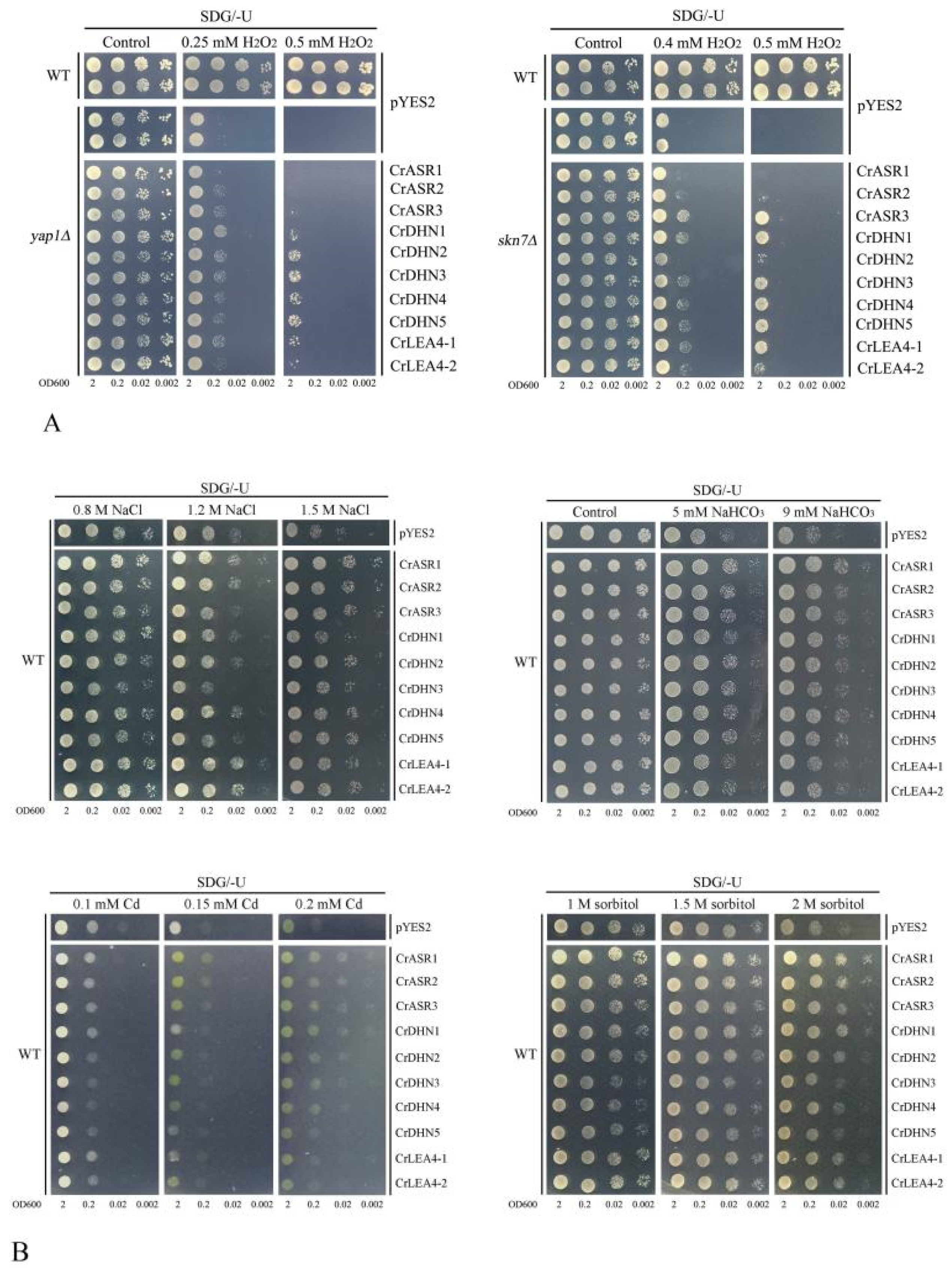
2.8. Abiotic Stress Tolerance of Yeast Heterologously Expressing CrLEAs and CrASRs
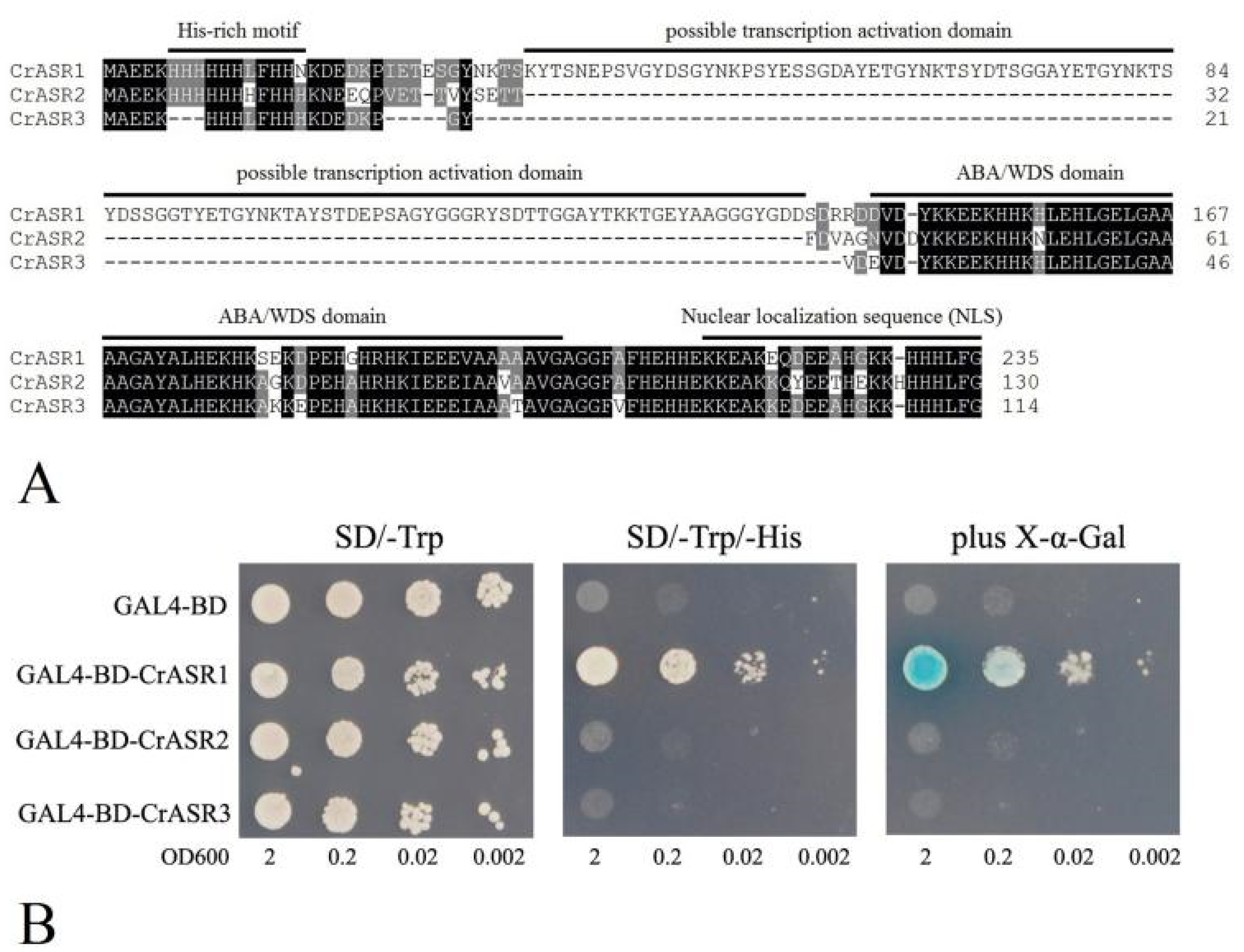
2.9. Analysis of the Transcriptional Activation Activity of CrASRs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Identification of LEA/ASR Genes in the C. rosea Genome and Phylogenetic Analysis of CrLEA/CrASR Superfamily Proteins
4.3. Analysis of Protein-Conserved Motifs and Biochemical Features of CrLEAs/ASRs
4.4. Gene Duplication and Collinearity Analysis of CrLEAs
4.5. Promoter Sequence Profiling of CrLEAs/ASRs
4.6. RNA-Seq of Different C. rosea Tissues
4.7. Expression Analysis by Quantitative Reverse Transcription (qRT) PCR of CrLEAs/ASRs under Different Stress Treatments
4.8. Cloning of CrLEA/ASR cDNAs and Heterologous Expression in Yeast
4.9. Transcriptional Activity Analysis of CrASRs in Yeast
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LEA | late embryogenesis abundant |
| ASR | abscisic acid-, stress-, and ripening-induced |
| LEAPdb | database for the late embryogenesis abundant proteins |
| DHN | dehydrin |
| SMP | seed maturation protein |
| ROS | reactive oxygen species |
| IDP | intrinsically disordered protein |
| WGD | whole-genome duplication |
| WT | wild type |
| YX | Yongxing Island |
| SCBG | South China Botanical Garden |
References
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2014, 241, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, A.M.; Martins, C.D.P.S.; Gonçalves, L.P.; Costa, M.G.C. Late Embryogenesis Abundant (LEA) Constitutes a Large and Diverse Family of Proteins Involved in Development and Abiotic Stress Responses in Sweet Orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0145785. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Gao, T.; Chen, J.; Yang, J.; Huang, H.; Yu, Y. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol. Biochem. 2019, 135, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xing, M.; Yang, W.; Mu, X.; Wang, X.; Lu, F.; Wang, Y.; Zhang, L. Genome-wide identification of and functional in-sights into the late embryogenesis abundant (LEA) gene family in bread wheat (Triticum aestivum). Sci. Rep. 2019, 9, 13375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hand, S.C.; Menze, M.A.; Toner, M.; Boswell, L.; Moore, D. LEA Proteins During Water Stress: Not Just for Plants Anymore. Annu. Rev. Physiol. 2011, 73, 115–134. [Google Scholar] [CrossRef] [Green Version]
- Reyes, J.L.; Campos, F.; Wei, H.; Arora, R.; Yang, Y.; Karlson, D.T.; Covarrubias, A.A. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008, 31, 1781–1790. [Google Scholar] [CrossRef]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wise, M.J.; Tunnacliffe, A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004, 9, 13–17. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef] [PubMed]
