Isolation of Small Extracellular Vesicles from Human Sera
Abstract
1. Introduction
2. Results
2.1. Nanoparticle Tracking Analysis of Serum-Derived sEV Purified by DUC or Exo-spin™
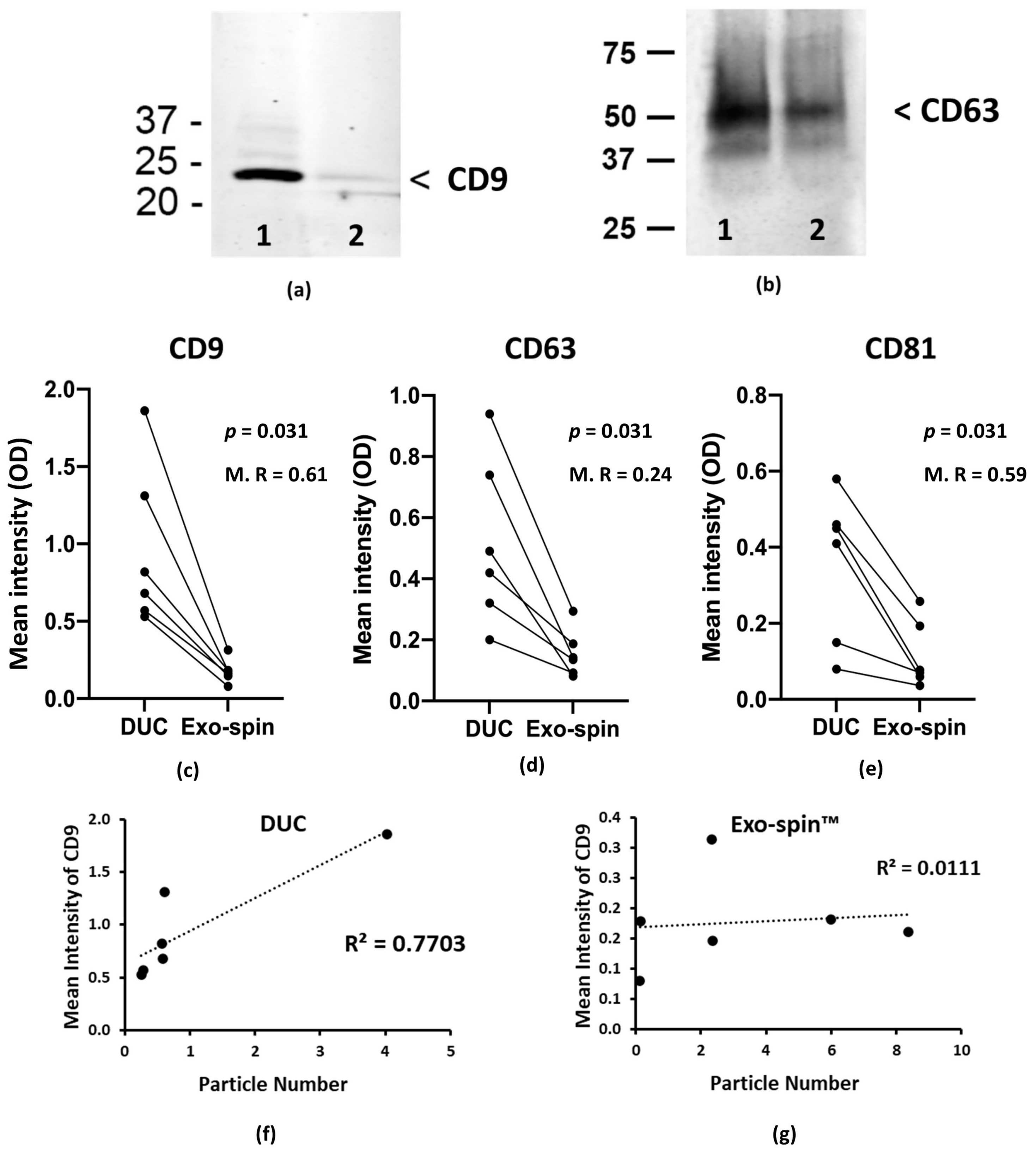
2.2. Analysis of the Abundance of Exosomal Markers by Western Blotting and ELISA
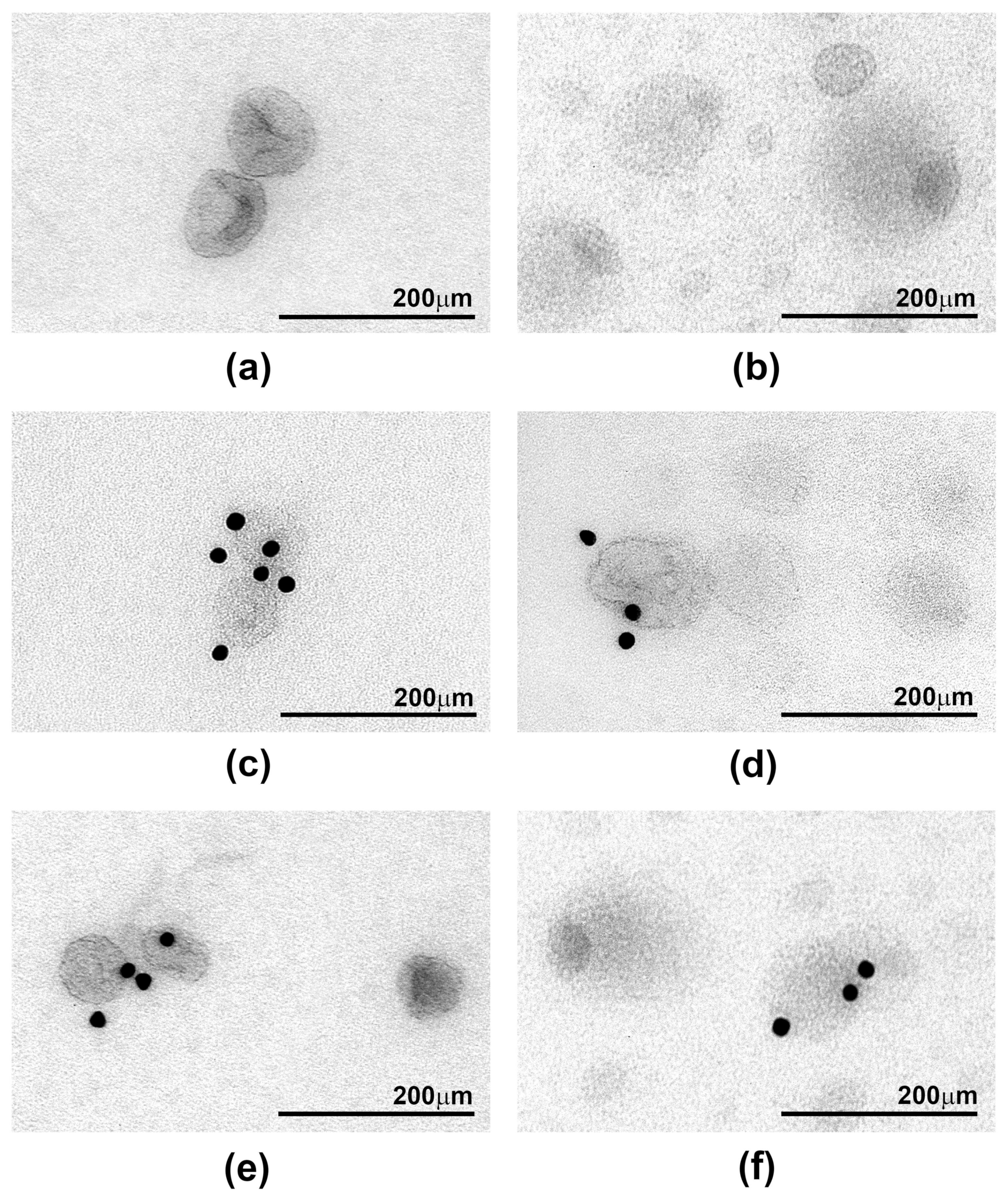
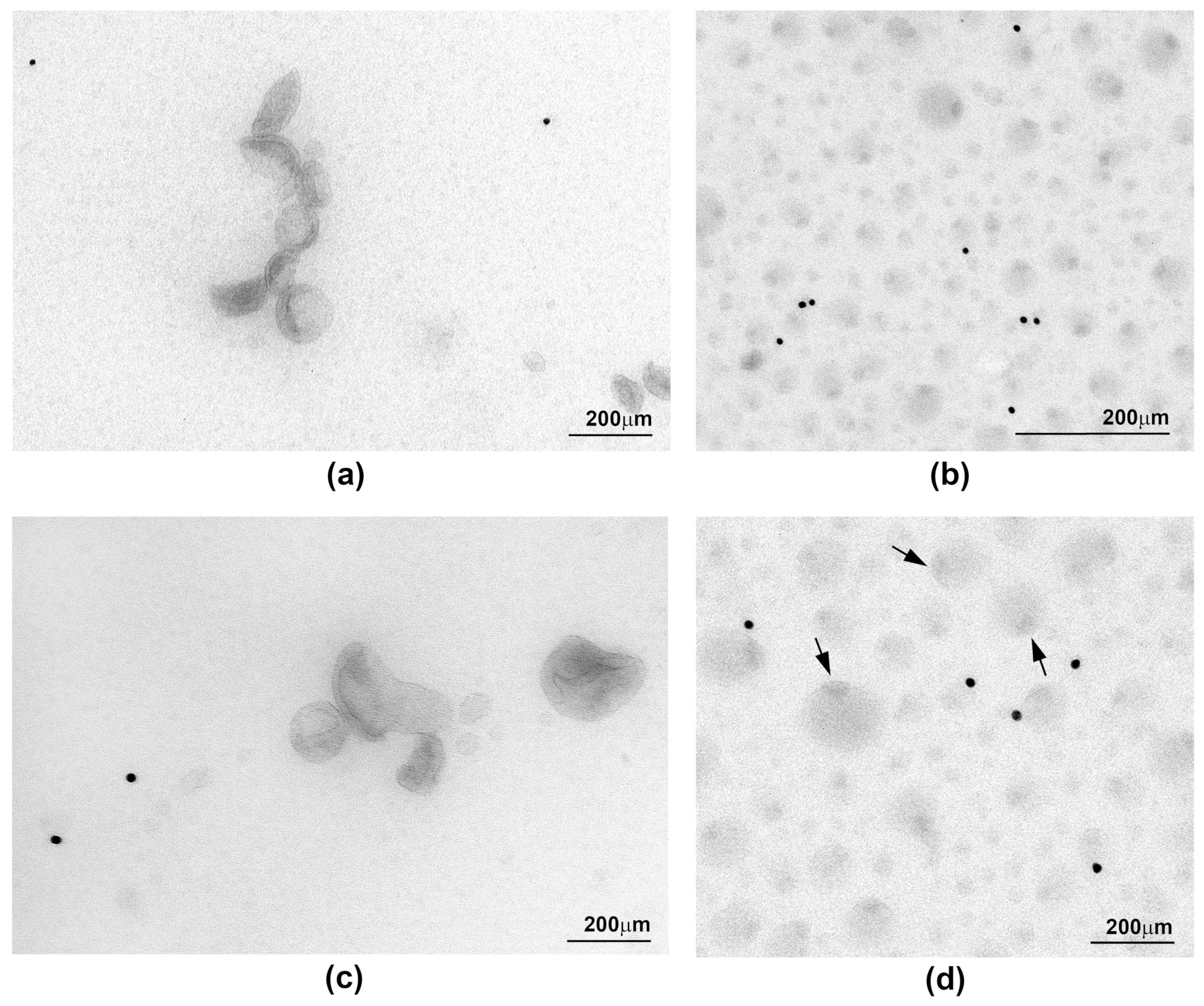
2.3. Ultrastructure of sEV Preparations Isolated by Exo-spin™ and DUC and Analysed by Transmission Electron Microscopy (TEM)
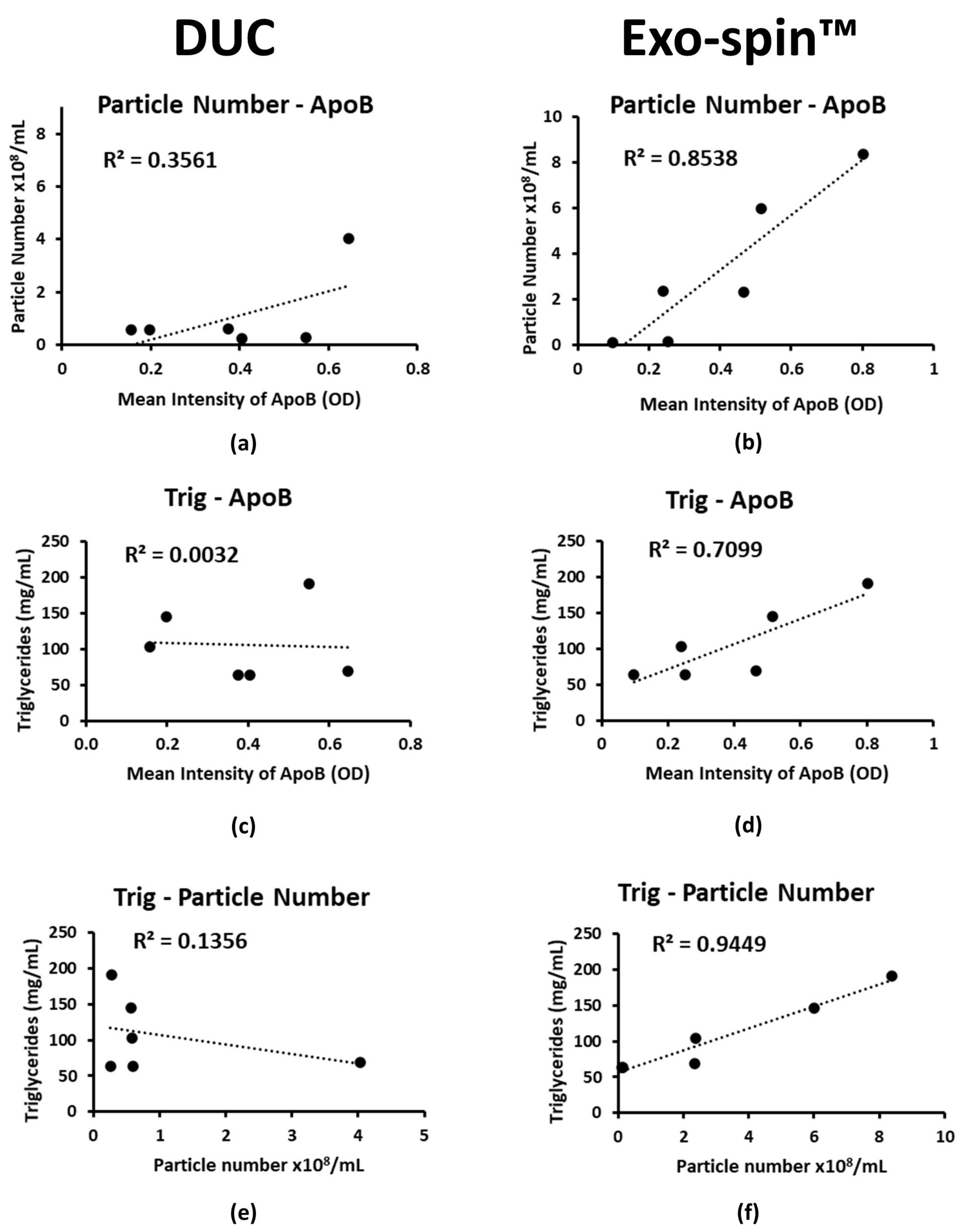
2.4. Co-Purified Lipoproteins Account for the Excess Particles in Exo-spin™ Preparations
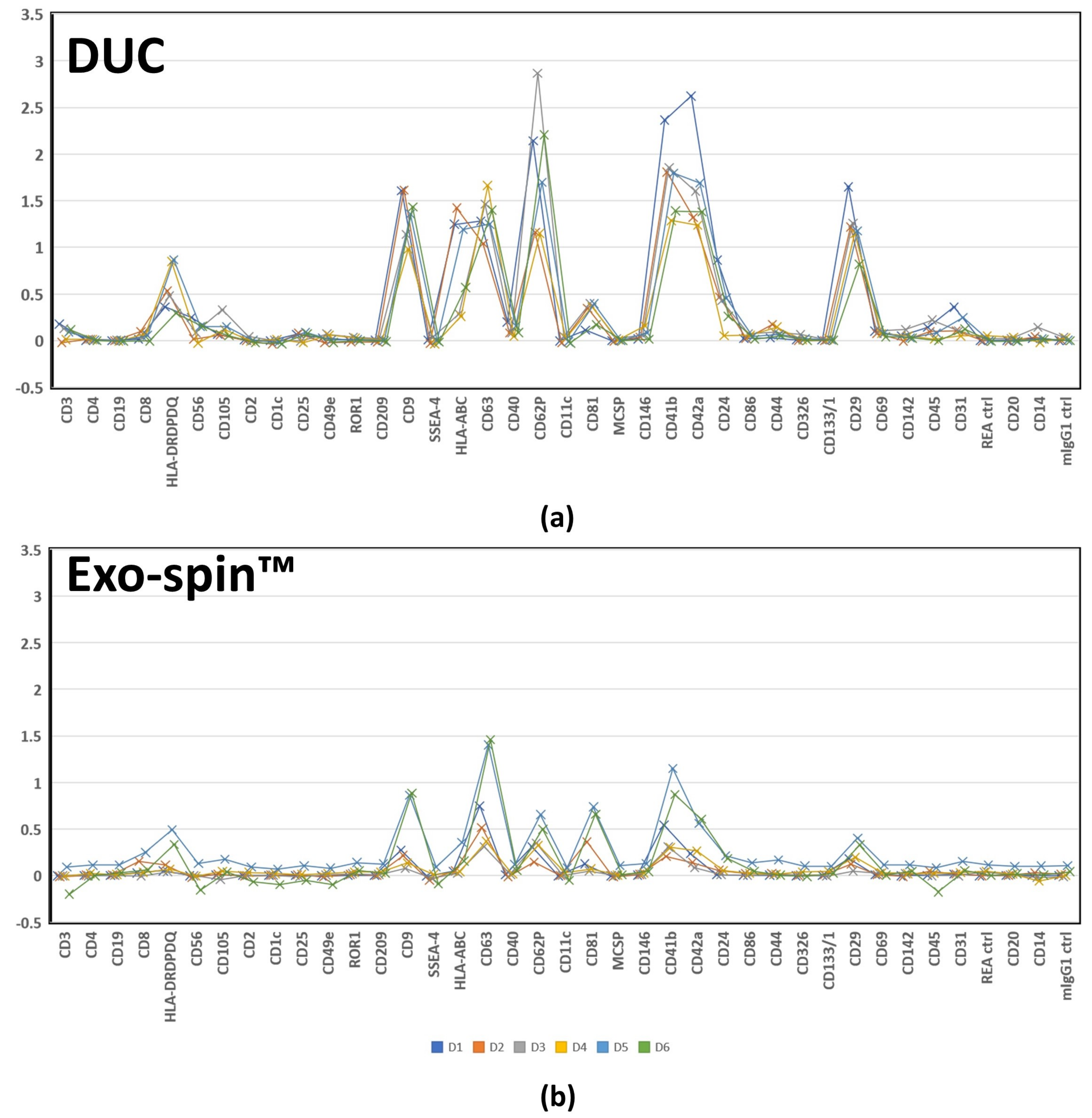
2.5. MACSPlex Analysis of Membrane Proteins on sEV Purified by DUC and Exo-spin™
3. Discussion
4. Materials and Methods
4.1. Blood Samples
4.2. Isolation of sEV by Differential Ultracentrifugation (DUC)
4.3. Isolation of sEV by Exo-spin™ Mini-HD Column EX05
4.4. Nanoparticle Tracking Analysis
4.5. Primary and Secondary Antibodies
4.6. Transmission Electron Microscopy and Immuno-Electron Microscopy
4.7. SDS-PAGE and Western Blotting
4.8. ELISA
4.9. MACPlex Human Exosome Pan Kit
4.10. Micro BCA™ Protein Assay Kit
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APC | Allophycocyanin |
| BCA | Bicinchoninic acid |
| BSA | Bovine serum albumin |
| DF | Dilution Factor |
| DUC | Differential ultracentrifugation |
| EDTA | Ethylenediaminetetraacetic acid |
| ELISA | Enzyme-linked immunoassay |
| EV | Extracellular vesicles |
| HDL | High-density lipoproteins |
