Bioinformatic Prediction of an tRNASec Gene Nested inside an Elongation Factor SelB Gene in Alphaproteobacteria
Abstract
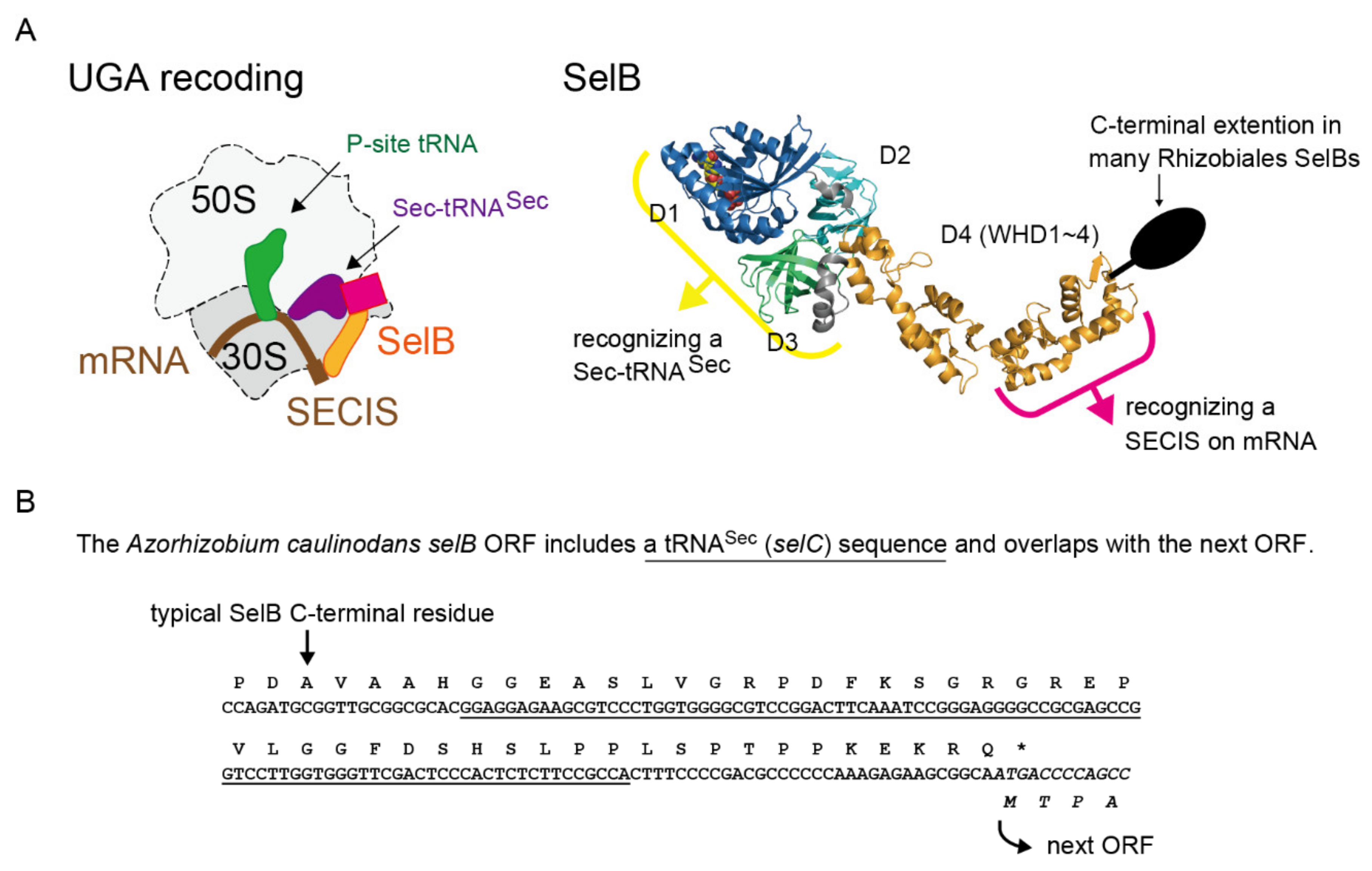
1. Introduction
2. Results
2.1. tRNASec is Encoded Inside the selB Gene in Diverse Alphaproteobacteria
2.2. tRNASec Remnant is Encoded inside the selB Gene in pSymA
2.3. tRNASec Partially Overlapping with the selB Gene in Gammaproteobacteria
2.4. Reversed tRNASec Overlapping with the selB Gene in Gammaproteobacteria
2.5. Alignment Analysis of the selB C-Terminal Amino Acid Extensions
2.6. A New Mechanism of Maintaining Homeostasis between selB and Sec-tRNASec?
3. Discussion
4. Materials and Methods
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar]
- Silva, I.R.; Serrão, V.H.; Manzine, L.R.; Faim, L.M.; da Silva, M.T.; Makki, R.; Saidemberg, D.M.; Cornélio, M.L.; Palma, M.S.; Thiemann, O.H. Formation of a Ternary Complex for Selenocysteine Biosynthesis in Bacteria. J. Biol. Chem. 2015, 290, 29178–29188. [Google Scholar]
- Fischer, N.; Neumann, P.; Bock, L.V.; Maracci, C.; Wang, Z.; Paleskava, A.; Konevega, A.L.; Schröder, G.F.; Grubmüller, H.; Ficner, R.; et al. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature 2016, 540, 80–85. [Google Scholar] [CrossRef]
- Itoh, Y.; Sekine, S.-I.; Yokoyama, S. Crystal structure of the full-length bacterial selenocysteine-specific elongation factor SelB. Nucleic Acids Res. 2015, 43, 9028–9038. [Google Scholar] [PubMed]
- Mansell, J.B.; Guévremont, D.; Poole, E.S.; Tate, W.P. A dynamic competition between release factor 2 and the tRNASec decoding UGA at the recoding site of Escherichia coli formate dehydrogenase H. EMBO J. 2001, 20, 7284–7293. [Google Scholar]
- Mukai, T.; Englert, M.; Tripp, H.J.; Miller, C.; Ivanova, N.N.; Rubin, E.M.; Kyrpides, N.C.; Söll, D. Facile Recoding of Selenocysteine in Nature. Angew. Chem. Int. Ed. Engl. 2016, 55, 5337–5341. [Google Scholar] [PubMed]
- Zhang, Y.; Romero, H.; Salinas, G.; Gladyshev, V.N. Dynamic evolution of selenocysteine utilization in bacteria: A balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006, 7, R94. [Google Scholar]
- Manian, S.S.; Gumbleton, R.; O’Gara, F. The role of formate metabolism in nitrogen fixation in Rhizobium spp. Arch. Microbiol. 1982, 133, 312–317. [Google Scholar]
- Pickering, B.S.; Oresnik, I.J. Formate-Dependent Autotrophic Growth in Sinorhizobium meliloti. J. Bacteriol. 2008, 190, 6409–6418. [Google Scholar] [CrossRef]