- González, R.M.; Iusem, N.D. Twenty years of research on Asr (ABA-stress-ripening) genes and proteins. Planta 2014, 239, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, P.G.; Carrari, F. ASR1 transcription factor and its role in metabolism. Plant Signal. Behav. 2015, 10, e992751. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Dong, Y.; Li, C.; Pan, Y.; Yu, J. SiASR4, the Target Gene of SiARDP from Setaria italica, Improves Abiotic Stress Adaption in Plants. Front. Plant Sci. 2017, 7, 2053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrari, F.; Fernie, A.R.; Iusem, N.D. Heard it through the grapevine? ABA and sugar cross-talk: The ASR story. Trends Plant Sci. 2004, 9, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Galau, G.A.; Chlan, C.A.; Dure, L. Developmental biochemistry of cottonseed embryogenesis and germination. Plant Mol. Biol. 1983, 2, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Hunault, G.; Jaspard, E. LEAPdb: A database for the late embryogenesis abundant proteins. BMC Genom. 2010, 11, 221. [Google Scholar] [CrossRef] [Green Version]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.-H.; Macherel, D. The Ubiquitous Distribution of Late Embryogenesis Abundant Proteins across Cell Compartments in Arabidopsis Offers Tailored Protection against Abiotic Stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [Green Version]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thali-ana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Tong, Q.; Wang, Y.; Wang, Z.; Xu, G.; Elias, G.K.; Li, S.; Liang, Z. Transcriptomic Analysis of the Grapevine LEA Gene Family in Response to Osmotic and Cold Stress Reveals a Key Role for VamDHN3. Plant Cell Physiol. 2020, 61, 775–786. [Google Scholar] [CrossRef]
- Artur, M.A.S.; Zhao, T.; Ligterink, W.; Schranz, E.; Hilhorst, H.W.M. Dissecting the genomic diversification of late embryo-genesis abundant (LEA) protein gene families in plants. Genome Biol. Evol. 2019, 11, 459–471. [Google Scholar] [CrossRef]
- Chatelain, E.; Hundertmark, M.; Leprince, O.; Le Gall, S.; Satour, P.; Deligny-Penninck, S.; Rogniaux, H.; Buitink, J. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012, 35, 1440–1455. [Google Scholar] [CrossRef]
- Verdier, J.; Lalanne, D.; Pelletier, S.; Torres-Jerez, I.; Righetti, K.; Bandyopadhyay, K.; Leprince, O.; Chatelain, E.; Vu, B.L.; Gouzy, J.; et al. A regulatory network-based approach dissects late maturation processes re-lated to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 2013, 163, 757–774. [Google Scholar] [CrossRef]
- Sun, X.; Rikkerink, E.H.; Jones, W.T.; Uversky, V.N. Multifarious Roles of Intrinsic Disorder in Proteins Illustrate Its Broad Impact on Plant Biology. Plant Cell 2013, 25, 38–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.P.; Giorgi, F.M.; Lohse, M.; Kvederaviciute, K.; Klages, S.; Usadel, B.; Meskiene, I.; Reinhardt, R.; Hincha, D.K. Tran-scriptome sequencing and microarray design for functional genomics in the extremophile Arabidopsis relative Thellungiella salsuginea (Eutrema salsugineum). BMC Genom. 2013, 14, 793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muvunyi, B.P.; Yan, Q.; Wu, F.; Min, X.; Yan, Z.Z.; Kanzana, G.; Wang, Y.; Zhang, J. Mining Late Embryogenesis Abundant (LEA) Family Genes in Cleistogenes songorica, a Xerophyte Perennial Desert Plant. Int. J. Mol. Sci. 2018, 19, 3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artur, M.A.S.; Rienstra, J.; Dennis, T.J.; Farrant, J.M.; Ligterink, W.; Hilhorst, H. Structural plasticity of intrinsically disor-dered LEA proteins from Xerophyta schlechteri provides protection in vitro and in vivo. Front. Plant. Sci. 2019, 10, 1272. [Google Scholar] [CrossRef] [Green Version]