| HRP | Horseradish peroxidase |
| LDL | Low-density lipoproteins |
| MHC | Major histocompatibility complex |
| NTA | Nanoparticle tracking analysis |
| OPD | O-Phenylenediamine dihydrochloride |
| RT | Room temperature |
| sEV | Small extracellular vesicles |
| TBS | Tris-buffered saline |
| TBST | Tris-buffered saline + Tween20 |
| TEM | Transmission electron microscopy |
| TRIS | Trisaminomethane |
References
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Hu, Q.; Su, H.; Li, J.; Lyon, C.; Tang, W.; Wan, M.; Hu, T.Y. Clinical applications of exosome membrane proteins. Precis. Clin. Med. 2020, 3, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Hromada, C.; Mühleder, S.; Grillari, J.; Redl, H.; Holnthoner, W. Endothelial extracellular vesicles-promises and challenges. Front. Physiol. 2017, 8, 275. [Google Scholar] [CrossRef]
- Simpson, R.J.; Lim, J.W.E.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta Gen. Subj. 2012. [Google Scholar] [CrossRef]
- Zhou, H.; Yuen, P.S.T.; Pisitkun, T.; Gonzales, P.A.; Yasuda, H.; Dear, J.W.; Gross, P.; Knepper, M.A.; Star, R.A. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006, 69, 1471–1476. [Google Scholar] [CrossRef] [PubMed]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C.; et al. Rapid Isolation and Detection of Exosomes and Associated Biomarkers from Plasma. ACS Nano 2017. [Google Scholar] [CrossRef] [PubMed]