- Barnett, M.J.; Fisher, R.F.; Jones, T.; Komp, C.; Abola, A.P.; Barloy-Hubler, F.; Bowser, L.; Capela, D.; Galibert, F.; Gouzy, J.; et al. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 2001, 98, 9883–9888. [Google Scholar] [CrossRef]
- Copeland, P.R. Making sense of nonsense: The evolution of selenocysteine usage in proteins. Genome Biol. 2005, 6, 221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vargas-Rodriguez, O.; Englert, M.; Merkuryev, A.; Mukai, T.; Söll, D. Recoding of the selenocysteine UGA codon by cysteine in the presence of a non-canonical tRNACys and elongation factor SelB. RNA Biol. 2018, 15, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Selmer, M.; Su, X.D. Crystal structure of an mRNA-binding fragment of Moorella thermoacetica elongation factor SelB. EMBO J. 2002, 21, 4145–4153. [Google Scholar] [CrossRef]
- Gulevich, A.Y.; Skorokhodova, A.Y.; Ermishev, V.Y.; Krylov, A.A.; Minaeva, N.I.; Polonskaya, Z.M.; Zimenkov, D.V.; Biryukova, I.V.; Mashko, S.V. A new method for the construction of translationally coupled operons in a bacterial chromosome. Mol. Biol. 2009, 43, 505–514. [Google Scholar] [CrossRef]
- Bloch, S.; Węgrzyn, A.; Węgrzyn, G.; Nejman-Faleńczyk, B. Small and Smaller—sRNAs and MicroRNAs in the Regulation of Toxin Gene Expression in Prokaryotic Cells: A Mini-Review. Toxins 2017, 9, 181. [Google Scholar] [CrossRef]
- Nakanishi, K.; Nureki, O. Recent progress of structural biology of tRNA processing and modification. Mol. Cells 2005, 19, 157–166. [Google Scholar] [PubMed]
- Doublet, V.; Ubrig, E.; Alioua, A.; Bouchon, D.; Marcadé, I.; Maréchal-Drouard, L. Large gene overlaps and tRNA processing in the compact mitochondrial genome of the crustacean Armadillidium vulgare. RNA Biol. 2015, 12, 1159–1168. [Google Scholar] [CrossRef]
- Yuan, Y.; Hwang, E.S.; Altman, S. Targeted cleavage of mRNA by human RNase P. Proc. Natl. Acad. Sci. USA 1992, 89, 8006–8010. [Google Scholar] [CrossRef]
- Paleskava, A.; Konevega, A.L.; Rodnina, M.V. Thermodynamic and kinetic framework of selenocysteyl-tRNASec recognition by elongation factor SelB. J. Biol. Chem. 2010, 285, 3014–3020. [Google Scholar] [CrossRef]
- Xu, Y.-W.; Jiang, Z.-H.; Mu, Y.; Zhang, L.; Zhao, S.-Q.; Liu, S.-j.; Wang, C.; Zhao, Y.; Lü, S.-W.; Yan, G.-L.; et al. Effects of combinatorial expression of selA, selB and selC genes on the efficiency of selenocysteine incorporation in Escherichia coli. Chem. Res. Chinese Univ. 2013, 29, 87–94. [Google Scholar] [CrossRef]