- Nagaraju, M.; Kumar, S.A.; Reddy, P.S.; Kumar, A.; Rao, D.M.; Kishor, P.B.K. Genome-scale identification, classification, and tissue specific expression analysis of late embryogenesis abundant (LEA) genes under abiotic stress conditions in Sorghum bicolor L. PLoS ONE 2019, 14, e0209980. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Lan, T. Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabu-liformis (Pinaceae) in Escherichia coli. Sci. Rep. 2016, 6, 14467. [Google Scholar]
- Golan, I.; Dominguez, P.G.; Konrad, Z.; Shkolnik-Inbar, D.; Carrari, F.; Bar-Zvi, D. Tomato ABSCISIC ACID STRESS RIP-ENING (ASR) gene family revisited. PLoS ONE 2014, 9, e107117. [Google Scholar] [CrossRef] [Green Version]
- Hamdi, K.; Salladini, E.; O’Brien, D.P.; Brier, S.; Chenal, A.; Yacoubi, I.; Longhi, S. Structural disorder and induced folding within two cereal, ABA stress and ripening (ASR) proteins. Sci. Rep. 2017, 7, 1–21. [Google Scholar] [CrossRef]
- Konrad, Z.; Bar-Zvi, D. Synergism between the chaperone-like activity of the stress regulated ASR1 protein and the osmolyte glycine-betaine. Planta 2008, 227, 1213–1219. [Google Scholar] [CrossRef]
- Dai, J.R.; Liu, B.; Feng, D.R.; Liu, H.Y.; He, Y.M.; Qi, K.B.; Wang, H.B.; Wang, J.F. MpAsr encodes an intrinsically unstruc-tured protein and enhances osmotic tolerance in transgenic Arabidopsis. Plant Cell Rep. 2011, 30, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, Q.; Yu, H.; Li, L.; Zhang, G.; Chen, X.; Jiang, M.; Tan, M. Comprehensive Analysis of the Cadmium Tolerance of Abscisic Acid-, Stress- and Ripening-Induced Proteins (ASRs) in Maize. Int. J. Mol. Sci. 2019, 20, 133. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2signalling in rice. Plant Biotechnol. J. 2016, 15, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Lin, R.; Su, H.; Chen, H.; Luo, M.; Yang, L.; Zhang, M. The functional identification of glycine-rich TtASR from Tetra-gonia tetragonoides (Pall.) Kuntze involving in plant abiotic stress tolerance. Plant Physiol. Biochem. 2019, 143, 212–223. [Google Scholar] [CrossRef]
- Hu, W.; Huang, C.; Deng, X.; Zhou, S.; Chen, L.; Li, Y.; Wang, C.; Ma, Z.; Yuan, Q.; Wang, Y.; et al. TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 2013, 36, 1449–1464. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Covarrubias, A.A. Late Embryogenesis Abundant (LEA) proteins in legumes. Front. Plant Sci. 2013, 4, 190. [Google Scholar] [CrossRef] [Green Version]
- Wetzler, D.E.; Fuchs Wightman, F.; Bucci, H.A.; Rinaldi, J.; Caramelo, J.J.; Iusem, N.D.; Ricardi, M.M. Conformational plas-ticity of the intrinsically disordered protein ASR1 modulates its function as a drought stress-responsive gene. PLoS ONE 2018, 13, e0202808. [Google Scholar] [CrossRef]