- Maroto, R.; Zhao, Y.; Jamaluddin, M.; Popov, V.L.; Wang, H.; Kalubowilage, M.; Zhang, Y.; Luisi, J.; Sun, H.; Culbertson, C.T.; et al. Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. J. Extracell. Vesicles 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in exosome isolation and analysis in health and disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Hong, C.S.; Stolz, D.B.; Watkins, S.C.; Whiteside, T.L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods 2014, 411, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pertierra, E.; Oliveira-Rodríguez, M.; Rivas, M.; Oliva, P.; Villafani, J.; Navarro, A.; Blanco-López, M.C.; Cernuda-Morollón, E. Characterization of plasma-derived extracellular vesicles isolated by different methods: A comparison study. Bioengineering 2019, 6, 8. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006. [Google Scholar] [CrossRef] [PubMed]
- Welton, J.L.; Webber, J.P.; Botos, L.A.; Jones, M.; Clayton, A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J. Extracell. Vesicles 2015, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef]
- Yu, L.L.; Zhu, J.; Liu, J.X.; Jiang, F.; Ni, W.K.; Qu, L.S.; Ni, R.Z.; Lu, C.H.; Xiao, M.B. A Comparison of Traditional and Novel Methods for the Separation of Exosomes from Human Samples. Biomed Res. Int. 2018, 1–9. [Google Scholar] [CrossRef]
- Tian, Y.; Gong, M.; Hu, Y.; Liu, H.; Zhang, W.; Zhang, M.; Hu, X.; Aubert, D.; Zhu, S.; Wu, L.; et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2020, 9. [Google Scholar] [CrossRef]
- Livshts, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Hvam, M.L.; Primdahl-Bengtson, B.; Boysen, A.T.; Whitehead, B.; Dyrskjøt, L.; Ørntoft, T.F.; Howard, K.A.; Ostenfeld, M.S. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef]