- Levi, O.; Garin, S.; Arava, Y. RNA mimicry in post-transcriptional regulation by aminoacyl tRNA synthetases. WIREs RNA 2020, 11, e1564. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Vargas-Rodriguez, O.; Englert, M.; Tripp, H.J.; Ivanova, N.N.; Rubin, E.M.; Kyrpides, N.C.; Söll, D. Transfer RNAs with novel cloverleaf structures. Nucl. Acids Res. 2016, 45, 2776–2785. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J. Transmutation of tRNA over time. Nat. Genet. 1999, 22, 8–9. [Google Scholar] [CrossRef]
- Suzuki, T.; Yashiro, Y.; Kikuchi, I.; Ishigami, Y.; Saito, H.; Matsuzawa, I.; Okada, S.; Mito, M.; Iwasaki, S.; Ma, D.; et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020, 11, 4269. [Google Scholar] [CrossRef]
- Zhang, X.; Leadbetter, J.R. Evidence for Cascades of Perturbation and Adaptation in the Metabolic Genes of Higher Termite Gut Symbionts. mBio 2012, 3, e00223-12. [Google Scholar] [CrossRef] [PubMed]
- Thanbichler, M.; Böck, A. The function of SECIS RNA in translational control of gene expression in Escherichia coli. EMBO J. 2002, 21, 6925–6934. [Google Scholar] [CrossRef]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R.; et al. The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucl. Acids Res. 2020, 49, D751–D763. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukai, T. Bioinformatic Prediction of an tRNASec Gene Nested inside an Elongation Factor SelB Gene in Alphaproteobacteria. Int. J. Mol. Sci. 2021, 22, 4605. https://doi.org/10.3390/ijms22094605
Mukai T. Bioinformatic Prediction of an tRNASec Gene Nested inside an Elongation Factor SelB Gene in Alphaproteobacteria. International Journal of Molecular Sciences. 2021; 22(9):4605. https://doi.org/10.3390/ijms22094605
Chicago/Turabian StyleMukai, Takahito. 2021. "Bioinformatic Prediction of an tRNASec Gene Nested inside an Elongation Factor SelB Gene in Alphaproteobacteria" International Journal of Molecular Sciences 22, no. 9: 4605. https://doi.org/10.3390/ijms22094605
APA StyleMukai, T. (2021). Bioinformatic Prediction of an tRNASec Gene Nested inside an Elongation Factor SelB Gene in Alphaproteobacteria. International Journal of Molecular Sciences, 22(9), 4605. https://doi.org/10.3390/ijms22094605