- Niveditha, V.R.; Sridhar, K.R. Antioxidant activity of raw, cooked and Rhizopus oligosporus fermented beans of Canavalia of coastal sand dunes of Southwest India. J. Food Sci. Technol. 2012, 51, 3253–3260. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Liu, N.; Ren, H.; Jian, S. Physiology and biochemical characteristics of Canavalia maritime under stress. J. Trop. Subtrop. Bot. 2019, 27, 157–163. [Google Scholar]
- Iusem, N.D.; Bartholomew, D.M.; Hitz, W.D.; Scolnik, P.A. Tomato (Lycopersicon esculentum) Transcript Induced by Water Deficit and Ripening. Plant Physiol. 1993, 102, 1353–1354. [Google Scholar] [CrossRef] [Green Version]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Feng, D.; Su, J.; Zhang, Y.; Dai, J.; Jin, H.; Liu, B.; He, Y.; Qi, K.; Wang, H.; et al. Cu2+ triggers reversible aggrega-tion of a disordered His-rich dehydrin MpDhn12 from Musa paradisiaca. J. Biochem. 2011, 150, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Li, R.H.; Liu, G.B.; Wang, H.; Zheng, Y.Z. Effects of Fe3+ and Zn2+ on the structural and thermodynamic properties of a soy-bean ASR protein. Biosci. Biotechnol. Biochem. 2013, 77, 475–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.-X.; Zhang, H.; Su, H.-X.; Xia, K.-F.; Jian, S.-G.; Zhang, M. Ipomoea pes-caprae IpASR Improves Salinity and Drought Tolerance in Transgenic Escherichia coli and Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, Y.; Liu, Y.; Zhang, J.; Song, L.; Guo, C. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant. Sci. 2016, 6, 1247. [Google Scholar] [CrossRef] [Green Version]
- Das Laha, S.; Dutta, S.; Schäffner, A.R.; Das, M. Gene duplication and stress genomics in Brassicas: Current understanding and future prospects. J. Plant Physiol. 2020, 255, 153293. [Google Scholar] [CrossRef]
- Wu, C.; Hu, W.; Yan, Y.; Tie, W.; Ding, Z.; Guo, J.; He, G. The late embryogenesis abundant protein family in cassava (Mani-hot esculenta Crantz): Genome-wide characterization and expression during abiotic stress. Molecules 2018, 23, 1196. [Google Scholar] [CrossRef] [Green Version]
- Ling, H.; Zeng, X.; Guo, S. Functional insights into the late embryogenesis abundant (LEA) protein family from Dendrobium officinale (Orchidaceae) using an Escherichia coli system. Sci. Rep. 2016, 6, 39693. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The role of the late embryogenesis-abundant (LEA) protein family in development and the abiotic stress response: A comprehensive expression analysis of potato (Solanum tu-berosum). Genes 2019, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Zhang, H.; Zheng, J.-X.; Mo, H.; Xia, K.-F.; Jian, S.-G. Functional Identification of Salt-Stress-Related Genes Using the FOX Hunting System from Ipomoea pes-caprae. Int. J. Mol. Sci. 2018, 19, 3446. [Google Scholar] [CrossRef] [Green Version]