- Szatanek, R.; Baran, J.; Siedlar, M.; Baj-Krzyworzeka, M. Isolation of extracellular vesicles: Determining the correct approach (review). Int. J. Mol. Med. 2015, 36, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Arlt, H.; Brock, K.P.; Lai, Z.W.; DiMaio, F.; Marks, D.S.; Liao, M.; Farese, R.V.; Walther, T.C. Cryo–electron microscopy structure of the lipid droplet–formation protein seipin. J. Cell Biol. 2018, 217, 4080–4091. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yan, J.; Shen, X.; Sun, Y.; Thulin, M.; Cai, Y.; Wik, L.; Shen, Q.; Oelrich, J.; Qian, X.; et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Larssen, P.; Wik, L.; Czarnewski, P.; Eldh, M.; Lof, L.; Goran Ronquist, K.; Dubois, L.; Freyhult, E.; Gallant, C.J.; Oelrich, J.; et al. Tracing cellular origin of human exosomes using multiplex proximity extension assays. Mol. Cell. Proteom. 2017, 16, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Trusted Exosome Purification. Available online: https://www.cellgs.com/products/exo-spinand8482-mini-hd-columns.html (accessed on 20 January 2021).
- Mørk, M.; Handberga, A.; Pedersen, S.; Jørgensen, M.M.; Bæk, R.; Nielsen, M.K.; Kristensen, S.R. Prospects and limitations of antibody-mediated clearing of lipoproteins from blood plasma prior to nanoparticle tracking analysis of extracellular vesicles. J. Extracell. Vesicles 2017. [Google Scholar] [CrossRef] [PubMed]
- Sódar, B.W.; Kittel, Á.; Pálóczi, K.; Vukman, K.V.; Osteikoetxea, X.; Szabó-Taylor, K.; Németh, A.; Sperlágh, B.; Baranyai, T.; Giricz, Z.; et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016. [Google Scholar] [CrossRef]