- Ricardi, M.M.; González, R.M.; Zhong, S.; Domínguez, P.G.; Duffy, T.; Turjanski, P.G.; Salgado Salter, J.D.; Alleva, K.; Carra-ri, F.; Giovannoni, J.J.; et al. Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol. 2014, 14, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Shen, L.; Yang, S.; Guan, D.; He, S. CaASR1 promotes salicylic acid- but represses jasmonic acid-dependent sig-naling to enhance the resistance of Capsicum annuum to bacterial wilt by modulating CabZIP63. J. Exp. Bot. 2020, 71, 6538–6554. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitu-tions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for in-teractive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Gietz, D.; Jean, A.S.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]



| Name | Gene Locus | Length (aa) | Mw (kD) | PI | Instability Index (II) | Aliphatic Index (AI) | GRAVY | Content of Disordered aa (%) | Subcellular Localization * |
|---|---|---|---|---|---|---|---|---|---|
| CrLEA1-1 | 03T007960 | 174 | 17.92 | 9.13 | 11.32 | 35.00 | −1.060 | 83.91 | mito: 10, nucl: 4 |
| CrLEA1-2 | 04T011772 | 192 | 20.00 | 5.38 | 45.87 | 49.53 | −0.725 | 83.33 | nucl: 7, cyto: 3 |
| CrLEA1-3 | 05T014731 | 136 | 14.98 | 9.03 | 54.57 | 60.44 | −0.992 | 94.12 | nucl: 9, mito: 3 |
| CrLEA2-1 | 01T000091 | 294 | 32.86 | 10.00 | 57.75 | 79.18 | −0.266 | 25.51 | chlo: 3, cyto: 3, E.R.: 3 |
| CrLEA2-2 | 01T000357 | 210 | 22.61 | 9.47 | 39.79 | 88.24 | 0.250 | 10.48 | chlo: 5, mito: 5 |
| CrLEA2-3 | 01T000402 | 221 | 24.59 | 9.21 | 37.80 | 103.57 | 0.078 | 18.55 | chlo: 5, cyto: 5 |
| CrLEA2-4 | 01T001888 | 207 | 22.93 | 9.32 | 34.25 | 95.12 | 0.038 | 12.08 | plas: 8, cyto: 4 |
| CrLEA2-5 | 01T003219 | 230 | 25.26 | 9.21 | 34.53 | 111.39 | 0.283 | 16.96 | plas: 4.5, vacu: 4, nucl_plas: 3 |
| CrLEA2-6 | 02T004727 | 259 | 28.57 | 9.88 | 35.46 | 82.39 | −0.232 | 18.53 | chlo: 10, mito: 3 |
| CrLEA2-7 | 02T004857 | 262 | 29.07 | 9.80 | 50.89 | 84.35 | −0.331 | 22.14 | cyto: 7 |
| CrLEA2-8 | 02T005159 | 207 | 23.10 | 9.78 | 33.03 | 100.24 | −0.048 | 13.04 | chlo: 4, plas: 4 |
| CrLEA2-9 | 02T006010 | 192 | 20.74 | 9.28 | 13.81 | 106.98 | 0.272 | 6.25 | vacu: 4, E.R.: 3 |
| CrLEA2-10 | 02T006879 | 190 | 21.16 | 6.83 | 28.11 | 96.42 | −0.112 | 6.32 | nucl: 8, chlo: 3 |
| CrLEA2-11 | 02T006881 | 248 | 27.67 | 10.05 | 33.42 | 94.68 | −0.042 | 14.52 | cyto: 8 |
| CrLEA2-12 | 02T007016 | 233 | 26.24 | 9.41 | 48.70 | 97.38 | −0.033 | 21.46 | cyto: 8 |
| CrLEA2-13 | 02T007186 | 250 | 27.17 | 9.34 | 36.97 | 90.88 | −0.004 | 16.4 | chlo: 8, mito: 4 |
| CrLEA2-14 | 03T008517 | 227 | 25.78 | 9.40 | 36.41 | 89.30 | −0.131 | 7.49 | golg: 4.5, golg_plas: 4.5, plas: 3.5, E.R.: 3 |
| CrLEA2-15 | 03T008518 | 228 | 26.55 | 9.48 | 37.63 | 81.18 | −0.291 | 10.09 | chlo: 10, pero: 3 |
| CrLEA2-16 | 03T008519 | 209 | 23.77 | 9.71 | 36.98 | 97.32 | 0.018 | 7.18 | cyto: 11 |
| CrLEA2-17 | 03T009222 | 197 | 21.48 | 9.76 | 31.71 | 110.71 | 0.356 | 7.61 | chlo: 6, E.R.: 4 |
| CrLEA2-18 | 03T009270 | 235 | 26.49 | 8.98 | 33.86 | 103.19 | 0.329 | 6.81 | cyto: 10 |
| CrLEA2-19 | 03T010271 | 222 | 24.12 | 9.64 | 24.57 | 121.08 | 0.232 | 15.77 | nucl: 6 |
| CrLEA2-20 | 03T010272 | 195 | 21.68 | 6.95 | 23.09 | 100.41 | 0.074 | 6.67 | E.R.: 4, mito: 3, extr: 3 |
| CrLEA2-21 | 03T010452 | 176 | 19.07 | 5.86 | 26.80 | 113.47 | 0.181 | 6.25 | plas: 6, vacu: 5 |
| CrLEA2-22 | 04T011627 | 200 | 23.20 | 9.40 | 35.70 | 104.65 | 0.115 | 0.5 | extr: 3, vacu: 3, E.R.: 3 |
| CrLEA2-23 | 04T011636 | 228 | 26.46 | 9.57 | 34.97 | 81.67 | −0.233 | 11.84 | chlo: 8 |
| CrLEA2-24 | 04T011638 | 210 | 23.82 | 9.63 | 47.73 | 101.52 | 0.099 | 6.19 | cyto: 7 |
| CrLEA2-25 | 04T012994 | 249 | 27.84 | 9.90 | 38.97 | 101.37 | −0.087 | 20.48 | cyto: 7, nucl: 3, E.R.: 3 |
| CrLEA2-26 | 04T013772 | 225 | 25.61 | 9.42 | 41.04 | 76.62 | −0.399 | 11.11 | chlo: 14 |
| CrLEA2-27 | 05T014592 | 322 | 35.41 | 10.11 | 58.09 | 71.71 | −0.333 | 31.68 | chlo: 3, vacu: 3 |
| CrLEA2-28 | 05T016584 | 272 | 29.42 | 9.60 | 42.56 | 84.19 | −0.214 | 32.35 | nucl: 5, chlo: 4, plas: 3 |
| CrLEA2-29 | 05T016770 | 202 | 23.15 | 9.75 | 44.59 | 107.13 | 0.049 | 11.39 | chlo: 5, mito: 3 |
| CrLEA2-30 | 06T017171 | 233 | 26.69 | 10.13 | 39.27 | 102.45 | −0.009 | 19.31 | cyto: 5, plas: 4 |
| CrLEA2-31 | 06T017436 | 320 | 35.70 | 4.80 | 25.33 | 91.97 | −0.437 | 16.25 | cyto: 9 |
| CrLEA2-32 | 06T017480 | 273 | 30.26 | 9.43 | 50.16 | 89.56 | −0.200 | 19.41 | cyto: 4, chlo: 3, plas: 3 |
| CrLEA2-33 | 06T018775 | 257 | 28.95 | 10.17 | 49.92 | 99.69 | −0.162 | 23.35 | nucl: 6, cyto: 5 |
| CrLEA2-34 | 07T020998 | 211 | 24.12 | 9.10 | 30.10 | 95.59 | 0.045 | 6.64 | extr: 5, chlo: 3, E.R.: 3 |
| CrLEA2-35 | 07T020999 | 499 | 57.65 | 9.51 | 31.38 | 89.80 | −0.086 | 2.2 | plas: 7.5, golg_plas: 6, golg: 3.5 |
| CrLEA2-36 | 07T021162 | 247 | 27.83 | 9.15 | 54.28 | 97.41 | −0.057 | 13.77 | plas: 4, golg: 3 |
| CrLEA2-37 | 07T021185 | 308 | 34.10 | 9.58 | 62.03 | 62.27 | −0.514 | 25.32 | chlo: 3, plas: 3, E.R.: 3 |
| CrLEA2-38 | 08T022453 | 223 | 24.65 | 9.50 | 27.00 | 97.89 | 0.155 | 15.25 | chlo: 5, cyto: 3 |
| CrLEA2-39 | 08T022523 | 219 | 24.64 | 9.74 | 53.37 | 106.80 | 0.185 | 5.94 | E.R.: 5 |
| CrLEA2-40 | 08T022549 | 252 | 27.66 | 6.83 | 38.09 | 110.32 | 0.311 | 5.56 | cyto: 7 |
| CrLEA2-41 | 08T023188 | 317 | 35.77 | 9.67 | 57.68 | 82.93 | −0.391 | 36.59 | chlo: 4, nucl: 3, mito: 3 |
| CrLEA2-42 | 09T024657 | 244 | 27.90 | 7.50 | 40.73 | 100.20 | 0.075 | 18.03 | chlo: 6 |
| CrLEA2-43 | 09T024732 | 184 | 19.82 | 9.68 | 21.51 | 121.25 | 0.497 | 4.89 | vacu: 5, extr: 3 |
| CrLEA2-44 | 09T024733 | 186 | 20.56 | 8.99 | 14.50 | 107.37 | 0.354 | 3.76 | extr: 4, vacu: 4 |
| CrLEA2-45 | 09T024798 | 220 | 24.57 | 8.97 | 38.64 | 104.95 | 0.119 | 6.82 | chlo: 4 |
| CrLEA2-46 | 09T025174 | 255 | 27.99 | 10.30 | 36.23 | 87.22 | −0.138 | 18.04 | chlo: 8, mito: 5 |
| CrLEA2-47 | 09T025255 | 308 | 34.13 | 9.93 | 58.73 | 72.37 | −0.347 | 29.55 | chlo: 4, E.R.: 3 |
| CrLEA2-48 | 09T025271 | 228 | 25.83 | 9.63 | 28.59 | 105.39 | 0.153 | 10.96 | vacu: 3 |
| CrLEA2-49 | 10T025889 | 190 | 21.52 | 9.52 | 31.74 | 88.79 | −0.412 | 9.47 | vacu: 5, extr: 3, E.R.: 3 |
| CrLEA2-50 | 10T025950 | 185 | 20.54 | 8.80 | 27.57 | 114.27 | 0.268 | 4.86 | extr: 3, vacu: 3, E.R.: 3 |
| CrLEA2-51 | 10T026128 | 183 | 20.17 | 9.67 | 34.72 | 89.40 | 0.371 | 5.46 | chlo: 7, vacu: 3 |
| CrLEA2-52 | 10T026178 | 192 | 21.44 | 8.41 | 50.06 | 97.40 | 0.068 | 18.23 | cyto: 7.5, cyto_nucl: 6, nucl: 3.5 |
| CrLEA2-53 | 10T026179 | 204 | 22.68 | 10.14 | 39.68 | 94.61 | −0.012 | 10.29 | chlo: 6, E.R.: 4 |