- Yuana, Y.; Levels, J.; Grootemaat, A.; Sturk, A.; Nieuwland, R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.P.; Hole, P.; Carr, B.; Redman, C.W.G.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788. [Google Scholar] [CrossRef]
- German, J.B.; Smilowitz, J.T.; Zivkovic, A.M. Lipoproteins: When size really matters. Curr. Opin. Colloid Interface Sci. 2006, 11, 171–183. [Google Scholar] [CrossRef]
- Lei, D.; Yu, Y.; Kuang, Y.L.; Liu, J.; Krauss, R.M.; Ren, G. Single-molecule 3D imaging of human plasma intermediate-density lipoproteins reveals a polyhedral structure. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 260–270. [Google Scholar] [CrossRef]
- Yu, Y.; Kuang, Y.L.; Lei, D.; Zhai, X.; Zhang, M.; Krauss, R.M.; Ren, G. Polyhedral 3D structure of human plasma very low density lipoproteins by individual particle cryo-electron tomography. J. Lipid Res. 2016, 57, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, J.; Cavigiolio, G.; Ishida, B.Y.; Zhang, S.; Kane, J.P.; Weisgraber, K.H.; Oda, M.N.; Rye, K.A.; Pownall, H.J.; et al. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. J. Lipid Res. 2011, 52, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Frühbeis, C.; Helmig, S.; Tug, S.; Simon, P.; Krämer-Albers, E.M. Physical exercise induces rapid release of small extracellular vesicles into the circulation. J. Extracell. Vesicles 2015, 4. [Google Scholar] [CrossRef]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Bachurski, D.; Schuldner, M.; Nguyen, P.H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M.; et al. Extracellular vesicle measurements with nanoparticle tracking analysis—An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8. [Google Scholar] [CrossRef]

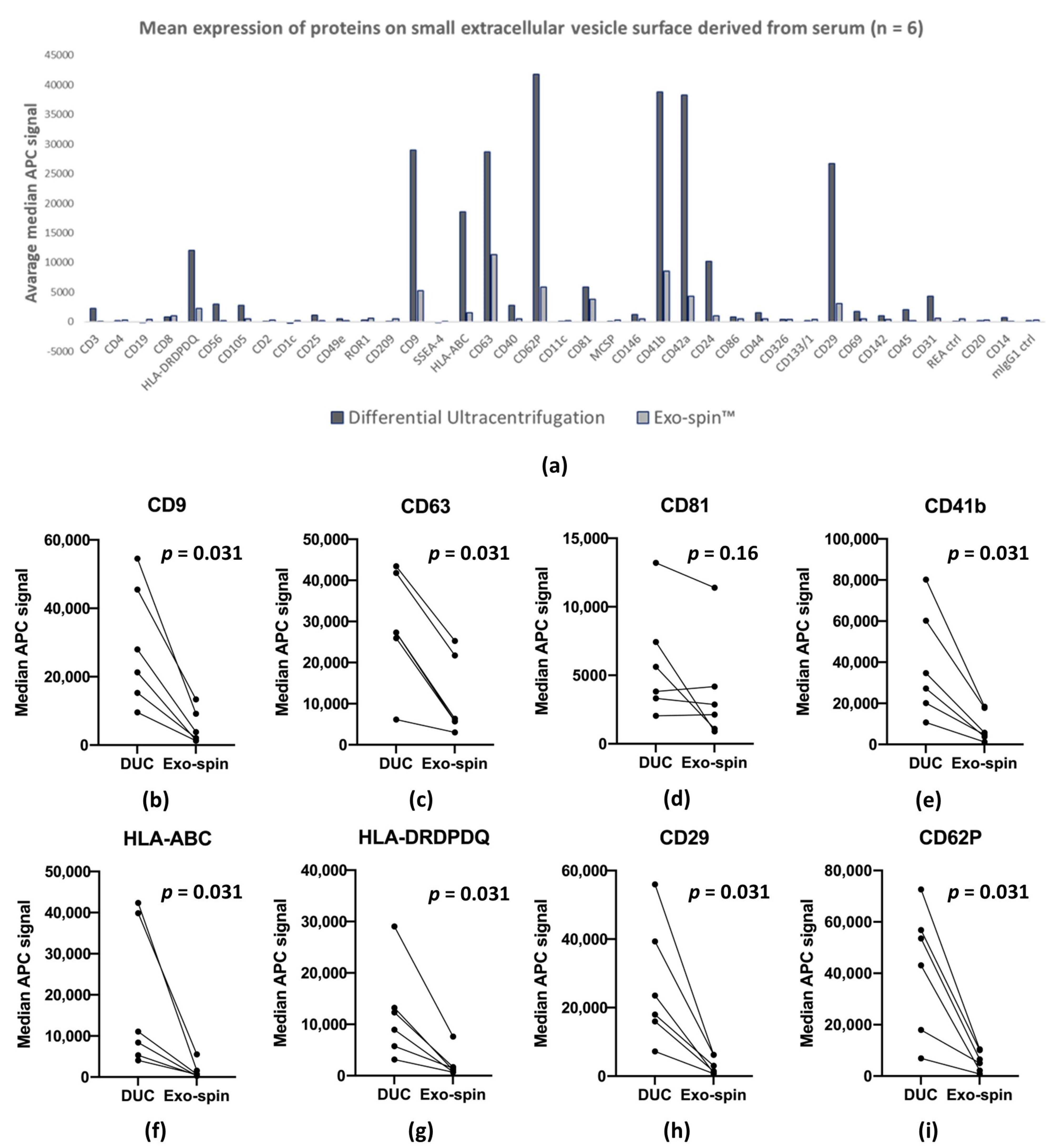
| Marker | DUC | Exo-spin™ |
|---|---|---|
| CD62P | 41,826 | 5839 |
| CD41b | 38,839 | 8594 |
| CD42a | 38,260 | 4270 |
| CD9 | 29,016 | 5195 |
| CD63 | 28,660 | 11,308 |
| CD29 | 26,669 | 3089 |
| HLA-ABC | 18,512 | 1518 |
| HLA-DRDPDQ | 12,069 | 2206 |
| CD24 | 10,192 | 957 |
| CD81 | 5905 | 3764 |
| CD31 | 4360 | 606 |
| CD56 | 2969 | 153 |
| CD105 | 2781 | 512 |
| CD40 | 2759 | 451 |
| CD3 | 2291 | 33 |
| CD45 | 2090 | 191 |
| CD69 | 1721 | 482 |
| CD44 | 1522 | 525 |
| CD146 | 1234 | 502 |
| CD25 | 1121 | 221 |
| CD142 | 1011 | 417 |
| CD86 | 788 | 481 |
| CD8 | 765 | 1052 |
| CD14 | 679 | 75 |
| CD49e | 466 | 155 |
| CD326 | 369 | 360 |
| ROR1 | 291 | 556 |
| mIgG1 ctrl | 210 | 281 |
| CD133/1 | 156 | 378 |
| CD4 | 139 | 320 |
| CD20 | 132 | 323 |
| REA ctrl | 128 | 504 |
| CD2 | 85 | 265 |
| MCSP | 55 | 258 |
| CD209 | 36 | 451 |
| CD11c | 0 | 233 |
| CD19 | −16 | 352 |
| SSEA-4 | −22 | 4 |
| CD1c | −198 | 161 |
| Characteristics | DUC | Exo-spin™ | Comments |
|---|---|---|---|
| Minimal serum volume for healthy individuals | 1 mL | 100 µL | DUC range: 1–5 mL |
| Exo-spin™ range: x–150 µL | |||
| Final volume of sEV resuspended in PBS | 200 µL | 400 µL | Volume of DUC can be adjusted for individual purposes |