| CrLEA2-54 | 10T026917 | 221 | 24.71 | 9.14 | 41.43 | 97.47 | −0.089 | 18.55 | cyto: 7, cysk: 4 |
| CrLEA2-55 | 11T027970 | 382 | 42.52 | 4.97 | 23.26 | 96.83 | −0.283 | 17.02 | cyto: 8, nucl: 3 |
| CrLEA2-56 | 11T028314 | 258 | 29.48 | 10.11 | 52.91 | 92.91 | −0.151 | 20.93 | chlo: 11 |
| CrLEA2-57 | 11T028577 | 244 | 27.78 | 10.10 | 47.89 | 104.18 | −0.164 | 18.85 | cyto: 5, nucl: 3 |
| CrLEA2-58 | 11T029314 | 264 | 29.23 | 9.27 | 46.36 | 88.98 | −0.062 | 18.56 | cyto: 8, chlo: 4 |
| CrLEA2-59 | 11T029316 | 264 | 29.19 | 9.11 | 47.73 | 90.45 | −0.031 | 18.18 | cyto: 8, chlo: 3 |
| CrLEA2-60 | 11T029318 | 264 | 29.43 | 9.23 | 48.32 | 83.45 | −0.146 | 18.18 | cyto: 7.5, cyto_nucl: 4.5, chlo: 4 |
| CrLEA3-1 | 01T003524 | 106 | 11.73 | 10.09 | 56.38 | 89.25 | −0.325 | 46.23 | chlo: 12 |
| CrLEA3-2 | 02T007058 | 84 | 9.69 | 9.34 | 40.26 | 68.57 | −0.664 | 39.29 | chlo: 5, mito: 4, nucl: 3 |
| CrLEA3-3 | 03T008662 | 97 | 10.57 | 9.91 | 43.60 | 76.39 | −0.300 | 47.42 | cyto: 7, chlo: 6 |
| CrLEA3-4 | 04T012522 | 97 | 10.20 | 9.70 | 27.95 | 77.63 | −0.252 | 46.39 | chlo: 13 |
| CrLEA3-5 | 04T013832 | 101 | 11.26 | 9.25 | 44.98 | 66.63 | −0.655 | 48.51 | mito: 8, chlo: 3 |
| CrLEA3-6 | 05T015908 | 105 | 11.32 | 9.88 | 60.57 | 69.62 | −0.456 | 54.29 | cyto: 8, mito: 5 |
| CrLEA4-1 | 03T008401 | 310 | 34.21 | 7.05 | 28.99 | 49.00 | −1.093 | 65.16 | chlo: 6, nucl: 6 |
| CrLEA4-2 | 04T011481 | 427 | 46.51 | 6.04 | 36.13 | 43.89 | −1.296 | 77.52 | nucl: 12 |
| CrLEA5-1 | 02T006409 | 95 | 10.26 | 5.87 | 48.75 | 41.16 | −1.355 | 87.37 | nucl: 6, cyto: 6 |
| CrLEA5-2 | 03T010121 | 158 | 17.54 | 9.44 | 49.86 | 48.73 | −1.237 | 72.78 | chlo: 9, nucl: 5 |
| CrLEA6-1 | 02T003983 | 82 | 9.20 | 9.20 | 47.60 | 54.76 | −1.194 | 73.17 | nucl: 6, mito: 5 |
| CrLEA6-2 | 05T014654 | 457 | 50.38 | 9.10 | 56.56 | 52.32 | −0.962 | 71.12 | nucl: 13 |
| CrDHN1 | 01T000101 | 80 | 8.58 | 6.49 | 24.98 | 51.12 | −1.068 | 48.75 | nucl: 5 |
| CrDHN2 | 02T003923 | 215 | 24.36 | 5.25 | 57.49 | 51.63 | −1.520 | 79.07 | nucl: 12 |
| CrDHN3 | 02T003929 | 190 | 19.95 | 8.81 | 30.05 | 39.11 | −1.043 | 82.63 | nucl: 13 |
| CrDHN4 | 06T019348 | 220 | 25.01 | 5.45 | 43.12 | 47.41 | −1.636 | 80.91 | nucl: 11 |
| CrDHN5 | 07T021123 | 194 | 19.47 | 6.28 | −5.30 | 16.65 | −1.215 | 72.16 | nucl: 9 |
| CrSMP1 | 04T011095 | 203 | 20.80 | 4.97 | 28.89 | 82.81 | −0.185 | 17.24 | cyto: 7, chlo: 6 |
| CrSMP2 | 04T012849 | 278 | 28.83 | 5.20 | 34.95 | 78.35 | −0.357 | 25.9 | cyto: 6, chlo: 4 |
| CrSMP3 | 04T013711 | 258 | 26.15 | 4.77 | 36.17 | 74.65 | −0.286 | 17.05 | cyto: 5, chlo: 4 |
| CrSMP4 | 09T023499 | 284 | 29.44 | 4.90 | 28.98 | 79.82 | −0.137 | 25 | chlo: 8 |
| CrASR1 | 04T013503 | 235 | 25.92 | 5.79 | 40.95 | 28.00 | −1.449 | 90.64 | nucl: 6, cyto: 3 |
| CrASR2 | 07T020512 | 130 | 14.98 | 6.34 | 42.06 | 47.46 | −1.321 | 80.77 | mito: 5, nucl: 4, cyto: 3 |
| CrASR3 | 07T020519 | 114 | 13.09 | 6.41 | 36.67 | 51.58 | −1.341 | 79.82 | mito: 5, nucl: 4, cyto: 3 |
| Duplicated Pair | Duplicate Type | Ka | Ks | Ka/Ks | Positive Selection |
|---|---|---|---|---|---|
| CrLEA1-1/CrLEA1-2 | Segmental | 0.391 | 1.260 | 0.310 | No |