| Total protein concentration | 32.4 ± 7.8 µg/mL | 40.6 ± 16.8 µg/mL | |
| Recovery (particle number/mL of serum) | 1.0 ± 1.1 × 108/mL | 497.6 ± 769.5 × 108/mL | High recovery of particles by Exo-spin is mostly associated with large numbers of lipoproteins being co-isolated; |
| Normalisation is essential since the initial applied volume of serum differs | |||
| Absolute number of particles | 2.7 ± 2.2 × 108 | 49.8 ± 76.9 × 108 | Absolute number of particles that is purified from heathy individuals according to the suited protocol |
| Particle size (diameter) ø | Mean = 148.1 ± 14.6 nm | Mean = 132.5 ± 12.1 nm | Large size of particles isolated by DUC may be associated with aggreagates induction by gravitational forces |
| Mode = 121.9 ± 20.6 nm | Mode = 110.9 ± 17.2 nm | ||
| Median = 138.0 ± 13.7 nm | Median = 123.1 ± 13.1 nm | ||
| % of particles < 150 nm = 57.66 ± 8.8% | % of particles < 150 nm = 69.6 ± 8.4% | ||
| Relative mean intensity by ELISA | CD81: 0.36 ± 0.18; | CD81: 0.12 ± 0.08; | Measured particle concentrations for these samples were: DUC = 10.54 × 108/mL; Exo-spin™ = 32.25 × 108/mL |
| CD63: 0.52 ± 0.25; | CD63: 0.16 ± 0.07; | ||
| CD9: 0.96 ± 0.48 | CD9: 0.18 ± 0.07 | ||
| Median APC fluorescence intensity by MACSPlex | Average: 7124.8 ± 12,203.1 | Average: 1462.4 ± 2451.1 | All samples were paired |
| CD81: 5905.2 ± 3686.9 | CD81: 3763.8 ± 3586.7 | ||
| CD63: 28,660 ± 12,303 | CD63: 11,308 ± 8743.4 | ||
| CD9: 29,016 ± 16,072 | CD9: 5194.5 ± 4542.6 | ||