| CrLEA2-2/CrLEA2-51 | Segmental | 0.125 | 0.992 | 0.126 | No |
| CrLEA2-4/CrLEA2-54 | Segmental | 0.235 | 0.911 | 0.258 | No |
| CrLEA2-5/CrLEA2-28 | Segmental | 0.267 | 1.221 | 0.219 | No |
| CrLEA2-9/CrLEA2-20 | Segmental | 0.343 | 0.858 | 0.400 | No |
| CrLEA2-10/CrLEA2-21 | Segmental | 0.268 | 0.981 | 0.273 | No |
| CrLEA2-12/CrLEA2-17 | Segmental | 0.215 | 0.704 | 0.305 | No |
| CrLEA2-13/CrLEA2-46 | Segmental | 0.232 | 0.967 | 0.240 | No |
| CrLEA2-15/CrLEA2-23 | Segmental | 0.253 | 0.857 | 0.295 | No |
| CrLEA2-16/CrLEA2-24 | Segmental | 0.105 | 1.042 | 0.101 | No |
| CrLEA2-30/CrLEA2-56 | Segmental | 0.187 | 0.761 | 0.246 | No |
| CrLEA2-31/CrLEA2-55 | Segmental | 0.102 | 0.449 | 0.227 | No |
| CrLEA2-33/CrLEA2-57 | Segmental | 0.157 | 0.564 | 0.278 | No |
| CrLEA2-38/CrLEA2-44 | Segmental | 0.320 | 0.591 | 0.541 | No |
| CrLEA2-39/CrLEA2-45 | Segmental | 0.174 | 0.757 | 0.230 | No |
| CrLEA3-1/CrLEA3-6 | Segmental | 0.278 | 0.692 | 0.402 | No |
| CrLEA3-3/CrLEA3-4 | Segmental | 0.138 | 0.669 | 0.206 | No |
| CrLEA4-1/CrLEA4-2 | Segmental | 0.419 | 1.700 | 0.246 | No |
| CrLEA5-1/CrLEA5-2 | Segmental | 0.086 | 0.702 | 0.123 | No |
| CrLEA2-14/CrLEA2-15 | Tandem | 0.488 | 1.489 | 0.328 | No |
| CrLEA2-19/CrLEA2-20 | Tandem | 0.906 | n/c a | n/c | No |
| CrLEA2-34/CrLEA2-35 | Tandem | 0.141 | 0.384 | 0.367 | No |
| CrLEA2-43/CrLEA2-44 | Tandem | 0.270 | 0.360 | 0.750 | No |
| CrLEA2-52/CrLEA2-53 | Tandem | 0.560 | 0.937 | 0.598 | No |
| CrLEA2-58/CrLEA2-59 | Tandem | 0.003 | 0.020 | 0.150 | No |
| CrLEA2-59/CrLEA2-60 | Tandem | 0.056 | 0.126 | 0.444 | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, R.; Zou, T.; Mei, Q.; Wang, Z.; Zhang, M.; Jian, S. Genome-Wide Analysis of the Late Embryogenesis Abundant (LEA) and Abscisic Acid-, Stress-, and Ripening-Induced (ASR) Gene Superfamily from Canavalia rosea and Their Roles in Salinity/Alkaline and Drought Tolerance. Int. J. Mol. Sci. 2021, 22, 4554. https://doi.org/10.3390/ijms22094554
Lin R, Zou T, Mei Q, Wang Z, Zhang M, Jian S. Genome-Wide Analysis of the Late Embryogenesis Abundant (LEA) and Abscisic Acid-, Stress-, and Ripening-Induced (ASR) Gene Superfamily from Canavalia rosea and Their Roles in Salinity/Alkaline and Drought Tolerance. International Journal of Molecular Sciences. 2021; 22(9):4554. https://doi.org/10.3390/ijms22094554
Chicago/Turabian StyleLin, Ruoyi, Tao Zou, Qiming Mei, Zhengfeng Wang, Mei Zhang, and Shuguang Jian. 2021. "Genome-Wide Analysis of the Late Embryogenesis Abundant (LEA) and Abscisic Acid-, Stress-, and Ripening-Induced (ASR) Gene Superfamily from Canavalia rosea and Their Roles in Salinity/Alkaline and Drought Tolerance" International Journal of Molecular Sciences 22, no. 9: 4554. https://doi.org/10.3390/ijms22094554
APA StyleLin, R., Zou, T., Mei, Q., Wang, Z., Zhang, M., & Jian, S. (2021). Genome-Wide Analysis of the Late Embryogenesis Abundant (LEA) and Abscisic Acid-, Stress-, and Ripening-Induced (ASR) Gene Superfamily from Canavalia rosea and Their Roles in Salinity/Alkaline and Drought Tolerance. International Journal of Molecular Sciences, 22(9), 4554. https://doi.org/10.3390/ijms22094554