| Morphology | Singles and aggregates of vesicles, cup-like shape, approximately half of vesicle were stained positively with exosomal markers | Whole image fully layered with vesicles, various morphologies that does not resemble exosomes, only few stained positively with exosomal markers | - |
| Co-purification | Low lipoprotein contamination | High lipoprotein contamination | Most of the particles isolated by Exo-spin™ from non-fasted patients were assigned as lipoproteins; |
| Albumin contamination approximately 12.85% | Non-detectable albumin contamination | Albumin contamination was evaluated by silver staining. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Małys, M.S.; Aigner, C.; Schulz, S.M.; Schachner, H.; Rees, A.J.; Kain, R. Isolation of Small Extracellular Vesicles from Human Sera. Int. J. Mol. Sci. 2021, 22, 4653. https://doi.org/10.3390/ijms22094653
Małys MS, Aigner C, Schulz SM, Schachner H, Rees AJ, Kain R. Isolation of Small Extracellular Vesicles from Human Sera. International Journal of Molecular Sciences. 2021; 22(9):4653. https://doi.org/10.3390/ijms22094653
Chicago/Turabian StyleMałys, Małgorzata S., Christof Aigner, Stefan M. Schulz, Helga Schachner, Andrew J. Rees, and Renate Kain. 2021. "Isolation of Small Extracellular Vesicles from Human Sera" International Journal of Molecular Sciences 22, no. 9: 4653. https://doi.org/10.3390/ijms22094653
APA StyleMałys, M. S., Aigner, C., Schulz, S. M., Schachner, H., Rees, A. J., & Kain, R. (2021). Isolation of Small Extracellular Vesicles from Human Sera. International Journal of Molecular Sciences, 22(9), 4653. https://doi.org/10.3390/ijms22094653







